Abstract
Large-scale pruning, the removal of long neuronal processes, is deployed widely within the developing nervous system and is essential for proper circuit formation. In Drosophila the dendrites of the class IV dendritic arborization sensory neuron ddaC undergo large-scale pruning by local degeneration controlled by the steroid hormone ecdysone. The molecular mechanisms that control such events are largely unknown.
To identify new molecules that orchestrate this developmental degeneration, we performed a genetic interaction screen. Our approach combines the strength of Drosophila forward genetics with detailed in vivo imaging of ddaC neurons. This screen allowed us to identify headcase (hdc) as a new gene involved in dendrite pruning. hdc is evolutionarily conserved, but the protein's function is unknown. Here we show that hdc is expressed just before metamorphosis in sensory neurons that undergo remodeling. hdc is required in a cell-autonomous manner to control dendrite severing, the first phase of pruning. Our epistasis experiments with known regulators of dendrite pruning reveal hdc as a founding member of a new pathway downstream of ecdysone signaling.
Introduction
In recent years our understanding of neural development has increased dramatically (Mason, 2009), but we still know relatively little about the cellular and molecular mechanisms that control neuron pruning (Luo and O'Leary, 2005). Large-scale pruning, where long neuronal processes are removed, is deployed widely within the developing nervous system and is essential for the proper construction of neural circuits. The best studied example occurs during the development of cortical layer 5 projection neurons whose subcortical axons undergo a branch-specific pruning, dependent on their region of origin (Stanfield et al., 1982). Data from retinal ganglion cells (Nakamura and O'Leary, 1989) and thalamocortical neurons (Portera-Cailliau et al., 2005) reveal that pruned axons are removed by local degeneration, not by distal to proximal branch retraction.
In insects that undergo a complete metamorphosis, a number of larval neurons are remodeled during the pupal–adult transition to become incorporated into adult circuits (Truman and Reiss, 1976; Truman, 1990). Pioneering studies of Drosophila mushroom body γ-neurons (γ-MB) reveal that both axons and dendrites prune by local degeneration (Watts et al., 2003). Detailed observations of these events show that branch thinning is coincident with changes in the microtubule cytoskeleton, followed by severing and fragmentation. These features are also evident in the remodeling of sensory neuron dendrites (Kuo et al., 2005; Williams and Truman, 2005).
During metamorphosis, pruning is gated by a nuclear hormone receptor complex containing the ecdysone receptor (EcR) and ultraspiricle (usp), the fly RXR homolog (Schubiger et al., 1998). This nuclear receptor complex detects changes in the titer of the steroid hormone 20-hydroxy ecdysone (referred to hereafter as ecdysone). The loss of either partner results in a complete block of the pruning (Schubiger et al., 1998; Lee et al., 2000). The transcription factor Sox14, a downstream target of EcR, has recently been identified as a regulator of branch severing in a pathway that includes the large multidomain cytosolic protein Mical (Kirilly et al., 2009). Alongside this, a small number of other molecules have been identified as players in the pruning pathway, including components of the ubiquitin proteasome system (UPS) (Watts et al., 2003; Kuo et al., 2006), caspases (Kuo et al., 2006; Williams et al., 2006), ik2, and Kat60L (Lee et al., 2009). Nevertheless, the relationship between these molecules remains unclear.
To identify new molecules that orchestrate large-scale pruning and find links between known players, we developed a genome-wide interaction screen. This combines the strength of Drosophila forward genetics with detailed in vivo imaging of class IV dendritic arborizing (da) sensory neurons. Using this approach, we identified a novel regulator of pruning encoded by the gene called headcase (hdc). By developing new tools to test the requirement of pruning regulators in class IV da neurons, we have shown that hdc acts in a cell-autonomous manner to control the severing of primary dendrites in sensory neurons. Epistatic experiments reveal that hdc belongs to a new pathway downstream of the ecdysone receptor, independent of the transcription factor Sox14.
Materials and Methods
Fly stocks.
In this study Drosophila of either sex were used.
The following GAL4 driver strains were used: C161-GAL4 (Shepherd and Smith, 1996), expressed in five dorsal da neurons; ppk1.9-GAL4 driver (Grueber et al., 2003), expressed in ddaC and occasionally in isolated epidermal cells; 201Y-GAL4 (Yang et al., 1995), expressed in γ-MB neurons; and elavc155-GAL4, a general neuronal driver (Lin et al., 1994).
For this study we used the following flies: hdc43 and hdc50 (Weaver and White, 1995); UAS-hdcΔ2 and UAS-hdc (Steneberg et al., 1998); UAS-EcR-RNAiCA104 (Schubiger et al., 2005); UAS-Sox14, Mical15256 (Kirilly et al., 2009); UAS-Sp1-RNAi and UAS-btd-RNAi (Estella et al., 2003); EsgG66B FRT40A (Hayashi, 1996); UAS-BrmDN (K804R) (Elfring et al., 1998); and CBP-ΔQ(Kumar et al., 2004). UAS-hdc-RNAi-15532R2 was obtained from the National Institute of Genetics (Japan). UAS-Sox14-RNAi-10856GD and UAS-Mical-RNAi-105837KK were obtained from the Vienna Drosophila RNAi Centre. The RNAi lines used for the screen were obtained from the National Institute of Genetics (Japan) and the Vienna Drosophila RNAi Centre (Dietzl et al., 2007). Deficiencies used for the screen were obtained from Bloomington Stock Center (http://flystocks.bio.indiana.edu), as well as the following flies, UAS-Dicer2, hs-FLP122, and Ecrm554fs.
Mosaic analysis with a repressible cell marker.
For mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999) of da sensory neurons, clones were induced in the embryo by a double heat shock method (Grueber et al., 2002). The following flies were generated: female: w, elavC155-GAL4, UAS-RedStinger, hs-FLP122; /+; FRT82B, tub-GAL80/TM6B Tb cross with male: w; ppk-eGFP; FRT82B hdc43/TM6B Tb and female: w, elavC155-GAL4, UAS-redstinger, hs-FLP122; FRT40A tub-GAL80/CyO to cross with male: w/w; esgG66B FRT40A/CyO;ppk-eGFP.
ddaC neurons were identified at 18 h after puparium formation (APF) by the ppk-eGFP, with MARCM clones expressing a nuclear RedStinger reporter protein.
For the MARCM analysis of the gamma MB neurons, the clones were induced in first instar larvae by applying a 1 h heat shock at 37°C as described previously (Lee et al., 1999; Lee and Luo, 1999) with female: y, w, hs-FLP; 201Y-GAL4, mCD8-GFP/CyO; FRT82B, tub-GAL80/TM6B Tb.
Staging of animals.
Individual animals were collected at pupariation and maintained at 25°C in a Petri dish with moist filter paper. Staging was denoted as hours after puparium formation or APF.
Immunocytochemistry.
Immunocytochemistry was performed as described by Truman et al. (2004)). Primary antibodies used were rabbit anti-GFP diluted 1/500 (Invitrogen), mouse-anti HDC mAb U33.7 diluted 1:5 and kindly provided by Robert White (University of Cambridge, Cambridge, UK) (Weaver and White, 1995), and mouse anti-Sox14 diluted 1:200 kindly provided by Fengwei Yu (National University of Singapore, Singapore) (Kirilly et al., 2009).
Secondary antibodies used were FITC donkey anti-rabbit IgG diluted 1:500 from Jackson ImmunoResearch Laboratories and Cy3-conjugated donkey anti-rat or anti-mouse IgG diluted 1:500 Stratech Scientific.
Imaging, image analysis, and quantification.
Confocal images were taken at 1 μm intervals using a Zeiss LSM 510 system.
Stacks were assembled in NIH ImageJ (http://rsb.info.nih.gov/ij/). Images were adjusted only for brightness and contrast using Adobe Photoshop (Adobe Systems).
Third-instar larvae, white pre-pupae, and pupae until 12 h APF were directly imaged using confocal microscopy. After 12 h APF, pupae were peeled out of the pupal case. Images of neurons were taken as maximum projections of 15–40 optical sections at 1 μm intervals. Quantification of all live images was carried out by counting the number of primary and secondary dendrites in a 230 × 230 μm region of the dendritic field of the ddaC neurons, originating from the second to fifth abdominal segments.
Statistical analyses.
Mann–Whitney and Kruskal–Wallis statistical tests were performed in Graphpad Prism 5 with values of p < 0.05 deemed to be significant.
Results
A genetic interaction screen to uncover molecules that control large-scale pruning
A number of larval sensory neurons in Drosophila undergo remodeling during metamorphosis to become components of the adult nervous system (Williams and Shepherd, 1999). This remodeling is achieved by the removal of larval arborizations followed by the outgrowth of adult-specific processes (Williams and Truman, 2004). Dorsal dendritic arborizing sensory neuron C (ddaC) is a class IV dendritic arborizing neuron located in the dorsal body wall of each larval hemisegment. ddaC pruning takes place in two distinct phases: severing and clearance. The first phase begins at the onset of metamorphosis at 0 h after puparium formation, APF. At this time, the proximal dendrites of ddaC begin to thin and form bead-like structures (Fig. 1A). By 10 h APF (Fig. 1B), 80% of the primary dendrites are severed (n = 50). In the second phase of pruning, these severed dendrites are removed. By 18 h APF, the dendrites of ddaC have been completely removed in 85% of the dorsal body wall territories examined (n = 50) (Fig. 1C). The remaining 15% of dorsal fields contain only a few small pieces of the severed dendrites that are soon removed (Fig. 1C). The timing of these events relative to puparium formation is highly stereotyped.
Figure 1.

A screen to uncover new players of ddaC pruning. Live imaging of ddaC neurons labeled by ppk-GAL4 expressing UAS-CD8-GFP. A–C, A timeline of ddaC pruning. Z-projections of dorsal abdominal body wall at 0 (A), 10 (B), and 18 h APF (C), respectively. At 10 h APF, almost all the branches are severed close to the soma. By 18 h APF, the severed dendrites are cleared, only the soma and axon remain. D, Scheme showing the genes known to be involved in ddaC pruning; arrows represent direct interactions. E, The expression of EcR-RNAiCA104 in ddaC robustly blocks the early stages of ddaC pruning. Dendritic branches are not severed at 18 h APF. F, Interaction screen: flies carrying ppk-GAL4, UAS-CD8-GFP, and a null mutation for EcR are crossed with balanced deficiencies. Pupal dominant markers on balancer chromosomes allow the selection of the appropriate genotypes. The F1 pupae trans-heterozygous for EcR and a deficiency are imaged at 18 h APF and assessed for defects in pruning. G–I, ddaC neurons from pupae trans-heterozygous for EcR and three different deficiencies at 18 h APF. G, In category i of F1 pupae, pruning is normal, meaning that there is no genetic interaction. H and I show F1 pupae in which genetic interactions are evident. H, In category ii, clearance defects occur and pieces of dendrite are still in the field at 18 h APF; severing does not appear to be affected. I, In category iii, severing and clearance defects are evident with some dendrites still attached to the soma. J–L, Identification of the gene responsible for pruning defects within the deficiency Df(3R)ED6332. J, Trans-heterozygous neuron for Df(3R)ED6332 and EcR at 18 h APF. K, RNAi-15532R2, which targets headcase (hdc), one of the genes removed by Df(3R)ED6332, at 18 h APF. L, Genetic interaction between EcR and hdc43, a null allele of hdc, at 18 h APF.
Relatively few molecules involved in large-scale pruning have been identified (Fig. 1D). To isolate novel regulators, we designed a genetic interaction screen. As the ecdysone receptor is at the apex of this pathway (Fig. 1E), we used it to generate a sensitized background. We recombined the null allele EcRM554fs with ppk-GAL4>CD8::GFP to visualize ddaC neurons directly in live animal. Flies heterozygous for EcR show normal pruning. Deficiencies were brought into this background, and the appropriate F1 pupae were imaged at 18 h APF to determine whether there were any changes in pruning (Fig. 1F). Using this approach, we screened 135 deficiencies that cover ∼80% of the third chromosome. On average, the deficiencies used removed ∼40 genes. Our trans-heterozygous F1s fell into three distinct categories. One group showed wild-type pruning at 18 h APF (57 deficiencies of 135). Importantly, in this group there were F1 genotypes in which a large number of genes, i.e., >140, were removed and pruning was found to be normal (Fig. 1G), suggesting that the loss of one copy of many different genes is not enough to generate pruning phenotypes. In the second category of deficiencies (34 deficiencies of 135), we observed clearance defects. In such trans-heterozygotes, severing appears to take place, and yet there is an increased frequency of hemisegments containing dendritic fragments (Fig. 1H). In the third category, we identified a number of deficiencies that resulted in severing defects (44 deficiencies of 135). In this group, dendrites were still found attached to the cell body at 18 h APF (Fig. 1I). Within the third group we identified the deficiency, Df(3R)ED5196, which removed kat-60L1, a gene that has been shown to be required for branch severing (Lee et al., 2009), suggesting that the screen is specific. We chose to focus on the third category, as severing is one of the first steps in pruning, and little is known about the molecular machinery that orchestrates it. Within this group we decided to further investigate Df(3R)ED6332 (Fig. 1J), as it removed only a small number of genes.
To identify the gene(s) responsible for the phenotype observed with Df(3R)ED6332, we expressed all of the available RNAi lines against the three genes covered by this deficiency and imaged the morphology of ddaC at 18 h APF. We found that two RNAi lines against the gene headcase (hdc), CG15532, were able to generate the severing defect (Fig. 1K). To confirm these results, we performed a genetic interaction test between EcRM554fs and two null alleles of headcase, hdc43 (Fig. 1L) and hdc50 (data not shown). Both alleles, when trans-heterozygous with EcRm554fs, recapitulate the severing defects (9%, n = 22 for hdc43 and 37%, n = 8 for hdc50) observed with either the deficiency (16.7%, n = 6) or RNAi (80%, n = 10 for 15532R2). Taken together, these data strongly suggest that the gene headcase is the causative factor within Df(3R)ED6332.
Spatial and temporal dynamics of hdc
To explore where and when hdc is expressed, we performed immunostaining with an antibody against HDC. headcase starts to be detectable in da sensory neurons only in late wandering third instar larvae (wL3) (Fig. 2B). Its expression becomes stronger following puparium formation (0 h APF) (Fig. 2C) and is maintained until at least 5 h APF (Fig. 2D).
Figure 2.
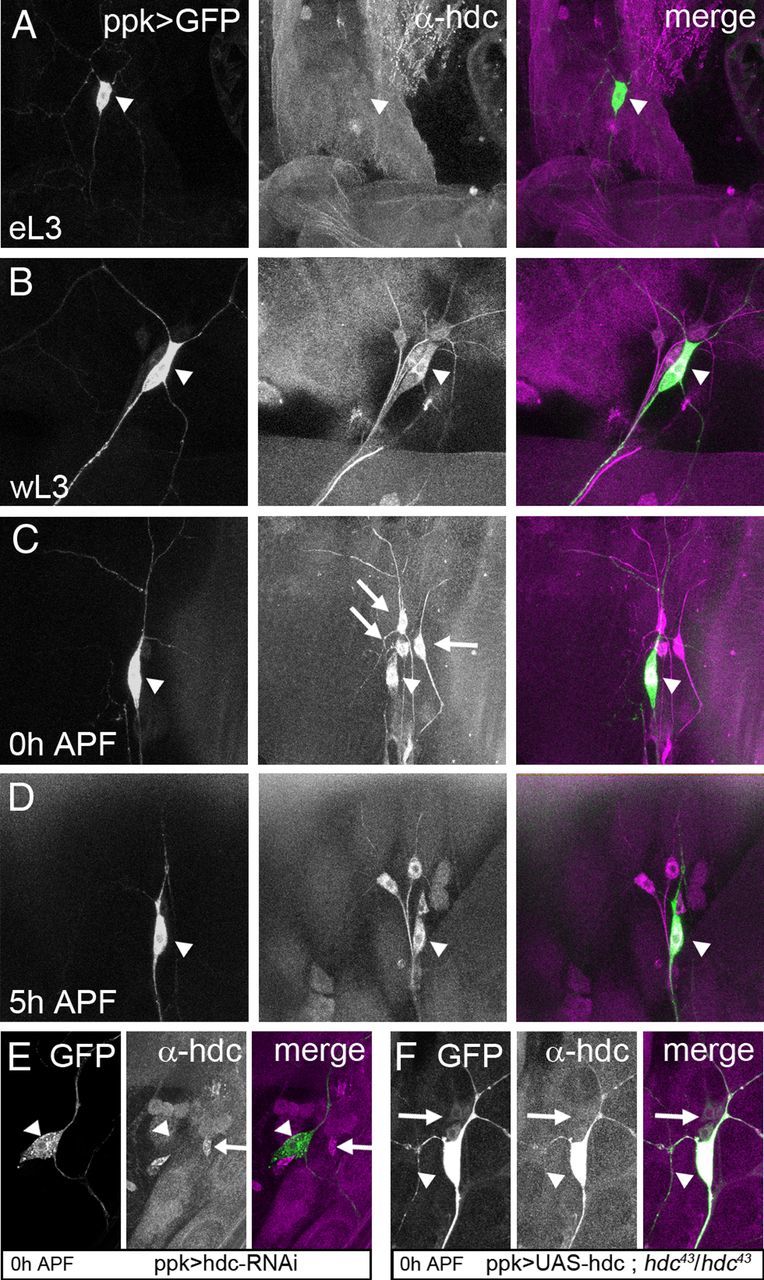
Spatial localization and temporal dynamics of hdc expression. A–D, Sensory neurons revealed by ppk-GAL4 driving UAS-CD8-GFP. These larval fillets are stained against HDC, and the GFP is observed without antibody. The arrowhead shows the position of ddaC neurons. A, hdc expression is not detectable in ddaC at early third instar larva (eL3). B–D, hdc expression starts during the wandering stage of L3 (wL3) (B), reaches a maximum at 0 h APF (C), and is still evident at 5 h APF (D). hdc is also expressed in some neighboring neurons (C, arrows). E and F are control experiments for HDC antibody. E, The specific expression of hdc-RNAi in ddaC neurons results in an absence of HDC staining (arrowhead), whereas this staining is still present in the neighboring da neurons (arrow). F, In hdc43 homozygous mutant background, HDC staining is absent in da neurons (arrows) except in ddaC (arrowheads), where we are overexpressing HDC.
The staining reveals a cytoplasmic localization of HDC in da neurons, with an enrichment close to the nucleus. Although this perinuclear staining is intense, HDC is also present at lower levels in the dendrites and the axon. Between 0 and 5 h APF there is no obvious change in the localization of HDC within ddaC. Staining was found to be completely absent in homozygous null mutants (data not shown). We also observed staining in three of the neighboring sensory neurons (Fig. 2C, arrows). We tested HDC antibody on flies expressing an RNAi against hdc specifically in ddaC neurons. In these individuals, we saw a loss of HDC staining in these neurons, whereas HDC was still detectable in the neighboring da neurons (Fig. 2E). As a further control, we overexpressed HDC in ddaC neurons in flies which were homozygous mutants for hdc. Under these conditions, we observed a strong HDC staining only in ddaC neurons (Fig. 2F). In summary, hdc is expressed at a time and in a location that is compatible with a role in sensory neuron pruning.
hdc is necessary for dendrite severing
The localization and onset of hdc expression in ddaC neurons and the targeted expression of RNAi tools designed to knock down hdc suggest that it may act in a cell-autonomous manner. To confirm this, we performed mosaic analysis with the MARCM system, generating ddaC neurons homozygous for a null allele of hdc.
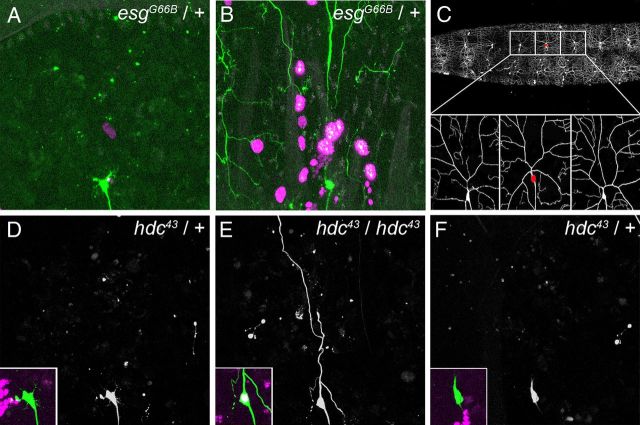
Although we, along with others, have routinely used the MARCM system to investigate the cell-autonomous requirement of gene products during pruning, we felt the need to advance this approach. We combined a ppk-eGFP reporter construct with a nuclear localized RedStinger reporter under the control of the enhancer trap elavC155-GAL4. ppk-eGFP labels all class IV da neurons whether clonal or nonclonal, whereas the homozygous mutant clones are marked by elavC155-GAL4, which expresses strongly in the nervous system and also at low level in other tissues, including the epidermis. We have thus optimized the MARCM system to unequivocally identify our clones as ddaC neurons, visualize both the location and frequency of clones in neighboring tissues, and, importantly, compare our homozygous mutant ddaC clones with adjacent heterozygous control ddaC neurons in the same animal (Fig. 3C). To test this system, we used flies heterozygous for the null allele esgG66B, where ddaC pruning is wild type (data not shown). However, in pupae with MARCM clones, we either found heterozygous esg control (nonclonal) neurons that prune normally (Fig. 3A) or heterozygous neurons where pruning was strongly delayed (Fig. 3B). With our enhanced system we can now visualize and remove pupae with global developmental defects from our future analysis.
Figure 3.
Cell-autonomous requirement of hdc in ddaC pruning. This figure shows live imaging of ddaC neurons labeled with ppk-eGFP; the MARCM clone is labeled with UAS-RedStinger, which provides a strong nuclear RFP signal, driven by elavC155-GAL4. A and B show two neurons from different 18 h APF pupae in which heat shocks were used to produce esgG66B MARCM clones. Neither of these neurons are MARCM clones, so they are heterozygous (esgG66B/+) and thus should not affect ddaC pruning. In A, the neuron undergoes wild-type pruning. In B, the neuron shows a strong pruning defect, not due to a cell autonomous effect but to a global developmental defect, as suggested by the high number of clonal epidermal cells expressing RedStinger (esgG66B homozygous; magenta nucleus). C, Modified MARCM strategy: selection of a MARCM clone ddaC neuron by its nuclear expression of UAS-RedStinger (red dot) and the two nonclonal neighboring neurons, which were used as controls. D–F show three adjacent ddaC neurons at 18 h APF belonging to the same pupa. D and F are not clonal, as they show no RedStinger expression (insets). The pruning of these neurons is wild type, and no pieces of dendrite are left in the field. The neuron in E is homozygous mutant for hdc, as revealed by RedStinger expression (inset). This neuron exhibits a severing defect, as a robust primary dendritic branch is still attached to the soma.
Using our modified MARCM technique, we performed clonal analysis with hdc43. We generated 16 pupae containing a single clonal (mutant) ddaC neuron and imaged the two adjacent nonclonal heterozygous ddaC neurons as controls (Fig. 3C). We observed severing defects in 50% of ddaC neurons homozygous mutant for hdc (Fig. 3E) where the neighboring control neurons were presenting no phenotype (Fig. 3D,F). Figure 3E is representative of how these severing phenotypes appear, with predominantly one or two dendrites remaining attached to the soma at 18 h APF. From these experiments, we conclude that hdc is required for the severing of dendrites in ddaC neurons and acts in a cell-autonomous manner.
hdc is required but not sufficient for ddaC branch severing
As the expression of hdc coincides with the onset of dendritic pruning and is required cell autonomously for this process, we wanted to establish whether the precocious expression of hdc is sufficient to initiate branch severing. The expression of wild-type headcase from early larval life using ppk-GAL4 did not disrupt the development of the proximal dendrites of ddaC, and there was no evidence of degeneration or cell death (Fig. 4A,C). We then looked at the same animals following pupariation, when native hdc is expressed, to determine whether overexpression can cause precocious severing. We quantified the number of I° (branches located between the soma and the first branch point within the arborization) and II° branches (those from the first to the second branch point) still attached to the soma at 6 h APF. This proximal region was chosen because it is where severing normally takes place. At 0 h APF, the average number of I° and II° dendrites were similar between ppk-GAL4/w1118 flies and ppk-GAL4>UAS-hdc flies with 10 (n = 21) and 9 (n = 20) dendrites, respectively. At 6 h APF, we found 6.2 and 5.2 dendrites in ppk-GAL4 (n = 38) and ppk>UAS-hdc (n = 19) flies, respectively. There was no statistically significant difference between the two groups (p = 0.3057). We conclude that hdc overexpression during either larval life or soon after puparium formation is not sufficient to induce or accelerate dendrite severing.
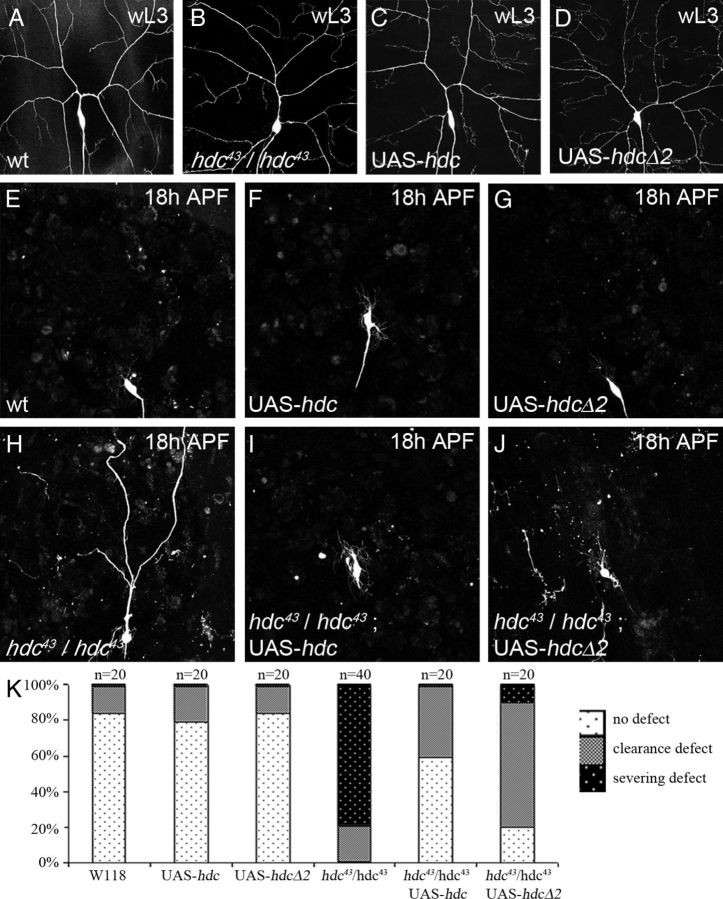
Figure 4.
Both hdc forms work in pruning. ddaC neurons labeled with UAS-CD8-GFP driven by ppk-GAL4. This combination is also used to drive UAS-hdc or UAS-hdcΔ2. A–D, Panel showing that the number of I° and II° dendrites in ddaC neurons in wandering larvae (wL3), before pruning, does not appear to be modified in the different genotypes used for the gain-of-function experiments. E–G, Panel showing that at 18 h APF, ddaC neurons are pruned normally when overexpressing UAS-hdc or UAS-hdcΔ2. H–J, At 18 h APF, the strong severing defect found in hdc43 homozygous background (H) is rescued by the expression of either UAS-hdc (I) or UAS-hdcΔ2 (J). There is more disruption of dendrite clearance in the UAS-hdcΔ2-rescued neurons (J). K, Quantification of both clearance and severing events confirming that both the short and the long forms of HDC can rescue hdc43/hdc43 pruning in a significant manner (p < 0.0001). Nevertheless, the short form (UAS-hdcΔ2) is less efficient than the expression of HDC full length.
The short form of hdc is able to rescue hdc loss of function during pruning
A curious feature of hdc is the presence of a stop codon in the middle of the gene, allowing the generation of two distinct proteins from the same transcript. headcase generates a short form termed hdcS (70 kDa) and a full-length form (120 kDa) referred to as hdcFL (Steneberg et al., 1998). To determine which form was important during pruning, we performed rescue experiments that took advantage of the fact that homozygous mutant hdc animals can survive until late pupal stages, after pruning is completed. We used two transgenes, UAS-hdcΔ2, which allows the generation of a protein equivalent to hdcS, and UAS-hdc, which can produce both the full-length and the short form in flies (Steneberg et al., 1998). In homozygous hdc43 animals we observed ∼80% of ddaC neurons with severing defects at 18 h APF (Fig. 4H,K). The specific expression of either the UAS-hdc or UAS-hdcΔ2 transgene in ddaC neurons was able to rescue these defects at 18 h APF, with only 0–10% of the neurons showing a failure in severing (Fig. 4I–K). Although the short form generated by UAS-hdcΔ2 was sufficient to rescue hdc function in the severing of ddaC neurons, qualitative analyses showed that in around 80% of neurons the rescue was incomplete, with severed pieces of dendrites in the field for UAS-hdcΔ2 pupae against only 40% for UAS-HDC pupae (Fig. 4 H–J). This data suggests that hdcΔ2 is sufficient to rescue ddaC pruning but is not as efficient as hdcFL.
headcase is also required for pruning in other neuronal classes
During metamorphosis, larval neurons undergo remodeling or programmed cell death (Truman et al., 1990). Within the dorsal group of sensory neurons there are 6 da neurons, dda A–F, which represent the four different classes of da neurons (Grueber et al., 2002). At the onset of metamorphosis, the class I da neurons, ddaD and ddaE, and class IV ddaC are remodeled, whereas the class II neuron ddaB and class III neurons ddaA and ddaF undergo apoptosis (Williams and Truman, 2005). To determine which of these neurons express hdc, we performed immunostaining on all the multidendritic sensory neurons in the dorsal group labeled by the enhancer trap line C161-GAL4. We found that the class I, II, and III da neurons were HDC positive, alongside the dorsal bipolar dendrite neuron dbd (Fig. 5A–C).
Figure 5.
Role of hdc in other neurons that undergo remodeling. A–C, We used C161-GAL4 to express UAS-CD8::GFP in 5 other da neurons and stain the wL3 fillets with HDC antibody. hdc is expressed in the other class of da neurons, dbd neuron (purple arrows), and ddaC (yellow arrowhead). D–F, We used C161-GAL4 expressing UAS-CD8::GFP to visualize dorsal sensory neurons (D). In wild-type pupae at 24 h APF (E), only two da neurons and dbd neuron remain in the field (arrows), and they have undergone pruning. The three other da neurons labeled in C161-GAL4 have undergo apoptosis by this time. F, In hdc43/hdc43 at 24 h APF, ddaA, B and F are absent, demonstrating that hdc is not required for the apoptosis of da neurons. The remaining neurons, ddaD, ddaE, and dbd, show consistent severing defects (arrows). G–I, Twenty-four hour APF γ-MB neurons labeled with 201Y-Gal4>UAS-mCD8-GFP. G, γ-MB neurons have almost completely pruned both their axons (arrows) and dendrites. H, In MARCM clones for hdc43, the pruning does not appear to be modified, and only a few axons remain (arrows). I, Pruning is suppressed by downregulating EcR with EcR-RNAiCA104, resulting in the persistence of axons at 24 h APF (arrows).
To determine whether hdc is deployed in the remodeling of these neurons, we used the enhancer trap C161-GAL4 to look at the effect of hdc loss of function in class I, II, and III ddaA B, D, E, and F neurons. At 0 h APF we saw no obvious differences between these neurons in wild-type and hdc homozygous mutant flies, suggesting that their development does not depend on hdc (Fig. 5D and data not shown). By 24 h APF in wild-type flies, the larval dendrites of ddaD, ddaE, and dbd neurons were completely pruned back, whereas in hdc43/hdc43 flies there were still a number of branches attached to the soma in 85% of the dorsal da groups (n = 20) (Fig. 5E,F). At 24 h APF, ddaA, B, and F were absent in both wild-type and hdc mutant flies, revealing that the hdc function was not required for the programmed cell death of these neurons. Moreover, we found that the increased expression of hdc in ddaA, B, or F did not prevent their death (data not shown).
Mushroom body γ-MB neurons remodel their axons and dendrites during metamorphosis before elaborating their adult-specific projections (Watts et al., 2003). To determine whether hdc is required during γ-neuron pruning, we used the driver 201Y-GAL4 to express hdc-RNAi. Alongside this, we generated homozygous hdc43 MB neuroblast clones and visualized the morphology of mutant (hdc/hdc) at 24 h APF, when axonal and dendritic pruning is largely complete (Fig. 5G). We found no evidence for a disruption in the development of γ-MB neurons mutant for hdc (Fig. 5H, n = 8), whereas expression of EcR RNAi with 201Y-GAL4 leads to a robust block of pruning (Fig. 5I). These data are consistent with a lack of HDC staining in γ-MB neurons (data not shown).
We conclude that hdc is involved in the pruning of different classes of sensory neurons but is not required in γ-MB neuron pruning, whereas EcR is required for pruning in both. We also find that removing hdc does not disrupt the programmed cell death of class II and III da neurons.
hdc is regulated by EcR but not by Sox14
Previous studies have shown that headcase can be regulated in imaginal tissues and the tracheal system by the transcription factors escargot (esg), buttonhead (btd), and Sp1 (Steneberg et al., 1998; Estella et al., 2003). To determine whether any of these play a role in pruning, we generated esg mutant MARCM clones n = 10 (Fig. 6A) and expressed RNAi against either btd or Sp1 using the ppk driver (Fig. 6B, C). None of these regulators disrupted pruning in ddaC, suggesting that hdc is controlled differently in remodeling neurons.
Figure 6.
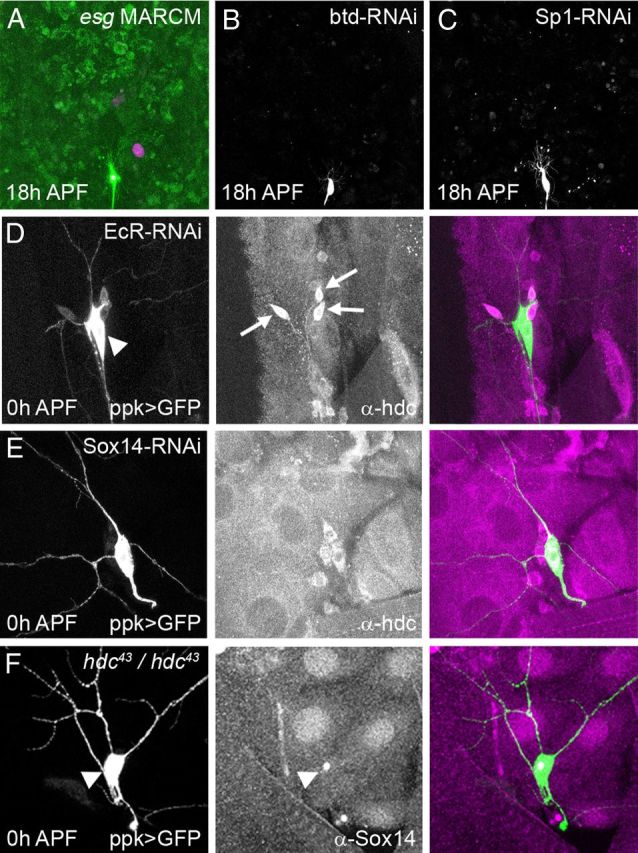
Regulation of hdc expression. A, MARCM clones for esgG66B show no defects in pruning at 18 h APF, (RedStinger expression in nucleus of homozygous mutant clone), B, C, ppk-GAL4>btd-RNAi (B) and ppk-GAL4>Sp1-RNAi (C) at 18 h APF; the pruning of ddaC neurons is not disrupted in either background. D–F, Fillets of 0 h APF pupa where ddaC neurons are labeled with UAS-CD8-GFP driven by ppk-GAL4, which is also used to drive EcR-RNAiCA104 or Sox14-RNAi when necessary. D, The expression of EcR-RNAiCA104 in ddaC neurons (arrowhead) suppresses hdc expression in a cell autonomous manner. HDC can still be seen in the neighboring da neurons (arrows). E, The expression of Sox14-RNAi in ddaC neurons does not affect hdc expression. F, In a hdc43/hdc43 background, Sox14 staining is still present in the nucleus of ddaC neurons (arrowhead)s.
As hdc expression closely follows the prepupal pulse of ecdysone, we asked whether hdc is regulated by ecdysone signaling. EcR was downregulated in ddaC by expressing EcR-RNAiCA104 with ppk-GAL4 (Fig. 1E). This cell-autonomous knockdown of EcR resulted in a lack of hdc expression at 0 h APF, when it is normally at its strongest (Fig. 6D). These data show that hdc is downstream of EcR and that it could be either a direct or indirect target. Recently, Brahma (Brm) and CREB binding protein (CBP) have been shown to be epigenetic factors that facilitate activation of downstream ecdysone response genes during pruning (Kirilly et al., 2011). The specific expression of a Brm or CBP dominant negative in ddaC results in a diminution of hdc expression at 0 h APF; this reduction is more pronounced with CBP dominant negative (data not shown). Thus far, Sox14 is the only direct target of EcR known to play a role in neuron pruning, and we wondered whether it could be controlling the expression of hdc. Previous work has demonstrated that RNAi against Sox14 leads to a loss of Sox14 staining and a strong severing phenotype in ddaC. However, this knockdown of Sox14 does not seem to affect hdc expression in ddaC neurons (Fig. 6E), suggesting that hdc expression is independent of Sox14. Conversely, we found that Sox14 expression was not affected in hdc mutant flies (Fig. 6F). Another possibility is that hdc could affect Sox14 function. We used the expression of Mical, a target of Sox14, as a read-out for Sox14 activity and found that Mical expression was also not modified in hdc mutant flies (data not shown).
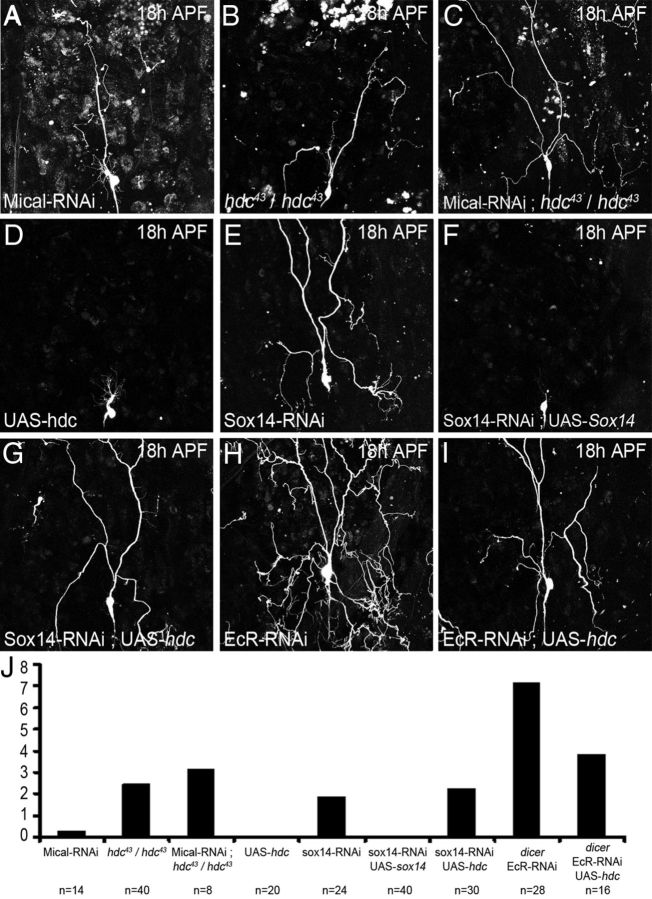
These experiments suggest that hdc and Sox14 belong to two parallel pathways downstream of EcR. To explore this idea further, we performed two additional functional assays. Firstly we expressed an RNAi against Mical in flies homozygous for hdc43 and found that 100% of ddaC neurons had severing phenotypes and an average of 3.125 I° and II° dendrites attached to the soma (Fig. 7C). This is compared to 20% severing defects and 0.3 dendrites attached with Mical-RNAi alone (Fig. 7A) or 80% severing defects with 2.45 dendrites attached in homozygous hdc43 alone (Fig. 7B). This reveals an additive phenotype for the double loss of function. Alongside this, the overexpression of Mical has previously been shown to partially rescue Sox14 loss of function (Kirilly et al., 2009). We found that the overexpression of hdc in a Sox14 loss-of-function background was unable to rescue the Sox14 phenotype (Fig. 7D–G,J). Interestingly, we found that hdc overexpression limits the severing defects induced by EcR-RNAiCA104 in ddaC at 18 h APF, even though they remain very strong (Fig. 7H–J). In a reverse experiment, we removed one copy of EcR in flies homozygous for hdc43 and found that 94% of ddaC neurons had severing phenotypes with an average of 3.5 I°and II° dendrites remaining attached to the soma (n = 16), compared to 80% with severing defects and 2.45 dendrites remaining in flies homozygous for hdc43 (n = 40). Taken together, these data strongly suggest that hdc and Sox14 belong to different parallel pathways downstream of EcR in pruning neurons.
Figure 7.
Epistasis experiments between hdc, Sox14, and EcR. ddaC neurons labeled with UAS-CD8-GFP driven by ppk-GAL4 at 18 h APF were also used to drive Mical-RNAi, UAS-hdc, Sox14-RNAi, UAS-Sox14, Mical-RNAi, and EcR-RNAiCA104 alone or in combination. A, The expression of Mical-RNAi can lead to severing defect in 20% of ddaC neurons. B, Severing defect observed in 80% of flies homozygous for hdc43. C, The expression of Mical-RNAi in ddaC neurons of homozygous hdc43 flies has an additive effect with 100% of the ddaC neurons presenting severing defect. D, hdc gain-of-function does not affect pruning of ddaC neurons. E, F, The strong severing defect observed with the expression of Sox14-RNAi (E) is completely rescued by the over expression of UAS-Sox14 (F). G, The expression of UAS-hdc is not sufficient to rescue the severing defect generated by Sox14-RNAi. H, I, The almost complete suppression of pruning in ddaC neuron expressing EcR-RNAiCA104 (H) is partially rescued by the overexpression of hdc (I). J, Quantification of I° and II° dendrites remaining attached to the soma. There is no significant difference between Sox14-RNAi and the combination of Sox14-RNAi and UAS-hdc. Although not total, the rescue of EcR-RNAiCA104 phenotype by UAS-hdc is significant (p < 0.0001).
Discussion
Large-scale pruning is deployed widely within the developing nervous system and yet little is known about the cellular and molecular mechanisms controlling it. This active removal of long neuronal processes is dynamic, with individual branches undergoing remodeling, severing, and clearance. Like programmed cell death, local branch-specific auto-destruction programs must be tightly regulated but also robust in their execution. We have exploited the metamorphic remodeling of the Drosophila sensory system to develop an interaction screen that allowed us to identify headcase, a molecule that controls branch severing.
A genetic interaction screen reveals a role for hdc in dendrite severing
Most of the genes involved in neuronal pruning have been identified using candidate-based approaches. With the exception of Sox14, these players are required for EcR expression (Lee et al., 1999; Schuldiner et al., 2008; Boulanger et al., 2011) or are relatively far downstream, i.e., direct regulators of the cytoskeleton (Lee et al., 2009). Our goal is to bridge this gap by identifying molecules that link EcR with these downstream targets. We established a genetic interaction screen that is both open ended and specific due to the detailed in vivo imaging of class IV dendritic arborizing sensory neurons. Interaction screens by their nature reveal targets that are in close genetic proximity. Here we used the null allele EcRM554fs as a sensitized background, as the ecdysone receptor sits at the apex of the pruning pathway. By focusing on disruptions to branch severing events, we have also biased the screen toward the discovery of molecules controlling the earliest steps of the pruning pathway. Using defined chromosomal deficiencies, we removed large numbers of genes at a time and could then locate the gene of interest with relative ease. This strategy allowed us to identify headcase (hdc), CG15532, as the gene responsible for the severing phenotype. Previous work in Drosophila revealed that a loss of hdc disrupts the differentiation of imaginal primordia during pupal development and that its expression closely prefigures the re-entry of imaginal cells into the cell cycle (Weaver and White, 1995). Headcase is an evolutionarily conserved protein, and growing evidence suggests that the human homolog HECA plays a key role in carcinogenesis (Makino et al., 2001; Chien et al., 2006; Dowejko et al., 2009; Dowejko et al., 2012). It is a highly basic protein (pI, 9.6) and could therefore be involved in protein–protein or protein–RNA interactions. Currently, there are no obvious domains that give an indication of how HDC acts within the cell. This shows one of the benefits of such an open-ended screen, as we would clearly not have selected hdc using candidate-based approaches. We decided to explore its biological function and establish where it fits within the pruning pathway.
hdc acts cell autonomously during dendrite pruning
We have improved MARCM-based approaches for studying pruning in the remodeling of sensory neurons by using a GAL4-independent tool (ppk-eGFP) to visualize all class IV da sensory neurons in combination with a nuclear localized RedStinger protein to reveal MARCM clones (GAL80 minus). This approach has a number of advantages. Firstly, it allows the easy selection of clones and unequivocal identification of ddaC neurons. Secondly, it provides the opportunity to compare mutant ddaC clones with adjacent heterozygous ddaC “control neurons” within the same pupa. This is a robust internal control, which is particularly important as the long embryonic heat shocks used to generate mitotic clones within the sensory system also induce clones in many other tissues. As elavC155-GAL4 also expresses GAL4 in epidermal cells, it gives us insight into the global frequency of mitotic clones. Such “invisible” non-neural clones can disrupt the overall timing of puparium development, leading to non-cell autonomous effects on neuron pruning. With this modified version of the MARCM, we were able to account for these issues and confirm a cell-autonomous role for HDC in branch severing.
hdc overexpression did not result in precocious or accelerated severing, indicating that it is not a limiting factor. These data and the lack of known catalytic domain points toward HDC acting in concert with other factors in a complex.
hdc expression is regulated by EcR but not by Sox14
Our expression data show that hdc is expressed in da sensory neurons from the end of larval life. This immunostaining shows that HDC is localized close to the nucleus, but not in it. It is also present at lower levels in proximal axons and dendrites. After branch severing begins, the majority of HDC remains perinuclear, suggesting that the specificity of HDC action is not likely to come from gross changes in its subcellular localization. The fact that headcase is not ubiquitously expressed but has a clear tissue-specific and temporal expression profile suggests that it is not a general factor but a notable component for gating the severing process. Normally, two forms of headcase are generated from a single transcript by a readthrough mechanism. Using transgenes, we find that both isoforms of HDC can rescue the pruning phenotype and that the long form is more effective. This mirrors the requirement for HDC function in the developing tracheal system (Steneberg et al., 1998; Steneberg and Samakovlis, 2001).
Previous studies have shown that hdc is regulated by buttonhead and Sp1 in imaginal discs or by escargot in the trachea. We wondered if one of these transcription factors could also regulate hdc expression in ddaC neurons. The loss of function of these genes does not lead to any pruning defects, demonstrating that they are not required for hdc regulation within the peripheral nervous system. Thus, depending on the tissue, hdc seems to be regulated by different transcription factors. Importantly, the removal of EcR abolishes hdc expression in ddaC neurons, confirming that it is under the control of ecdysone signaling, downstream of EcR in the pruning pathway. It is unlikely that hdc is a direct target of EcR since it was never found in previous microarray analyses looking for EcR targets (Lee et al., 2003; Li and White, 2003; Beckstead et al., 2005). As Sox14 is the only downstream transcription factor known to be involved in neuron pruning, we wanted to determine whether it could regulate hdc expression. Both immunostaining and rescue experiments lead us to the conclusion that there is no cross-regulation between Sox14 and hdc in ddaC. Altogether, this data strongly supports the proposal that hdc and Sox14 belong to two parallel pathways downstream of EcR. This observation of two and probably more independent pathways is consistent with the need for the precise and active control of this auto-destructive machinery in different neuron types and different compartments within the cell.
hdc function is conserved in other classes of sensory neurons
To explore the role of hdc in other cell types, we looked at different neurons that remodel during metamorphosis. Our immunostaining revealed that hdc is expressed in all of the multidendritic (md) neurons we looked at, apart from dmd1. We found that loss of hdc function leads to a severing defect in class I and IV da sensory neurons and the dorsal bipolar dendrite neuron, dbd. Moreover, hdc loss or gain of function in class II and III da neurons does not change their fate, as they still undergo apoptosis (data not shown). We also found that headcase does not play a role in the γ-MB neuron pruning. This observation is interesting as we are looking at three distinct types of remodeling decisions within the fly nervous system: the loss of dendrites in ddaC pruning, the removal of both axons and dendrites in γ-MB, and cell death for class II and III da neurons. None of these appear to be a default state, because suppression of the pruning does not trigger apoptosis as seen in this study, nor does the blockage of cell death initiate branch severing (Williams and Truman, 2005). All three outcomes are triggered by the same ecdysone signal via EcR and one of its targets, Sox14. Here, we discovered a downstream target of the ecdysone signaling specific to dendrite pruning in a subset of remodeling neurons. This target is the founding member of a new pathway parallel to Sox14. We have exploited the metamorphic development of neurons and ecdysone signaling, which is of course specific to the phylum Ecdyzoa, but our expectation is that the batteries of downstream genes deployed during pruning are likely to be evolutionarily conserved and, thus, a useful entry point for investigating general principles of neurite pruning.
Conclusion
We have revealed for the first time that hdc is required in a cell-autonomous manner at the severing step during md neuron pruning. Importantly, our data also reveals that hdc acts independently of Sox14, despite both being under the control of EcR. These observations highlight that pruning is an association of multiple parallel pathways downstream of EcR.
Furthermore, we show through the identification of hdc that we have developed a genetic interaction screen that is an efficient and elegant way to populate the pruning pathway. This is a fundamental step toward understanding the complex regulation of this active and tightly regulated auto-destruction process.
Footnotes
This work was funded by the Medical Research Council and the Wellcome Trust. We thank Shigeo Hayashi, Ginés Morata, Christos Samakovlis, Rob White, Fengwei Yu, the NIG-FLY (Fly Stocks of National Institute of Genetics, Japan), the Vienna Drosophila RNAi Center, and the Bloomington Stock Center for generously providing fly stocks and other reagents. We also thank David Brierley, Muriel Boube, Matthias Landgraf, and Margrit Schubiger for helpful comments and suggestions on the manuscript.
References
- Beckstead RB, Lam G, Thummel CS. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger A, Clouet-Redt C, Farge M, Flandre A, Guignard T, Fernando C, Juge F, Dura JM. ftz-f1 and Hr39 opposing roles on EcR expression during Drosophila mushroom body neuron remodeling. Nat Neurosci. 2011;14:37–44. doi: 10.1038/nn.2700. [DOI] [PubMed] [Google Scholar]
- Chien CC, Chang CC, Yang SH, Chen SH, Huang CJ. A homologue of the Drosophila headcase protein is a novel tumor marker for early-stage colorectal cancer. Oncol Rep. 2006;15:919–926. [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dowejko A, Bauer RJ, Muller-Richter UD, Reichert TE. The human homolog of the Drosophila headcase protein slows down cell division of head and neck cancer cells. Carcinogenesis. 2009;30:1678–1685. doi: 10.1093/carcin/bgp189. [DOI] [PubMed] [Google Scholar]
- Dowejko A, Bauer R, Bauer K, Muller-Richter UD, Reichert TE. The human HECA interacts with cyclins and CDKs to antagonize Wnt-mediated proliferation and chemoresistance of head and neck cancer cells. Exp Cell Res. 2012;318:489–499. doi: 10.1016/j.yexcr.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR, Daubresse G, DePace A, Kennison JA, Tamkun JW. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C, Rieckhof G, Calleja M, Morata G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development. 2003;130:5929–5941. doi: 10.1242/dev.00832. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Hayashi S. A Cdc2 dependent checkpoint maintains diploidy in Drosophila. Development. 1996;122:1051–1058. doi: 10.1242/dev.122.4.1051. [DOI] [PubMed] [Google Scholar]
- Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, Low BC, Kolodkin AL, Wang H, Yu F. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci. 2009;12:1497–1505. doi: 10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilly D, Wong JJ, Lim EK, Wang Y, Zhang H, Wang C, Liao Q, Wang H, Liou YC, Yu F. Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron. 2011;72:86–100. doi: 10.1016/j.neuron.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Jamal T, Doetsch A, Turner FR, Duffy JB. CREB binding protein functions during successive stages of eye development in Drosophila. Genetics. 2004;168:877–893. doi: 10.1534/genetics.104.029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci U S A. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13:350–357. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci U S A. 2009;106:6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Lin DM, Fetter RD, Kopczynski C, Grenningloh G, Goodman CS. Genetic analysis of Fasciclin II in Drosophila: defasciculation, refasciculation, and altered fasciculation. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Luo L, O'Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- Makino N, Yamato T, Inoue H, Furukawa T, Abe T, Yokoyama T, Yatsuoka T, Fukushige S, Orikasa S, Takahashi T, Horii A. Isolation and characterization of the human gene homologous to the Drosophila headcase (hdc) gene in chromosome bands 6q23–q24, a region of common deletion in human pancreatic cancer. DNA Seq. 2001;11:547–553. doi: 10.3109/10425170109041340. [DOI] [PubMed] [Google Scholar]
- Mason C. The development of developmental neuroscience. J Neurosci. 2009;29:12735–12747. doi: 10.1523/JNEUROSCI.4648-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, O'Leary DD. Inaccuracies in initial growth and arborization of chick retinotectal axons followed by course corrections and axon remodeling to develop topographic order. J Neurosci. 1989;9:3776–3795. doi: 10.1523/JNEUROSCI.09-11-03776.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portera-Cailliau C, Weimer RM, De Paola V, Caroni P, Svoboda K. Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 2005;3:e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M, Wade AA, Carney GE, Truman JW, Bender M. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development. 1998;125:2053–2062. doi: 10.1242/dev.125.11.2053. [DOI] [PubMed] [Google Scholar]
- Schubiger M, Carré C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–5248. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd D, Smith SA. Central projections of persistent larval sensory neurons prefigure adult sensory pathways in the CNS of Drosophila. Development. 1996;122:2375–2384. doi: 10.1242/dev.122.8.2375. [DOI] [PubMed] [Google Scholar]
- Stanfield BB, O'Leary DD, Fricks C. Selective collateral elimination in early postnatal development restricts cortical distribution of rat pyramidal tract neurones. Nature. 1982;298:371–373. doi: 10.1038/298371a0. [DOI] [PubMed] [Google Scholar]
- Steneberg P, Samakovlis C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001;2:593–597. doi: 10.1093/embo-reports/kve128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneberg P, Englund C, Kronhamn J, Weaver TA, Samakovlis C. Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the Drosophila trachea. Genes Dev. 1998;12:956–967. doi: 10.1101/gad.12.7.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW. Metamorphosis of the central nervous system of Drosophila. J Neurobiol. 1990;21:1072–1084. doi: 10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- Truman JW, Reiss SE. Dendritic reorganization of an identified motoneuron during metamorphosis of the tobacco hornworm moth. Science. 1976;192:477–479. doi: 10.1126/science.1257782. [DOI] [PubMed] [Google Scholar]
- Truman JW, Fahrbach SE, Kimura K. Hormones and programmed cell death: insights from invertebrate studies. Prog Brain Res. 1990;86:25–35. doi: 10.1016/s0079-6123(08)63164-7. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- Weaver TA, White RA. headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development. 1995;121:4149–4160. doi: 10.1242/dev.121.12.4149. [DOI] [PubMed] [Google Scholar]
- Williams DW, Shepherd D. Persistent larval sensory neurons in adult Drosophila melanogaster. J Neurobiol. 1999;39:275–286. [PubMed] [Google Scholar]
- Williams DW, Truman JW. Mechanisms of dendritic elaboration of sensory neurons in Drosophila: insights from in vivo time lapse. J Neurosci. 2004;24:1541–1550. doi: 10.1523/JNEUROSCI.4521-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132:3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]