Abstract
Adipose differentiation is a complex process controlled by a network of transcription factors and co-regulators. We compared the global gene expression patterns of adipogenic and nonadipogenic clones of 3T3-F442A preadipocytes and identified the transcriptional cofactor, vestigial-like 3 (Vgll3), as an inhibitor of adipogenesis. Vgll3 expression is down-regulated during terminal adipocyte differentiation in vitro and negatively correlates with weight and total fat mass in vivo. Furthermore, enforced Vgll3 expression inhibits the differentiation of preadipocytes in vitro, whereas shRNA-mediated knockdown of Vgll3 expression promotes differentiation. Expression of Vgll3 promoted the expression of genes associated with bone and chondrocyte formation, suggesting that Vgll3 participates in the decision of mesenchymal cells to proceed down the adipocyte, bone, or cartilage lineages. The elucidation of factors involved in specification of the adipocyte phenotype may aid in the identification of new strategies for the treatment of metabolic disease.
Keywords: mesenchymal differentiation, PPAR, lipid metabolism, nuclear receptor
The prevalence of metabolic disorders such as diabetes and obesity is increasing in developing and industrialized societies. Unraveling the intricate network of biological pathways and factors that govern adipogenesis, the process of development and differentiation of adipose tissue, is expected to provide insight into the physiology and pathophysiology underlying these disorders. A number of regulatory factors that participate in the positive and negative control of this process have been identified. Many established mediators of adipogenesis are involved in transcriptional regulation, including PPARγ, the CEB/Ps, and members of the KLF family of transcription factors (1, 2). More recently, transcription co-regulators, such as PRDM16, PGC-1α, and TLE3, have been found to be important for the fine-tuning of particular features of the adipogenic program (3–5). However, it is likely that additional factors that have yet to be identified and characterized contribute to this complex process.
The lipid-activated transcription factor PPARγ is the master regulator of adipocyte differentiation and is necessary and sufficient for the development of adipose tissue (6, 7). Free fatty acids and eicosanoids are naturally occurring ligands capable of activating PPARγ however, a specific and biologically relevant endogenous ligand for PPARγ has yet to be identified. Clinically, PPARγ is the molecular target of the thiazolidinedione class of therapeutic drugs that act as ligands capable of inducing PPARγ activity and the adipogenic program. Thiazolidinediones are highly effective insulin sensitizers in humans, but their use has been limited by side effects and safety concerns (8). The elucidation of novel factors that regulate the adipocyte differentiation program could provide the foundation for the potential identification of new therapeutic targets.
Since the discovery of PPARγ, a variety of approaches have been used in an effort to uncover additional regulatory factors involved in the conversion of progenitor preadipocytes cells into fully mature, lipid-laden adipocytes. Such strategies have included transcriptional profiling of the preadipocyte cell line, analysis of knockout/transgenic mouse models, high-throughput cDNA screening, and the identification of adipogenic small molecules (9–12). Alternative strategies offer the potential of uncovering additional novel determinants of adipocyte differentiation not identified by prior approaches. For example, a recent study used analysis of gene expression in clonal sublines derived from Swiss3T3 fibroblasts to identify zfp423 as a new regulator of adipogenesis (13).
In the current study, we compared global gene expression in clonal sublines of committed 3T3-F442A preadipocytes in an effort to uncover adipogenic modulators. We identified the mRNA encoding vestigial-like 3 (Vgll3) as a transcript differentially regulated between adipogenic and nonadipogenic clones. Further analysis revealed that expression Vgll3 is down-regulated during adipocyte differentiation. Constitutive expression of Vgll3 in differentiating preadipocytes potently inhibits adipocyte differentiation and up-regulates the expression of osteogenic genes, whereas knockdown of Vgll3 promotes differentiation. These studies demonstrate that Vgll3 acts as a negative regulator of terminal adipocyte differentiation and support further investigation of Vgll3 as a regulator of mesenchymal-derived cellular differentiation programs.
MATERIALS AND METHODS
Reagents and plasmids
GW7845 was kindly provided by T. Willson (GlaxoSmithKline). Insulin (#12585-014) was from Gibco® (Life Technologies). Dexamethasone (#D2915) and 3-isobutyl-1- methylxanthine (IBMX, #17018) were from Sigma-Aldrich. Human Vgll3 was obtained by using plasmid clone #30528902 obtained from Open Biosystems (catalog #MHS1010-98050653; Thermo Scientific). Primers used to obtain a full-length human Vgll3 PCR product (per NCBI Reference Sequence NM_016206) were 5′-GGGGACAAGTTT GTACAAAAAAGCAGGCTCTGCCACCATGAGTTGTGCGGAGGT-3′ (forward [fwd]) and 5′-TCAGTACCACGGTGATTCCT TACTCTTGTCTTGATGCTGTAGACCTGTA TCGAA-3′ (reverse [rev]). This PCR product was used as a template to produce an amplified Gateway®-adapted, full-length human Vgll3 PCR product using the forward primer described above and the primer 5′-GGGGACCACTTTGTACAAGAAA GCTGGGTCTCAGTACCACGGTGATTCCTTAC-3′ (rev). BP and LR recombination was used to clone full length human Vgll3 into a Gateway®-adapted pCDNA-DEST47 mammalian expression vector (Invitrogen) and pBABEpuromycin retroviral expression vector (14). LacZ and human Vgll3 adenoviral particles were generated using the pAd⁄CMV⁄V5-DEST Gateway®-adapted adenoviral vector (Invitrogen). All descriptions of Vgll3 mRNA overexpression refer to human Vgll3 unless specifically indicated otherwise.
Mammalian cell culture and retro/adenovirus production
3T3-F442A and 3T3-L1 preadipocytes were maintained in DMEM supplemented with 10% calf serum. To prepare for adipocyte differentiation, 3T3-F442A and 3T3-L1 cells were grown to confluence in a 6-well plate or in a 10 cm culture dish in DMEM supplemented with 10% FBS. 3T3-L1 cells were stimulated to differentiate (1 day after confluence) by treating with dexamethasone (1 μM), IBMX (0.5 mM), and insulin (5 μg/ml) for 2 days after confluence followed by either insulin (5 μg/ml) and GW7845 (10 nM or 20 nM when indicated) or insulin (5 μg/ml) alone. 3T3-F442A cells were stimulated to differentiate (1 day after confluence) by treating with insulin (5 μg/ml) with or without GW7845 (10 nM). Growth medium was exchanged every 2 days during the course of adipogenic differentiation. When treating 3T3-L1 with osteogenic media, cells were allowed to reach confluence and treated with 10% FBS, β-glycerophophate (1 M) + ascorbic acid (50 μg /ml). Retrovirus was obtained by overnight transfection of Phoenix E cells with pBABEpuro-empty vector and pBABEpuro-hVgll3 using Fugene Transfection Reagent (Promega) followed by growth media exchange and harvesting of retrovirus 48 h later. Adenovirus was amplified, purified, and titered by Viraquest Inc.
shRNA plasmids and endogenous Vgll3 knockdown
Vgll3 shRNA constructs were designed using BLOCK-IT RNAi designer tool (Invitrogen). Sense and antisense oligos were annealed and cloned into the pENTR/U6 plasmid (Invitrogen). Using LR recombination (Invitrogen), shRNA constructs were subcloned into a Gateway®-adapted pBabe-Puromycin plasmid and transfected into Phoenix E cells. Oligos used in this study are LacZ shRNA 5′ -CACCGGGCCAGCTGTATAGACATC TCGAAAGATGTCTATACAGCTGGCCC-3′, mVgll3 sh1 5′-CACCGAAAGAGCTG AGCTGTCTCGCCCGAAGGCGAGACAG CTCAGCTC-3′, mVgll3 sh2 5′-CACCGG AACTTTAGCATCCAGATAACGAATTATCTGG ATGCTAAAGTTCC-3′. Only the sense strands are indicated here. Differentiation of 3T3-L1 preadipocytes expressing shRNAs was carried out using dexamethasone (1 μM), IBMX (0.5 mM), and insulin (3.5 μg/ml) for 48 h followed by stimulation with insulin (3.5 μg/ml) only in 10% FBS.
Gene expression and microarrays
Total RNA was isolated using Trizol reagent (Invitrogen) and reverse transcribed using the iScript cDNA synthesis kit (Biorad). cDNA was quantified by real-time PCR using SYBR Green (Diagenode) and an ABI 7900 instrument. Gene expression levels were determined by using a standard curve. All data from the genes analyzed were normalized to the housekeeping gene 36B4 and were performed in duplicate. Primers used for real-time PCR are: (vgll3: fwd 5′-CCGGAACCCCTGGCAG-3′ rev 5′-CTTGTCCTGATGCTGAAGACC-3′, human Vgll3 fwd 5′-CTACAGTCACCTCTGCT ACCT-3′ rev 5′-CTTGTCTTGATGCTGTAGACC-3′, PPARγ fwd 5′-TGGTAATTTCTTGTGAAGTGC-3′ rev 5′- TGGTAATTTCTTGTGAAGTGC-3′, aP2 fwd 5′-CACCGCAGACGACAGGAAG-3′ rev 5′-GCACCTGCACCAGGGC-3′, Adiponectin fwd 5′-CCGGAACCCCTGGCAG-3′ rev 5′-CTGAACGCTGAGCGATA CACA-3′, CD36 fwd 5′-GGCCAAGCTATTGCGACAT-3′ rev 5′-CAGATCCGAACACAGCGTAGA-3′, Chop fwd 5′-GCGACAGAGCCAGAATAACA-3′ rev 5′-GATGCACTTCCTTCTGGAACA-3′, Klf2 fwd 5′-CTAAAGGCGCATCTGCGTA-3′ rev 5′-TAGTGGCGGGTAAGCTCGT-3′, 36B4 fwd 5′-ACTGGTCTAGGACCCGAGAAG-3′ rev 5′-TCCCACCTTGTCTCCAGTCT-3′, Ankrd1fwd 5′-GCTGGAGCCCAGATTGAA-3′ rev 5′-CTCCACGACATGCCCAGT-3′, Col1A1fwd 5′-CCGCTGGTCAAGATGGTC-3′ rev 5′-CTCCAGCCTTTCCAGGTTCT-3′, Col1A2 fwd 5′-CGGAGAAGCTGGATCTGC-3′ rev 5′-CAGGAGGACCCATTACACCA-3′, Snai2 fwd 5′-TGCAAGATCTGTGGCAAG G-3′ rev 5′-CAGTGAGGGCAAGAGAAAGG -3′, Opn fwd 5′-CCCGGTGAAAGTGACTGATT-3′ rev 5′-TTCTTCAGAGGACACAGCATT-3′, Ptprv fwd 5′-AACACCACAGGCTGGACAC-3′ rev 5′-GGGCTTCACTGGTCACATTTA-3′ Sox9 fwd 5′-CAGCAAGACTCTGGGCAAG-3′ rev 5′-TCCACGAAGGGTCTCTTCTC-3′, Thbs1 fwd 5′-CCCCAACCTTCCCAACTC-3′ rev 5′-GGGTTGTAATGGAATGGAC AG-3′, MMP3 fwd 5′-TTGTTCTTTGATGCAGTCAGC-3′ rev 5′-GATTTGCGCCAAAAGTGC-3′, Col12A1 fwd 5′-ACTGGGGAGGAGACCACTG-3′ rev 5′-TGGTCTGTATCTAATCCGATA-3′, Adm fwd 5′-TTCGCAGTTCCGAAAGAAGT-3′ rev 5′-GGTAGCTGCTGGATGCTTGT-3′, Gadd45β fwd 5′-CTGCCTCCTGGTCACGAA-3′ rev 5′-TTGCCTCTGCTCTCTTCACA-3′, Osr2 fwd 5′-CCAGGCAGACATCGGTTC-3′ rev 5′-GGGTGTGAGGGGGAAAAG-3′, Alp fwd 5′-AAACCCAGAACACAAGCATTC-3′ rev 5′-TCCACCAGCAAGAAGAAGCC-3′). Isolated sublines were prepared for microarray analysis by growing 3T3-F442A cells in 10% FBS to confluence and treating cells with insulin (5 μg/ml) for 48 h. For each cell subline, RNA was pooled from six biological replicates and analyzed using a whole mouse genome array from Illumina (MouseWG-6 BeadChip). Analysis was carried out by the Southern California Genotyping Consortium at UCLA. Results were analyzed using GenomeStudio (Illumina). For adenovirus experiments, 3T3-L1 cells stably expressing the Coxsackie and Adenovirus Receptor were grown in 10% FBS and insulin (5 μg/ml). Upon reaching confluence, cells were treated with LacZ and hVgll3 expressing adenovirus overnight (12 h) using an multiplicity of infection of 50 in 10% FBS with insulin (5 μg/ml) and GW7845 (20 nM). Cells were then exchanged with fresh 10% FBS containing insulin (5 μg/ml) and GW7845 (20 nM), and RNA was harvested 48 h later. Three biological replicates were used to pool collected RNA, and samples were processed by the UCLA Clinical Microarray Core using GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix). Results were analyzed using GenespringGX (Affymetrix). Vgll3 transcript expression in adipose tissue in vivo was correlated with quantifiable clinical traits using data obtained from studies as described previously (16, 17).
Luciferase reporter assay
3T3-L1 cells were grown in 10% FBS in 24-well culture plates until roughly 80% confluence. Cells were cotransfected with 100 ng of pGL3-aP2-luciferase (6), 100 ng of pCMX-PPARγ, 20 ng of pCMX-RXR, 100 ng of pCDNA-hVGll3, and 1 ng of Renila Luciferase control vector using Fugene Transfection Reagent (Promega). Cells were transiently transfected for 6 h and then exchanged with fresh 10% FBS and allowed to incubate overnight. The medium was then exchanged with 10% FBS with or without PPARγ ligand GW7845 (100 nM). Cells were allowed to incubate for 48 h, and Luciferase activity was measured with the Dual-Luciferase® Reporter Assay System (Promega) and a GLOMAX® Luminometer (Promega). Firefly Luciferase activity was normalized to Renila Luciferase.
RESULTS
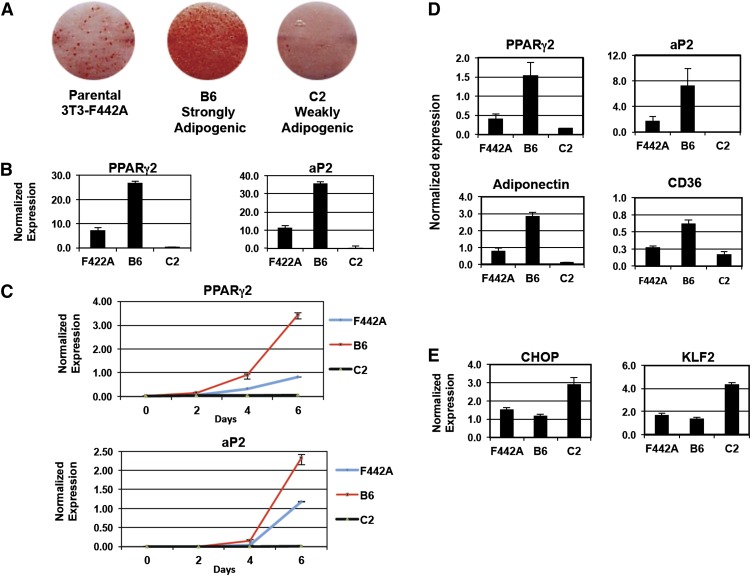
We isolated sublines of 3T3-F442A preadipocytes with varying capacities for adipogenic differentiation through clonal selection (supplementary Fig. I). Two clonal sublines, designated as “B6” and “C2,” were found to be highly divergent in their potential to become mature adipocytes. B6 displayed a very high propensity for adipocyte differentiation, whereas C2 exhibited almost no ability to differentiate (Fig. 1A). In accordance with this observation, expression levels of PPARγ2 and its target gene aP2 were up- and down-regulated in B6 and C2 sublines compared with the parental 3T3-F442A cell line, respectively (Fig. 1B, C). To uncover novel genes affecting adipogenesis, we performed global gene expression profiling in the B6 and C2 sublines using cDNA microarrays (supplementary Fig. II). This analysis was performed on cells 48 h after confluence to identify gene expression differences presenting early in the differentiation process. A large number of genes were differentially regulated between these two sublines in these experiments. As expected, transcript levels of terminal adipocyte marker genes (Adiponectin, CD36) were highest in the B6 subline, whereas levels of expression of known antiadipogenic genes (CHOP, KLF2) were highest in the C2 subline (Fig. 1D, E).
Fig. 1.
Isolation and characterization of preadipocyte sublines. A: Oil Red O staining of 3T3-F442A parental, B6, and C2 sublines 8 days after stimulation with 10% FBS containing insulin (5 μg/ml) and PPARγ agonist GW7845 (10 nM). B: Gene expression in 3T3-F442A cells on day 8 was analyzed by real-time qPCR and normalized to 36B4 control. C: Time course of PPARγ2 and aP2 expression in F442A cell lines cultured in 10% FBS analyzed by real-time PCR. D: Expression of adipogenic genes in 3T3-F442A cell lines isolated sublines 48 h after confluence (10% FBS + insulin). E: Expression of antiadipogenic genes in 3T3-F442A cell lines isolated sublines 48 h after confluence (10% FBS + insulin). Experiments were repeated at least twice with qualitatively similar results.
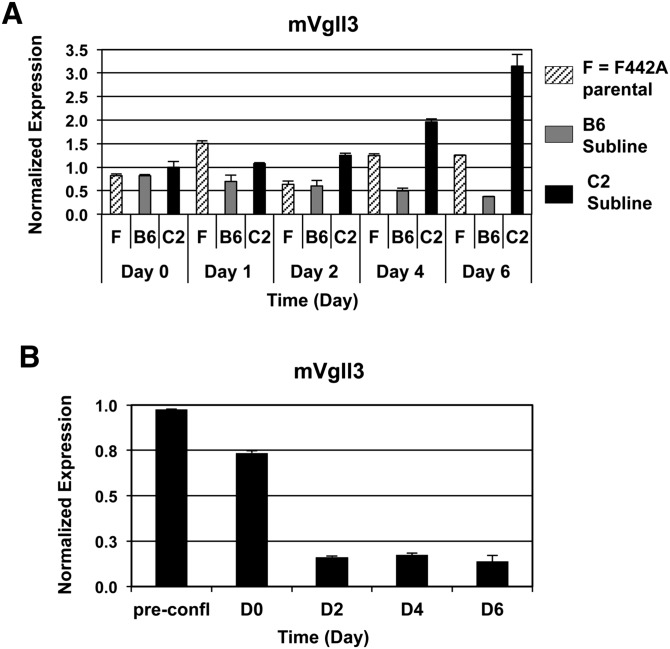
Many genes previously reported to be important mediators of adipocyte differentiation are known to function as transcription factors or transcriptional cofactors (1–6) Therefore, we focused our subsequent analysis on established or putative transcriptional regulators of gene expression that also displayed at least a 2-fold differential expression signal ratio between the B6 and C2 sublines. Based on these criteria, Vgll3 was identified as a candidate gene for further analysis. Upon further examination, we observed that endogenous Vgll3 gene expression was inversely correlated with the degree of mature adipocyte formation. In a time course of adipogenesis, Vgll3 expression steadily declined in the highly differentiated B6 cells at day 6 (Fig. 2A). By contrast, there was a gradual escalation in Vgll3 expression in the antiadipogenic C2 cell line (Fig. 2A). Furthermore, Vgll3 expression levels were strongly reduced in adipogenically differentiating 3T3-L1 cells stimulated with PPARγ ligand (GW7845) (Fig. 2B).
Fig. 2.
Vgll3 mRNA expression is down-regulated during adipogenesis. A: Expression of endogenous of murine Vgll3 mRNA during a time course of adipogenesis in 3T3-F442A parental, B6, and C2 isolated sublines (10% FBS + insulin). B: Expression of murine Vgll3 mRNA during a time course of adipogenesis [10% FBS + DMI + GW7845 (20 nM)]. Experiments were repeated at least twice with qualitatively similar results.
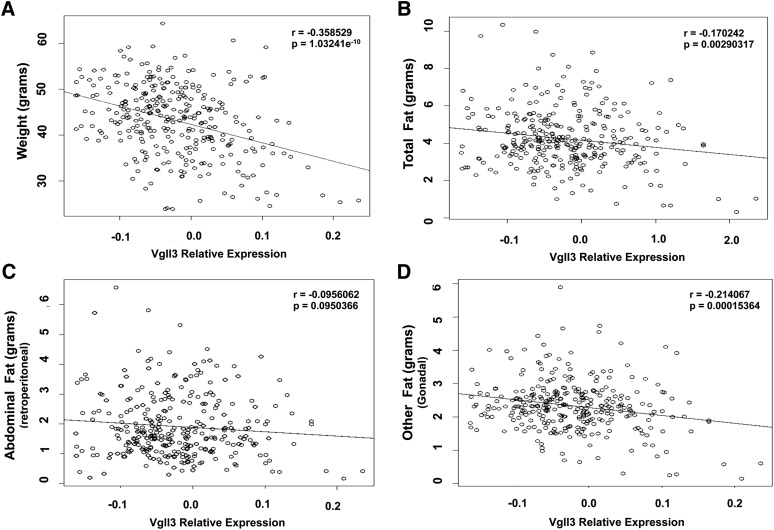
We next analyzed the expression of Vgll3 in adipose tissue in vivo. Vgll3 transcript levels were correlated with clinical traits obtained from previous studies of expression quantitative trait loci in a mouse F2 population produced by the intercrossing of F1 mice (15, 16). We found that expression of Vgll3 was inversely correlated with total body weight and adipose tissue mass (Fig. 3A–D and supplementary Fig. III). This correlation was particularly strong in mesenteric and gonadal fat depots. However, correlation in abdominal retroperitoneal and subcutaneous depots did not reach statistical significance. These in vivo findings were consistent with our in vitro results and suggested that down-regulation of Vgll3 gene expression could play a role in adipocyte development.
Fig. 3.
Correlation of adipose Vgll3 mRNA expression with total weight (A), total fat content (B), retroperitoneal fat content (C), and gonadal fat content (D) (n = 313) (15).
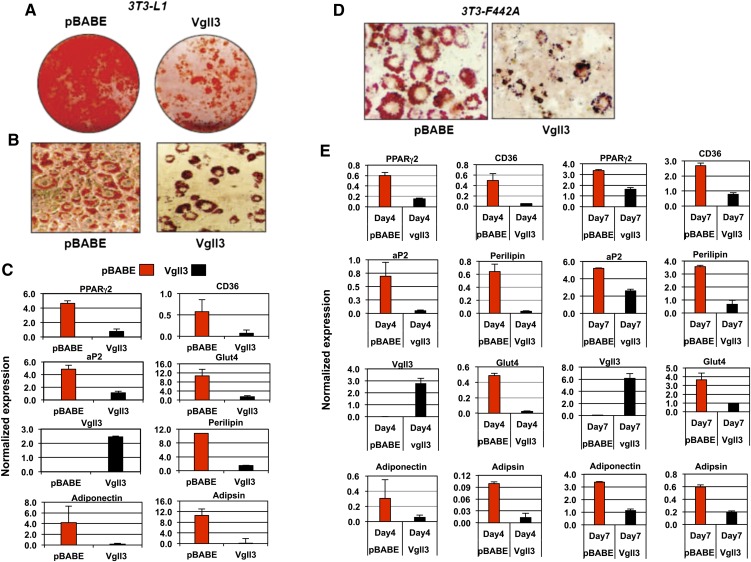
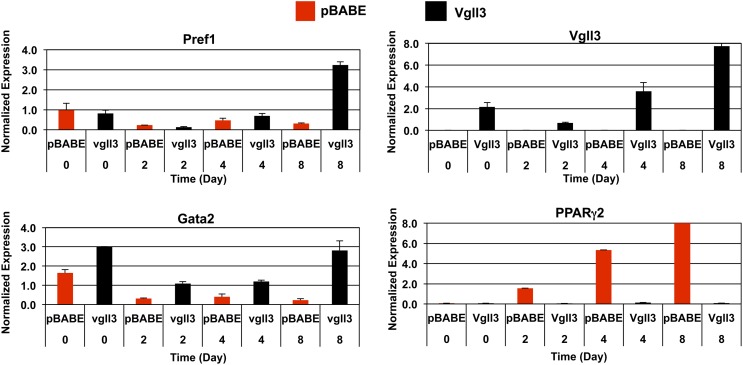
To explore this possibility, we ectopically expressed Vgll3 in committed preadipocyte cell lines using a retroviral vector. Multiple independent stable pools of control and Vgll3-expressing 3T3-F442A and 3T3-L1 cells were selected and analyzed for their differentiation capacity. In 3T3-F442A cells, enforced expression of Vgll3 resulted in a modest but reproducible decrease in lipid accumulation as assessed by Oil Red O staining at Day 7 of differentiation compared with vector controls (Fig. 4D). In 3T3-L1 cells, Vgll3 strongly inhibited lipid accumulation (Fig. 4A, B). Real-time qPCR analysis showed that the expression of adipocyte differentiation markers was also reduced in response to Vgll3 expression, confirming that Vgll3 was affecting differentiation per se. The expression of PPARγ2, aP2, and a panel of other adipocyte marker genes was reduced in cells constitutively expressing Vgll3 (Fig. 4C, E). Furthermore, Vgll3 overexpression was found to up-regulate the expression of Gata-2 and Pref-1, two genes expressed in adipocyte precursors and previously reported to be strongly down-regulated during adipogenesis (Fig. 5) (17, 18).
Fig. 4.
Overexpression of Vgll3 inhibits adipocyte differentiation. A: Oil-red-O staining of 3T3-L1 overexpressing Vgll3 or pBABE vector control at Day 8 of differentiation. B: Microscopic view of Oil-Red-O stained 3T3-L1 cells overexpressing Vgll3 or pBABE control at day 8 of differentiation. C: Expression of adipocyte gene markers in 3T3-L1 cells overexpressing Vgll3 or pBABE control at day 8 of adipocyte differentiation (n = 2). D: Microscopic view of Oil-Red-O stained 3T3-F442A cells overexpressing Vgll3 or pBABE control at day 7 of differentiation. E: Expression of adipogenic gene markers at day 4 and day 7 of adipocyte differentiation in 3T3-F442A cells overexpressing Vgll3 versus pBABE control. Experiments were repeated at least twice with qualitatively similar results.
Fig. 5.
Overexpression of Vgll3 during adipogenesis up-regulates the expression of antiadipogenic genes. Expression of the adipogenic genes Pref-1 and Gata-2 was analyzed by real-time PCR during a time course of 3T3-L1 differentiation. Similar results were obtained in two independent experiments.
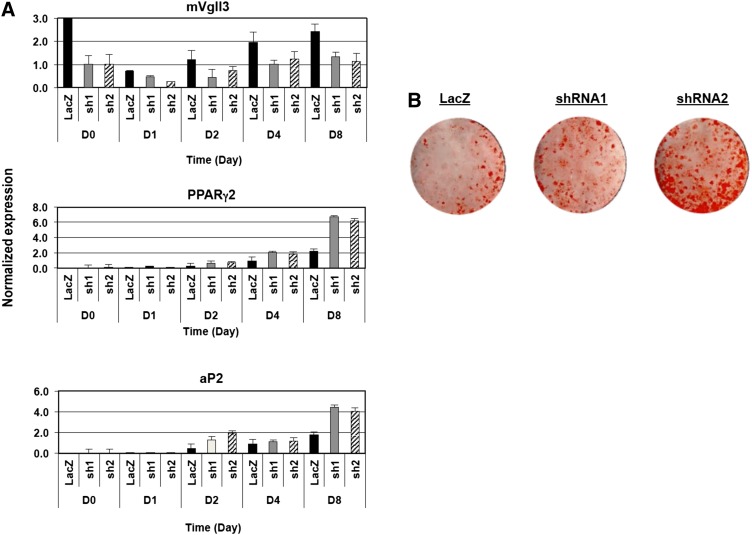
To assess whether blocking endogenous Vgll3 expression affected the progression of adipogenesis, retroviruses expressing inhibitory shRNAs were used to obtain multiple independent 3T3-L1 cell pools with reduced levels of Vgll3 transcripts. Vgll3 knockdown was validated by real-time qPCR (Fig. 6A), and the ability of the knockdown cells to differentiate was evaluated. To allow the detection of subtle changes in adipogenic capacity, these studies were carried out under conditions minimally required for adipogenic stimulation (10% FBS, dexamethasone [1 μM], IBMX [0.5 mM], insulin 3.5 μg/ml). An increase in morphological adipocyte differentiation and lipid accumulation was observed in cell lines expressing Vgll3-specific shRNAs compared with control shRNA (Fig. 6B). Consistent with this result, the expression of the adipogenic markers PPARγ and aP2 was also increased (Fig. 6A). These data further support the hypothesis that the decrease in Vgll3 expression during adipogenesis may be important for proper conversion of committed preadipocytes to mature adipocytes.
Fig. 6.
Knockdown of endogenous Vgll3 in 3T3-L1 preadipocytes promotes adipocyte differentiation. A: Expression of murine Vgll3, PPARγ2, and aP2 in 3T3-L1 preadipocytes transduced with retrovirus expressing control or one of two different shRNAs targeting the Vgll3 transcript was determined by real-time PCR. B: Oil-Red-O staining of 3T3-L1 preadipocytes expressing control or Vgll3 shRNAs on Day 8 of adipocyte differentiation. Differentiation of 3T3-L1 preadipocytes expressing shRNAs was carried out using dexamethasone (1 μM), IBMX (0.5 mM), and insulin (3.5 μg/ml) for 48 h followed by stimulation with insulin (3.5 μg/ml) only in 10% FBS. Similar results were obtained in at least two independent experiments.
To investigate the mechanism for Vgll3 effects on adipogenesis, we tested the ability of Vgll3 to affect PPARγ transcriptional activity. We transfected PPARγ and RXR into 3T3-L1 preadipocytes combination with vector or Vgll3 expression plasmid and assayed the induction of the −5.4 kb aP2-luciferase reporter. In contrast to the recently identified PPARγ cofactor TLE3 (5), Vgll3 had no effect on PPARγ activity in this assay (supplementary Fig. IV). This result suggested that Vgll3 was unlikely to be acting by directly inhibiting PPARγ transcriptional activity.
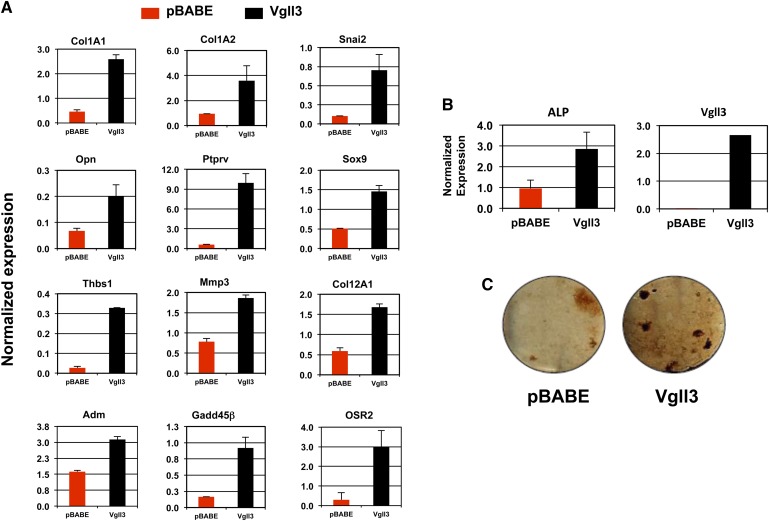
We next endeavored to determine the gene set acutely responsive to Vgll3 expression. We transduced 3T3-L1 cells expressing the coxsackie adenovirus receptor with human Vgll3-expressing and LacZ-expressing adenoviral vectors. To identify differentially regulated genes, we used transcriptional profiling with cDNA arrays (data not shown). A number of genes previously reported to be associated with other mesenchymal differentiation programs were found to be up-regulated in response to potent Vgll3 activity. In particular, several genes associated with bone formation were induced in response to Vgll3 expression, including Adm, Opn, Mmp3, Thbs1, Col12a1, and Osr2. We confirmed that these and other genes linked with bone and chondrocyte differentiation were induced in differentiating 3T3-L1 cells stably expressing human Vgll3 by real-time PCR (Fig. 7A). Furthermore, when 3T3-L1 cells expressing Vgll3 were cultured in osteogenic differentiation media for 21 days, an increase in alkaline phosphatase gene expression and von Kossa staining was observed (Fig. 7B, C). Together, these results indicate that inappropriate Vgll3 expression in adipogenically differentiating preadipocytes leads to the expression of genes associated with bone and chondrocyte differentiation.
Fig. 7.
Overexpression of Vgll3 up-regulates genes associated with nonadipose tissue differentiation programs and promotes expression of markers of bone differentiation in 3T3-L1 cells grown under osteogenic conditions. A: Expression of osteogenic- and chondrogenic-associated genes was analyzed in 3T3-L1 preadipocytes at day 8 of adipocyte differentiation. B: mRNA expression of alkaline phosphatase in 3T3-L1 preadipocytes cultured for 21 days under osteogenic conditions. C: Von Kossa staining in 3T3-L1 preadipocytes cultured for 21 days under osteogenic conditions. Similar results were obtained in at least two independent experiments.
To assess whether Vgll3 overexpression enhances osteogenesis in vitro, C310T1/2 cells ectopically expressing Vgll3 or pBABE control were grown under osteogenic conditions until Day 30. These experiments demonstrated that constitutive expression of Vgll3 promoted the expression of some osteogenic markers examined (e.g., Opn and Ocn) but not others (supplementary Fig. V). In addition, enhanced expression of Adm, Col1A2, and Runx2 was observed at Day 0 in undifferentiated C310T1/2 cells overexpressing Vgll3. Expression of endogenous Vgll3 was elevated during the course of osteogenic differentiation, in contrast to its down-regulation during adipogenesis. These results therefore suggest that regulation of Vgll3 expression and function is likely to be highly context dependent. Future investigations of Vgll3 are necessary to further clarify the mesenchymal-based gene pathways and cellular differentiation programs that are regulated by Vgll3.
DISCUSSION
The development of adipose tissue is a process that involves the coordinated action of genes that positively and negatively regulate this process. In this present study, we documented an inverse relationship between the degree of adipogenic capacity and expression of the putative transcriptional cofactor Vgll3. We also showed that body weight and adipose tissue mass was inversely correlated with Vgll3 expression in adipose tissue in vivo. Enforced expression of Vgll3 in preadipocytes inhibits adipocyte differentiation in association with the induction of bone and chondrocyte markers. Our results suggest that down-regulation of Vgll3 expression during adipogenesis may be important for the specification of adipocyte program and the suppression of genes associated with other mesenchymal cell fates.
Mammalian Vgll3 is related to the transcriptional cofactor Vestigial (Vg), originally described in Drosophila melanogaster. Drosophila Vestigial is involved in determining cell fate in the developing fly wing and muscle (19–21). In mammals, there are four highly conserved “vestigial-like” genes. Previous reports have suggested that members of this protein family are associated with muscle development and function (22, 23). For example, Vgll3 has been reported to be expressed in developing muscle tissues of the mouse embryo (24). We showed that the suppression of adipogenesis by Vgll3 was accompanied by the induction of a panel of genes associated with other mesenchymal cell fates, including bone and cartilage. Thus, whether Vgll3 is a regulator of muscle, bone, or chondrocyte development and differentiation are important questions that future studies will need to address.
We also found that stable overexpression of Vgll3 induced the expression of the well-characterized inhibitors of adipocyte differentiation Pref-1 and Gata-2. This observation suggests that suppression of Vgll3 expression during differentiation may be important for the suppression of Pref-1 and Gata-2 expression. Previously published reports have suggested that Pref-1 and Gata-2 are positive regulators of other mesenchymal differentiation programs (25). Furthermore, Sonic hedgehog has been shown to up-regulate Gata-2 and genes associated with osteogenesis, similar to our observations with Vgll3 (26). Additional analysis of Vgll3 as a potential interacting player with Pref-1, Gata-2, and other known regulatory pathways that suppress formation of mature fat tissue is warranted.
In humans, the amount of intra-abdominal and visceral fat has been linked to the progression of the metabolic syndrome. More recently, it has been suggested that visceral mesenteric fat in particular may be causally associated with the development of insulin resistance and type 2 diabetes (27, 28). Not surprisingly, specific adipose depots display stark differences in gene expression (29). For example, genes associated with cell development have been reported to be differentially expressed between various fat depots (30). In the current study, we found that Vgll3 expression is inversely correlated with mesenteric and gonadal adipose content. This result is reminiscent of previous observations with Tbx15. This developmental transcription factor is strongly differentially regulated between subcutaneous and visceral fat depots in rodents and humans and was shown to impair adipogenesis in 3T3-L1 cells when overexpressed (31). It remains an open question whether differences in Vgll3 expression exist between different fat depots in humans.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health grants HL066088 and HL030568. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Rosen E. D., MacDougald O. A. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7: 885–896 [DOI] [PubMed] [Google Scholar]
- 2.Rosen E. D., Walkey C. J., Puigserver P., Spiegelman B. M. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14: 1293–1307 [PubMed] [Google Scholar]
- 3.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H. M., Erdjument-Bromage H., et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 454: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kajimura S., Seale P., Tomaru T., Erdjument-Bromage H., Cooper M. P., Ruas J. L., Chin S., Tempst P., Lazar M. A., Spiegelman B. M. 2008. Regulation of the brown and white fat gene programs through PRDM16/CtBP transcriptional complex. Genes Dev. 22: 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanueva C. J., Waki H., Godio C., Nielsen R., Chou W. L., Vargas L., Wroblewski K., Schmedt C., Chao L. C., Boyadjian R., et al. 2011. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 13: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tontonoz P., Hu E., Spiegelman B. M. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 79: 1147–1156 [DOI] [PubMed] [Google Scholar]
- 7.Barak Y., Nelson M. C., Ong E. S., Jones Y. Z., Ruiz-Lozano P., Chien K. R., Koder A., Evans R. M. 1999. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell. 4: 585–595 [DOI] [PubMed] [Google Scholar]
- 8.Nissen S. E., Wolski K. 2007. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N. Engl. J. Med. 356: 2457–2471 [DOI] [PubMed] [Google Scholar]
- 9.Mori T., Sakaue H., Iguchi H., Gomi H., Okada Y., Takashima Y., Nakamura K., Nakamura T., Yamauchi T., Kubota N., et al. 2005. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 280: 12867–12875 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka T., Yoshida N., Kishimoto T., Akira S. 1997. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 16: 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moitra J., Mason M. M., Olive M., Krylov D., Gavrilova O., Marcus-Samuels B., Feigenbaum L., Lee E., Aoyama T., Eckhaus M., et al. 1998. Life without white fat: a transgenic mouse. Genes Dev. 12: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waki H., Park K. W., Mitro N., Pei L., Damoiseaux R., Wilpitz D. C., Reue K., Saez E., Tontonoz P. 2007. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARγ expression. Cell Metab. 5: 357–370 [DOI] [PubMed] [Google Scholar]
- 13.Gupta R. K., Arany Z., Seale P., Mepani R. J., Ye L., Conroe H. M., Roby Y. A., Kulaga H., Reed R. R., Spiegelman B. M. 2010. Transcriptional control of preadipocyte determination by Zfp423. Nature. 464: 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hummasti S., Tontonoz P. 2006. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol. Endocrinol. 20: 1261–1275 [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Yehya N., Schadt E. E., Wang H., Drake T. A., Lusis A. J. 2006. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Nas A., Ingram-Drake L., Sinsheimer J. S., Wang S. S., Schadt E. E., Drake T., Lusis A. J. 2010. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics. 185: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong Q., Dalgin G., Xu H., Ting C. N., Leiden J. M., Hotamisligil G. S. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 290: 134–138 [DOI] [PubMed] [Google Scholar]
- 18.Smas C. M., Sul H. S. 1993. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 73: 725–734 [DOI] [PubMed] [Google Scholar]
- 19.Bernard F., Lalouette A., Gullaud M., Jeantet A. Y., Cossard R., Zider A., Ferveur J. F., Silber J. 2003. Control of apterous by vestigial drives indirect flight muscle development in Drosophila. Dev. Biol. 260: 391–403 [DOI] [PubMed] [Google Scholar]
- 20.Williams J. A., Bell J. B., Carroll S. B. 1991. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 5: 2481–2495 [DOI] [PubMed] [Google Scholar]
- 21.Paumard-Rigal S., Zider A., Vaudin P., Silber J. 1998. Specific interactions between vestigial and scalloped are required to promote wing tissue proliferation in Drosophila melanogaster. Dev. Genes Evol. 208: 440–446 [DOI] [PubMed] [Google Scholar]
- 22.Maeda T., Chapman D. L., Stewart A. F. 2002. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem. 277: 48889–48898 [DOI] [PubMed] [Google Scholar]
- 23.Chen H. H., Mullett S. J., Stewart A. F. 2004. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J. Biol. Chem. 279: 30800–30806 [DOI] [PubMed] [Google Scholar]
- 24.Mielcarek M., Piotrowska I., Schneider A., Günther S., Braun T. 2009. VITO-2, a new SID domain protein, is expressed in the myogenic lineage during early mouse embryonic development. Gene Expr. Patterns. 9: 129–137 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Sul H. S. 2009. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 9: 287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh J. M., Gao X., McKay J., McKay R., Salo Z., Graff J. M. 2006. Hedgehog signaling plays a conserved role in inhibiting fat formation. Cell Metab. 3: 25–34 [DOI] [PubMed] [Google Scholar]
- 27.Yang Y. K., Chen M., Clements R. H., Abrams G. A., Aprahamian C. J., Harmon C. M. 2008. Human mesenteric adipose tissue plays unique role versus subcutaneous and omental fat in obesity related diabetes. Cell. Physiol. Biochem. 22: 531–538 [DOI] [PubMed] [Google Scholar]
- 28.Catalano K. J., Stefanovski D., Bergman R. N. 2010. Critical role of the mesenteric depot versus other intra-abdominal adipose depots in the development of insulin resistance in young rats. Diabetes. 59: 1416–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atzmon G., Yang X. M., Muzumdar R., Ma X. H., Gabriely I., Barzilai N. 2002. Differential gene expression between visceral and subcutaneous fat depots. Horm. Metab. Res. 34: 622–628 [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto Y., Gesta S., Lee K. Y., Tran T. T., Saadatirad P., Kahn C. R. 2010. Adipose depots possess unique developmental gene signatures. Obesity (Silver Spring). 18: 872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gesta S., Bezy O., Mori M. A., Macotela Y., Lee K. Y., Kahn C. R. 2011. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc. Natl. Acad. Sci. USA. 108: 2771–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.