Abstract
The impact of malaria intervention measures (insecticide-treated net use and artemisinin combination therapy) on malaria genetics was investigated at two sites in western Kenya: an endemic lowland and an epidemic highland. The genetic structure of the parasite population was assessed by using microsatellites, and the prevalence of drug-resistant mutations was examined by using the polymerase chain reaction–restriction fragment length polymorphism method. Two years after intervention, genetic diversity remained high in both populations. A significant decrease in the prevalence of quintuple mutations conferring resistance to sulfadoxine-pyrimethamine was detected in both populations, but the mutation prevalence at codon 1246 of the Plasmodium falciparum multidrug resistance 1 gene had increased in the highland population. The decrease in sulfadoxine-pyrimethamine–resistant mutants is encouraging, but the increase in P. falciparum multidrug resistance 1 gene mutations is worrisome because these mutations are linked to resistance to other antimalarial drugs. In addition, the high level of genetic diversity observed after intervention suggests transmission is still high in each population.
Introduction
The World Health Organization estimates that malaria causes 300–500 million clinical cases annually, with more than one million deaths per year.1 Recently, major funding from the President's Malaria Initiative and the Global Fund to Fight against AIDS, Tuberculosis and Malaria has significantly enhanced malaria control efforts in sub-Saharan Africa where malaria morbidity and mortality are highest.2 The main malaria control tools include treating infected persons with antimalarial drugs and reducing human-mosquito contact rates through mosquito control.3–6 The large-scale distribution of insecticide-treated nets (ITNs) and a change in the first-line antimalarial drug from sulfadoxine-pyrimethamine (SP) or chloroquine (CQ) to artemisinin combination therapy (ACT) marked the beginning of a new era of malaria control campaign in sub-Saharan Africa.4,5
The goal of any malaria control program is to reduce overall transmission of the parasite and consequently malaria-induced morbidity and mortality. In Kenya, the current strategy involves a two-tiered system. First, more than seven million ITNs were distributed free of charge in 2006 to children and pregnant women.1,5–10 The ITNs minimize contact rates of the mosquito vector, thereby reducing transmission and malaria incidence.11–13 This reduced transmission may have an important effect on the parasite's genetic variability, a factor that might influence its adaptive ability. With reduced transmission, the malaria parasite has fewer opportunities to sexually recombine, and therefore lose favorable drug-resistant combinations.14 This may result in lower population genetic diversity. Second, the use of ACT to treat uncomplicated malaria targets the transmission stages of malaria and the asexual stages.1 The prescription of a drug that affects both blood stages of the parasite could also lead to reduced transmission.
The previous long-term usages of CQ and SP have resulted in a rapid spread of drug resistant malaria genotypes.4,15 In 2004, artemether-lumefantrine (Coartem™; Roche, Basel, Switzerland) was introduced to replace SP and in 2006 was provided free of charge in health facilities with the support from the Global Fund to Fight AIDS, Tuberculosis and Malaria.2 The introduction of artemether-lumefantrine has relaxed the selection pressure on resistance to CQ and SP, but has posed additional selection pressure on the genes related to artemether-lumefantrine resistance.
Drug resistance in malaria parasites is associated with genetic mutations in target genes and can be monitored using molecular methods. Chloroquine resistance is determined by the major point mutation at codon 76 of the P. falciparum CQ resistance transporter (pfcrt) gene.16 This mutation highly correlates with increased clinical CQ tolerance and treatment failure.16–19 In addition, point mutations in the P. falciparum multidrug resistance 1 (pfmdr1) gene (e.g., N86Y, Y183F, S1034C, N1042D, and D1246Y) have been shown to modulate CQ resistance.20 Resistance to antifolates is associated with point mutations in the dihydrofolate reductase (pfdhfr) and dihydropteroate synthetase (pfdhps) genes.21,22 The quintuple mutations in pfdhfr and pfdhps (triple S108N/N51I/C59R mutations in dhfr and double A437G/K540E mutations in dhps) are associated with the clinical failure of SP treatment for P. falciparum malaria.21,22 Resistance to artemisinins has been reported in Southeast Asia, but it has not been detected in Africa.23,24
The objective of this study was to determine the effects of the new two-tiered control program on the genetic diversity and the prevalence of malaria drug-resistant mutations in western Kenya. Two sentinel sites, one in an endemic lowland (Kombewa) and one in an epidemic highland (Kakamega), were examined. Data on malaria prevalence during this period have been published.5 In brief, at the beginning of the study period in 2005, the prevalence of P. falciparum was 41% in Kakamega and this significantly decreased throughout the four-year study period; the prevalence in 2008 was 6.8%.5 The prevalence of P. falciparum in 2005 in Kombewa was 48%; the prevalence decreased to 31% in 2007, but increased in 2008, and the prevalence surpassing pre-intervention levels (49%).5 We hypothesized that malaria parasite genetic diversity would be significantly reduced in Kakamega because of decreases in overall transmission as measured by malaria prevalence, but this effect would not be as noticeable in Kombewa because of a lack of a sustained reduction in malaria prevalence. In addition, as SP has been phased out, we expected to see a reduction in the point mutation frequencies in the codons of pfdhfr and pfdhps genes coding for resistance to this drug in both study populations.
Methods
Study areas.
This study was conducted in two sites in western Kenya with differing levels of transmission. Kakamega, a highland site 1,500–1,600 meters above sea level, is characterized by valleys and depressions surrounded by densely populated hills and is hyperendemic for malaria. Kombewa, a lowland site with a mean altitude of 1,200 meters above sea level, has a rolling terrain bisected by small streams and is holoendemic for malaria. Plasmodium falciparum is the primary malaria parasite species, with the predominant malaria mosquito vector species being Anopheles gambiae, An. arabiensis, and An. funestus.25,26 The estimated entomologic inoculation rate during 2003–2004 was 16.6 and 31.1 infectious bites per person per year in Kakamega and Kombewa, respectively.26
Parasitologic survey.
Monthly parasitologic surveys were conducted at both sites beginning in January 2004. The parasite surveys sampled school children 6–15 years of age who were enrolled in one of our long-term malaria prevalence and transmission studies.5 An average of 100–200 children at each site were tested regularly each month. Blood samples were collected by the standard finger prick method, and thick and thin blood smears were prepared for microscopic observation. Blood dots were made on filter paper for genetic analysis. Monthly malaria prevalence data has been published5; this study focuses on changes of malaria parasite population genetics during this period.
Surveys of antimalarial drug use.
We collected information on antimalarial drug prescriptions during 2004–2008 from hospitals/clinics in both study sites. Antimalarial drugs prescribed or used by the residents included CQ, SP, amodiaquine (AQ), the AQ + SP combination, quinine, and ACT, including artemether-lumefantrine (Coartem™).
DNA extraction and microsatellite genotyping.
DNA of positive blood samples from Kakamega (August 2005 and June–September 2008) and Kombewa (January 2005 and June–July 2008) were extracted by using the Saponin/Chelex method.27 A nested DNA polymerase chain reaction (PCR) was used to confirm all infections as P. falciparum, P. ovale, P. vivax, or P. malariae.28 Any samples that failed to amplify after re-extraction were discarded from further analyses.
All P. falciparum positive samples were analyzed at 10 polymorphic microsatellite loci (Poly α, Pfg377, 2490, TA 81, TA 87, Ara2, TA1, PfPK2, Ta109, and TA42) with modifications.29 We used the M13 tailed primer method to fluorescently label our primers.30,31 The PCRs were conducted in a total volume of 16 μL with 14.4 μL of 1.1× PCR master mix (Abgene, Rochester, NY), 0.16 μL of each 25 μM primer (forward primer with M13 tail), 0.3 μL of 1 μM M13 forward primer and 1 μL of DNA template. Samples were initially denaturized at 95°C for 5 minutes before 45 amplification cycles (94°C for 30 seconds, 45°C for 30 seconds, and 65°C for 45 seconds), followed by a final extension of 65°C for 7 minutes. Samples were analyzed on an automated 4300 DNA analyzer (Li-Cor, Lincoln, NE), and alleles were quantified by using the Gene ImagIR 4.33 software (Li-Cor).
Because Plasmodium parasites are haploid, each allele on the pherogram represents a different clone of the parasite. Multiple alleles were counted if they were at least on-third the height of the predominant allele in the infection. The proportion of multiclonal infections was calculated as the number of infections with more than one allele at ≥ 8 of the 10 microsatellite loci scored. Infection complexity (no. of clones within a single infection) was calculated as the minimum number of clones within an infection. To make this calculation, the multilocus genotypes for each sample were examined and the highest number of alleles observed for any one locus in that infection determined the infection complexity.32,33 For population genetic analyses, such as expected heterozygosity (He), the number of alleles per locus, the effective number of alleles per locus (Ne), population bottleneck, and population differentiation (FST), only the predominant allele (the highest peak on the pherogram) for each infection was used.31,32,34
Molecular identification of drug-resistant mutations.
Mutations in genes associated with drug resistance were analyzed by using a nested PCR and mutation-specific restriction enzyme digest protocol.35–37 We analyzed nine codons in four genes for resistance to CQ and SP: pfcrt (K76T), pfmdr1 (N86Y, N1042D, and D1246Y), pfdhfr (N51I, C59R, and S108N), and pfdhps (A437G and K540E). Genomic DNA from P. falciparum clones HB3, W2, and Dd2 (MR4, Manassas, VA) were used as positive controls. Restriction enzyme digests were subjected to electrophoresis on a 3% agarose gel stained with ethidium bromide. Infections that showed mixed-genotypes (cut and uncut bands) were re-analyzed to confirm the findings. Samples exhibiting mixed infections were scored separately from the wild and mutant genotypes. As drug selection pressure decreases, we expected to see an influx of the wild genotypes in circulation. Thus, by keeping mixed infection counts separate from the wild and mutant genotypes, we might detect a prevalence change in each genotype pattern.
Data analysis.
All samples positive for P. falciparum were included in this study, and as such, our sample sizes from each site/year differ from each other. The impact of malaria interventions within a site was determined by dividing surveys into two time periods: pre-intervention (2005) and post-intervention (2008). Changes in microsatellite allele frequencies and the number of multiclonal infections were analyzed by using the chi-square test. Changes in the prevalence of drug-resistant mutations were compared between the two survey periods, and the prevalence of infections carrying double, triple, quadruple, or quintuple mutations for SP resistance were analyzed by using Fisher's exact test. To analyze the number of mutations for SP resistance, the number of mutants within an infection was summed up over the five genes examined (three in pfhdfr and two in pfdhps). Therefore, a double mutant means an infection had any two resistant mutations for SP, not a specific pairing; a triple mutant had any three SP-resistant mutations and so forth. A quintuple mutant had all five key resistant mutations (108N/51I/59R/437G/540E). Likewise, a wild-type parasite had only wild-type alleles at all five of the SP-resistant genes examined. For this analysis, mixed infections were scored as mutants because they were carrying the resistant genotypes.
The genetic diversity of P. falciparum infections was examined by calculating He and Ne by using GenAlEx6.1.38 The genetic differentiation between the two populations was estimated in two ways: between sites within years (Kakamega versus Kombewa in 2005 and 2008) and within sites between years (Kakamega 2005 versus 2008 and Kombewa 2005 versus 2008) by using the FST analysis in the program GDA.39 Bootstrap resampling was used to determine if the FST values calculated were significantly different from zero.
Sustained reductions in malaria transmission could result in a parasite population bottleneck, leading to a dramatic reduction in the genetic diversity at microsatellite loci over time. However, initially, the parasite population may show signs of short-term increases in genetic diversity when compared with that expected under mutation-drift equilibrium. The computer program Bottleneck was used to determine if any reduction in transmission intensity had sustained effects on parasite population genetic diversity by analyzing the data for significant changes in the level of microsatellite diversity.40,41 We used two mutational models for microsatellites, the infinite alleles model (IAM) and the stepwise mutational model (SMM), to examine patterns of heterozygote deficiency/excess at each locus.
Multilocus linkage disequilibrium for each population was tested by using LIAN version 3.5 (http://adenine.biz.fh-weihenstephan.de/cgi-bin/lian/lian.cgi.pl).42 Monte Carlo simulations (with 10,000 reiterations) and parametric analyses were performed. Linkage disequilibrium detects nonrandom associations between alleles at different microsatellite loci and is typically, but not always, a sign of low transmission intensity.32,43
Ethics.
Ethical approval for this study was granted by the Ethical Review Committee of Kenya Medical Research Institute and by the Institutional Review Board of University of California at Irvine. Written assent for children (< 18 years of age) and consent for adults were obtained before enrollment. Inclusion criteria were provision of informed assent/consent and age > 6 months. Exclusion criteria included unwillingness to participate in the study.
Results
Genetic diversity alterations.
The genetic diversity of malaria populations pre-intervention and post-intervention were examined by using 10 polymorphic microsatellite markers. The He value remained high between the two time periods for both sites (Kombewa: He = 0.737 versus 0.802, 2005/2008 respectively; Kakamega: He = 0.764 versus 0.820, 2005/2008 respectively) (Table 1). For Kakamega and Kombewa populations, the proportion of multiclonal infections increased from 75.7% (53 of 70) in 2005 to 97.9% (46 of 47) in 2008 (P = 0.001) in Kakamega and from 79.5% (35 of 44) in 2005 to 86.9% (86 of 99) in 2008 (P > 0.05) in Kombewa. The effective number of alleles in Kakamega in 2005 increased from 5.8 to 6.9 in 2008, and the effective number of effective alleles in Kombewa increased from 5.5 in 2005 to 6.6 in 2008.
Table 1.
Microsatellite allele data for Plasmodium falciparum for the four study populations, Kenya*
| Population, year | N | % Multiclonal | No. clones | Na | Ne | He |
|---|---|---|---|---|---|---|
| Kakamega 2005 | 70 | 76 | 1.99 ± 0.09 | 11.30 ± 1.18 | 5.82 ± 0.96 | 0.76 ± 0.05 |
| Kakamega 2008 | 47 | 98 | 3.32 ± 0.13 | 12.10 ± 1.27 | 6.89 ± 1.09 | 0.82 ± 0.04 |
| Kombewa 2005 | 44 | 80 | 1.04 ± 0.16 | 8.60 ± 0.97 | 5.45 ± 0.82 | 0.74 ± 0.07 |
| Kombewa 2008 | 99 | 87 | 2.38 ± 0.08 | 13.50 ± 1.39 | 6.62 ± 0.99 | 0.80 ± 0.04 |
Values are the mean ± SE. N = sample size; % multiclonal = proportion of infections with > 1 allele at more than 2 loci; No. clones = mean number of clones per infection; Na = no. of observed alleles; Ne = no. of effective alleles; He = unbiased expected heterozygosity.
When we examined the allelic make-up at each individual locus, we found a lack of overall change in allele frequencies for either population between two sampling years. In Kakamega, the allele frequencies only significantly differed for two loci between 2005 and 2008 at loci ARA2 (P = 0.008) and TA 42 (P = 0.0001). The Kombewa populations in 2005 and 2008 were also similar, differing only at one microsatellite locus TA 81 (P = 0.03). With all 10 loci combined, we found that the parasite populations were not genetically differentiated between sites within years, or within sites between years (Table 2).
Table 2.
FST values with 95% confidence interval for all pairwise comparisons (between sites within years or within sites between years) for Plasmodium falciparum, Kenya*
| Comparison | FST (95% confidence interval) |
|---|---|
| Kakamega 2005 vs. Kombewa 2005 | 0.002 (–0.004 to –0.009) |
| Kakamega 2008 vs. Kombewa 2008 | 0.011 (–0.001 to –0.033) |
| Kakamega 2005 vs. Kakamega 2008 | 0.014 (–0.0005 to –0.039) |
| Kombewa 2005 vs. Kombewa 2008 | 0.007 (–0.002 to –0.019) |
The inclusion of zero in all confidence intervals indicates no significant differentiation.
Alterations in drug-resistance profiles.
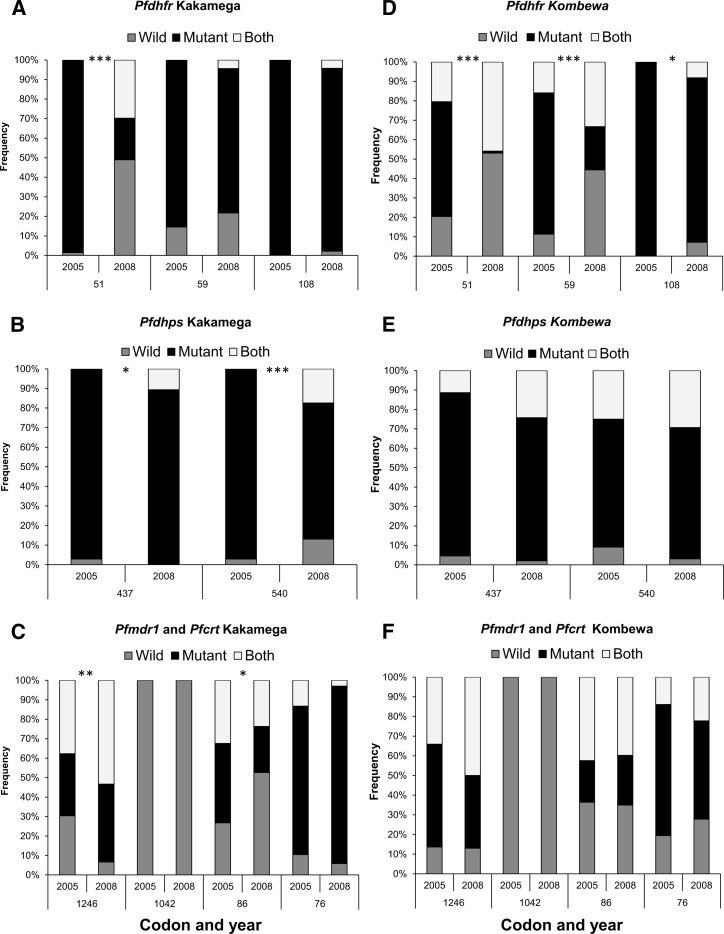
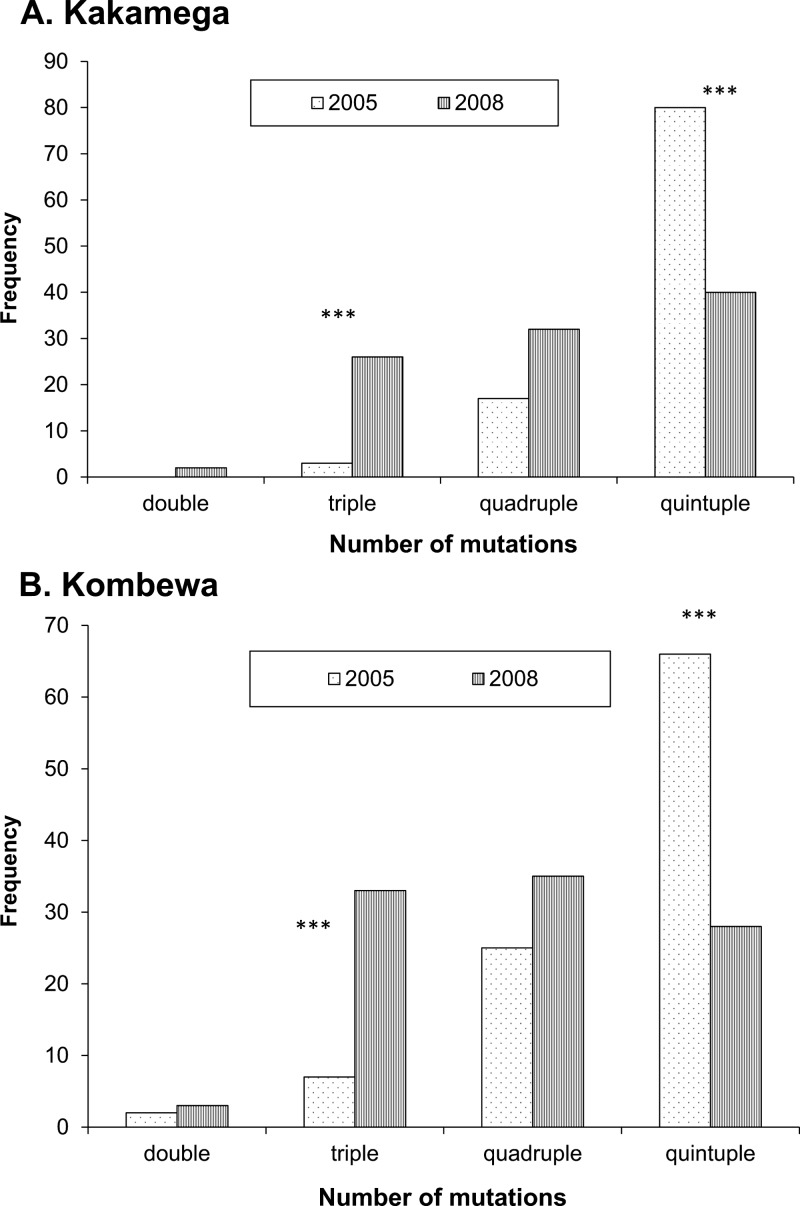
The prevalence of SP-resistant mutants at the pfdhfr codon 51 and the pfdhps codons 437 and 540 were significantly different in the Kakamega 2008 parasite population compared with the 2005 parasite sample (P < 0.001, P = 0.011, and P < 0.001 respectively). For these codons, a significantly higher number of wild genotypes were observed either alone or in mixed infections with mutant genotypes (Figure 1A and B). A significant increase in the prevalence of triple SP mutations (P < 0.001) was observed, but the prevalence of infections carrying quintuple SP mutations decreased from 80% in 2005 to 40% in 2008 (P < 0.001; Figure 2A). A noticeable increase for CQ wild genotypes was also found at pfmdr1 codon 86 (27% in 2005 compared with 52% in 2008; P = 0.021) (Figure 1C), and a significant increase in the mutant allele prevalence for codon 1246 of the pfmdr1 gene was also found (P = 0.009) (Figure 1C). No significant differences were detected for codon 76 of the pfcrt gene, codon 1042 of the pfmdr1 gene, or codons 59 and 108 of the pfdhfr gene.
Figure 1.
Prevalence of wild, mutant, and mixed-genotype infections for drug-resistant genes in Plasmodium falciparum, Kenya. A–C, Kakamega (highland) samples from 2005 and 2008. D–F, Kombewa (lowland) samples from 2005 and 2008. Pfdhfr = P. falciparum dihydrofolate reductase; Pfdhps = P. falciparum dihydropteroate synthase; Pfmdr1 = P. falciparum multidrug resistance gene 1; Pfcrt = P. falciparum chloroquine resistance transporter. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 2.
Prevalence of double, triple, quadruple, and quintuple mutations for sulfadoxine-pyrimethamine resistance (measured as the total number of mutations present over all five genes examined) in Plasmodium falciparum, Kenya. A, Kakamega (highland) and B, Kombewa (lowland). ***P < 0.001.
In Kombewa, a significant reduction in SP-resistant mutations for all three pfdhfr codons was detected (P < 0.001 for codons 51 and 59, and P = 0.023 for codon 108) (Figure 1D), but no other significant alterations were observed for mutations at pfdhps, or the CQ genes pfmdr1 and pfcrt (Figure 1E and F). The proportion of quintuple mutants resistant for SP significantly decreased from 66% in 2005 to 28% in 2008 (P < 0.001) (Figure 2B), and the prevalence of triple mutations increased (P < 0.001).
Testing for population bottlenecks and linkage disequilibrium.
Both populations showed an initial decrease in overall malaria prevalence, and Kakamega continued to have reduced prevalence compared with pre-intervention estimates.5 Analysis using the program Bottleneck suggests that the Kakamega parasite population had experienced a significant bottleneck because of this sustained reduction, with 9 of 10 loci showing heterozygote excess under the IAM (P < 0.05) (Table 3). The Kombewa site did not have a genetic bottleneck because no heterozygote excess was found with the IAM test, and under the SMM, only 2 of 10 loci had heterozygote excess (P = 0.016) (Table 3).
Table 3.
Number of microsatellite loci showing an excess in heterozygosity for Plasmodium falciparum when compared with expected values under mutation-drift equilibrium, Kenya*
| Population, year | SMM | IAM | ||
|---|---|---|---|---|
| No. loci in excess | Significance | No. loci in excess | Significance | |
| Kombewa 2008 | 2 | P = 0.016 | 8 | P = 0.189 |
| Kakamega 2008 | 5 | P = 0.401 | 9 | P = 0.049 |
Two mutational models were used to determine expected results: the stepwise mutational model (SMM) and the infinite allele model (IAM). The Wilcoxon signed-rank test assigns a significance level for the expected number of loci with heterozygote excess.
Three of the four study populations showed no signs of linkage disequilibrium (P > 0.05 for Kombewa 2005 (index of association [ISA] = –0.0058), Kombewa 2008 (ISA = –0.0049), and Kakamega 2008 (ISA = 0.007). Kakamega 2005, the highland sample before changes in the malaria control program in 2006, showed significant linkage disequilibrium (P < 0.001, ISA = 0.0218).
Dynamics of antimalarial drug use.
The two sentinel sites showed different antimalarial drug prescription patterns. In Kakamega, the primary drug prescriptions were SP and quinine in 2005, accounting for 38% and 35% of all medicines prescribed, respectively. After the distribution of ACT to healthcare facilities in 2006, prescriptions for ACT increased from 1% in 2005 to 50% in 2007, and then decreased to 29% in 2008. In 2008, the primary drug prescribed for malaria treatment was quinine (51%) and not ACT, possibly because of a lack of ACT stock. Chloroquine was only prescribed in 2004 in 2% of the cases examined. In contrast, the clinics in Kombewa frequently prescribed SP and AQ (49% and 43%, respectively) in 2005, and quinine made up the remaining prescriptions. In 2006, 100% of malaria infections were treated with ACT, but this quickly decreased to 60% in 2008, with a return to the prescription of SP in 40% of the cases.
Discussion
The development of new malaria control regimens warrant investigations as to their effectiveness at reducing overall malaria transmission and alterations in the malaria parasite's genetic structure. In addition, consistent monitoring of these programs can highlight areas where the new protocols are not being followed, so that interventions can occur sooner rather than later. This report examined how two newly implemented control efforts (increased ITN use and the new drug combination therapy ACT) affected parasite population structure in terms of overall genetic diversity and fluctuations in the prevalence of drug resistant mutations in two regions of western Kenya. Samples taken before the new control measures were implemented in 2005 were compared with samples taken two years after increased ITN distribution and a drug policy change to Coartem™ in 2008.
A sustained reduction in the transmission of malaria parasites, such as that observed in Kakamega, could reduce the level of diversity in the parasite gene pool. An interesting finding from our study illustrates that even with a decrease in transmission, parasite populations remained highly diverse over time (Table 1). In Kakamega, the diversity at microsatellite markers was higher post-intervention (He = 0.75 versus 0.81) (Table 1). This finding could be attributed to a bottleneck effect in which a rapid and prolonged reduction in transmission of the parasite results in an initial overabundance of diversity, as rare alleles are lost first from the population, leading to a lower expected heterozygosity estimate.41 This initial higher diversity is temporary and lasts only until a new equilibrium is reached.41 In Kakamega, nine of 10 microsatellite loci had higher observed heterozygosity measures than expected under the infinite alleles model of mutation (Table 3).
In addition, the proportion of multiclonal infections also increased in 2008 (from 77% in 2005 to 98% in 2008) (Table 1). This finding is perplexing when considering the drastic reduction in overall malaria prevalence in the area. One explanation could be that more parasite strains were producing gametocytes, leading to multiple parasite strains being transmitted within a single infectious bite. Plasmodium falciparum genotypes differ in their gametocyte production, and they might even adjust their gametocyte production when faced with altered transmission dynamics.44 If each strain within an infection was producing gametocytes at the same time during periods of lower transmission intensity, as a life history strategy to ensure transmission, a surge in multiclonal infections may occur.44 Sexual recombination in the insect vector could also play a role in this increase in genetic diversity and multiclonal infections.
Increased sexual recombination could also be the reason why we noticed no linkage disequilibrium in the Kakamega 2008 samples compared with those from 2005. The non-random association between alleles in 2005 could be caused by the high proportion of drug-tolerant genotypes in circulation, or a higher frequency of inbreeding. As selection pressure for these genotypes waned in 2006 and afterwards, these associations may have broken down, leading to no linkage observed in 2008.
Kombewa also experienced an increase in diversity after intervention (He = 0.74 versus 0.80) (Table 2), but this does not appear to have been caused by a bottleneck. Within six months of the distribution of ITNs and ACT, the prevalence began to increase and in 2008 reached pre-intervention levels.5 In this instance, a bottleneck is not as likely because we found that only two loci showed heterozygote excess when we analyzed the data under the SMM (Table 3). No linkage disequilibrium was observed for either Kombewa population (before or two years after control program implementation).
Gene flow between populations may account for the maintained diversity we observed in our study populations because the populations were genetically similar (Table 2). Bonizzoni and others examined the population genetics of lowland and highland sites in Kenya and found that many of the highland genotypes were similar to those found in the lowlands.25 Their results provide strong evidence to the theory that infections in the highlands are continuously fueled by incoming genotypes from lowland population.26,31,37
The maintenance of diversity and lack of linkage disequilibrium in both populations after the introduction of ITNs suggests that the any reductions in transmission were not substantial enough to reduce the parasite gene pool. Gatei and others found a similar trend in a near-by area of Kenya.13 Although overall microsatellite diversity did not substantially differ after large-scale implementation of ITNs, individual loci did differ, including one associated with gametocyte stages (locus 377).13 Our data support this finding because a few individual microsatellite loci significantly differed between years (ARA2 and TA42 for Kakamega and TA 81 for Kombewa), but these were not the same loci identified by the study of Gatei and others. A diverse population suggests that recombination is occurring, and that drug-resistant mutations have the opportunity to be lost during each transmission event.14,45 Overall, our data suggest that the transmission of P. falciparum in these regions is still high, although the highland region had shown sustained reductions in parasite prevalence.
Two-years after the change from SP as the first-line drug to the ACT formulation Coartem™, a significant decrease in the prevalence of SP-resistant mutants in both populations was observed (Figures 1 and 2). In 2005, most infections had the quintuple mutations for SP and in 2008, we saw more triple mutations in both populations, showing that drug resistance to SP may be lost in a stepwise manner (Figure 2).14 Despite SP still being available for prescription, and the similar acting antibiotic trimethoprim/sulfamethoxazole being prescribed in the area, the quintuple mutants for SP decreased in our study populations. This finding suggests that resistance to SP is costly and may be quickly lost with sustained limitations of SP use. It could also imply that increased sexual recombination is breaking apart the favorable drug combinations, accounting for the high level of diversity observed.
Unfortunately, because we were unable to collect and analyze samples from each study site in 2006 and 2007, we can only infer that any changes we see between 2005 and 2008 were sustained throughout the entire study period. This assumption is supported when we compare our drug resistance data from Kakamega 2008 to those from previously published data from Iguhu in 2006.25 Iguhu is a village within the Kakamega highland district and a good portion of our highland samples also came from this village.5,25,35 In 2006, 63% of the infections examined had the quintuple mutations for SP resistance, and in 2008, only 40% had all five key mutations (both < 80% of infections reported in 2005). Interestingly, we found that most infections in both years still had mutations to CQ, although this drug has not been commonly prescribed in the study area. The pfcrt mutant 76T, the major mutation contributing to CQ treatment failure, increased from 78% in 2006 to 92% in 2008. Comparing our data from 2008 to the 2006 sample, we saw a decrease in the prevalence of mutants at codon 1246 of the pfmdr1 gene (54% in 2006 versus 40% in 2008), but this was still higher than what was reported in 2005 (32%). The prevalence of mutants for codon 86 remained similar (24% for 2006 and 2008), which is lower than the prevalence found in 2005 (41%).5,25,35
The maintenance of these CQ drug-resistant mutations within a population after cessation of drug use suggests a few possibilities (provided there is a fitness cost for the drug-resistant mutant). First, low transmission intensity could limit the rate at which favorable drug mutations are lost. This possibility does not appear to be the case because the parasite populations are genetically diverse and we observed a significant decrease in SP-resistant mutants within two years. Second, a second drug with a similar function as the withdrawn drug may be in use. Quinine resistance has been proposed to be attributed to mutations in the pfmdr1 gene, most notably the mutant alleles 1042D, 86Y and 1246Y (also selected by CQ treatment).46–48 Therefore, the maintenance of these mutations in the study population, even after the cessation of CQ use, could be attributed to the continued use of quinine.
Some of the changes we observed in allele prevalence during 2005–2008 in our study populations could have been caused by selection for genotypes tolerant to ACT, although resistance to ACT has not been detected in Africa.49 Field studies have highlighted the wild alleles N86 and D1246 and the mutant allele 184F of the pfmdr1 gene and the wild allele K76 of the pfcrt gene to play key roles in treatment failure to lumefantrine, one of the drugs in the ACT treatment Coartem™.47,50–52 Three of these alleles (N86, D1246, and K76) are considered susceptible alleles to CQ.35 This antagonistic selection may enable these susceptible alleles for CQ resistance to reenter a population. In this study, we did not find any significant changes these allele frequencies in the Kombewa population. However, we observed some changes in allele frequencies at the pfmdr1 gene in Kakamega. In 2008, we saw significant increases in the frequencies of the wild genotype (N86) and the mutant genotype (1246Y); because these two alleles confer antagonistic resistance, the increase in their frequencies is perplexing. This finding could have been caused by the increase in the use of quinine as a drug after 2006 in Kakamega, but these genes should be monitored to determine what, if any, tolerance to Coartem™ they confer.
The drug-resistance profiles in this study may have been different had Coartem™ been prescribed consistently throughout the study period (2006–2008). Our data from hospital clinics suggest that Coartem™ prescriptions increased in 2006 (2007 for Kakamega), but then quickly decreased, and drugs such as SP or quinine became the most commonly prescribed in 2008. This finding could have been caused by a lack of subsidized Coartem™ available after 2006 to these clinics.53,54 Our study did not survey shops in the private sector to determine what drugs were being purchased and the prices for each. Costs for the drug is a primary concern for patients, and an increase in self-medication for malaria episodes could negatively affect the changes we have thus far seen in the SP-resistant mutant prevalence.54,55 However, as costs for ACT decrease, it is unclear whether patients will continue to choose SP over the more effective antimalarial drug.
This study enabled us to obtain insights of how the genetics of P. falciparum have changed during new control measure implementation. These results show the importance for consistent monitoring of malaria control programs to determine the effects on both the parasite's genetic diversity and level of resistance to current and previously relinquished drugs. After two years, the new ACT drug Coartem™ is not as widely used as SP. The significant reduction of quintuple mutants resistant to SP after just two years is promising, but for control efforts to enable reintroduction of this drug for future use, the new drug policy must be more strictly adhered to. In addition, the enforcement of proper bed net use and re-treatment is necessary to continue reducing malaria transmission in these and other malaria-prone regions of Africa.
ACKNOWLEDGMENTS
We thank the field team for their years of hard work and for their technical assistance in the field and laboratory; the communities and hospitals in the study areas for their support and willingness to participate in this research; M. Bonizzoni for reading and providing comment on an earlier version of this manuscript; and M. Wijesinha for statistical assistance. This paper is published with the permission of the Director of the Kenya Medical Research Institute.
Footnotes
Financial support: This study was supported by the National Institute of Health grants R01 AI050243 and D43 TW001505 to Guiyun Yan.
Authors' addresses: Anne M. Vardo-Zalik, The Pennsylvania State University, York, PA, E-mail: amv12@psu.edu. Guofa Zhou, Daibin Zhong, and Guiyun Yan, Program in Public Health, College of Health Sciences, University of California Irvine, Irvine, CA, E-mails: zhoug@uci.edu, dzhong@uci.edu, and guiyuny@uci.edu. Yaw A. Afrane and Andrew K. Githeko, Climate and Human Health Research Unit, Kenya Medical Research Institute, Kisumu, Kenya, E-mails: yaw_afrane@yahoo.com and githeko@yahoo.com.
References
- 1.World Health Organization . World Malaria Report. WHO Library Cataloguing-in-Publication Data 25. Geneva: World Health Organization; 2008. [Google Scholar]
- 2.Global Fund to Fight AIDS, Tuberculosis and Malaria. 2011. http://www.theglobalfund.org/en/partnershipforum/2011/ Available at.
- 3.Amin AA, Walley T, Kokwaro GO, Winstanley PA, Snow RW. Commentary: reconciling national treatment policies and drug regulation in Kenya. Health Policy Plan. 2007;22:111–112. doi: 10.1093/heapol/czl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin A, Zurovac D, Kangwana B, Greenfield J, Otieno D, Akhwale W, Snow R. The challenges of changing national malaria drug policy to artemisinin-based combinations in Kenya. Malar J. 2007;6:72. doi: 10.1186/1475-2875-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou G, Afrane YA, Vardo-Zalik A, Atieli H, Zhong D, Wamae P, Himeidan YE, Minakawa N, Githeko AK, Yan G. Changing patterns of malaria epidemiology between 2002 and 2010 in western Kenya: the fall and rise of malaria. PLoS ONE. 2011;6:e20318. doi: 10.1371/journal.pone.0020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . Insecticide Treated Mosquito Nets: A Position Statement. Geneva: World Health Organization; 2007. [Google Scholar]
- 7.Aregawi M, Cibulskis R, Otten M, Williams R, Dye C. World Malaria Report, 2008. Geneva: World Health Organization; 2008. [Google Scholar]
- 8.Roll Back Malaria Malaria Elimination in Africa. 2009. http://www.rbm.who.int/globaladvocacy/eventsarchive.html Available at.
- 9.Nafo-Traore F, Judd EJ, Okwo-Bele J. Protecting Vulnerable Groups in Malaria-Endemic Areas in Africa through Accelerated Deployment of Insecticide-Treated Nets: A Joint WHO-UNICEF Statement. Geneva: World Health Organization; 2005. WHO/HTM/RBM/2005.57: 2. [Google Scholar]
- 10.Roll Back Malaria . Scaling Up Insecticide Treated Netting Programmes in Africa: A Strategic Framework for Coordinated National Action. Geneva: World Health Organization; 2005. [Google Scholar]
- 11.Bhattarai A, Ali AS, Kachur SP, Martensson A, Abbas AK, Khatib R, Al-Mafazy AW, Ramsan M, Rotllant G, Gerstenmaier JF, Molteni F, Abdulla S, Montgomery SM, Kaneko A, Bjorkman A. Impact of artemisinin-based combination therapy and insecticide-treated nets on malaria burden in Zanzibar. PLoS Med. 2007;4:e309. doi: 10.1371/journal.pmed.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Meara WP, Bejon P, Mwangi TW, Okiro EA, Peshu N, Snow RW, Newton CR, Marsh K. Effect of a fall in malaria transmission on morbidity and mortality in Kilifi, Kenya. Lancet. 2008;372:1555–1562. doi: 10.1016/S0140-6736(08)61655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatei W, Kariuki S, Hawley W, ter Kuile F, Terlouw D, Phillips-Howard P, Nahlen B, Gimnig J, Lindblade K, Walker E, Hamel M, Crawford S, Williamson J, Slutsker L, Shi Y. Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J. 2010;9:353. doi: 10.1186/1475-2875-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laufer MK, Plowe CV. Withdrawing antimalarial drugs: impact on parasite resistance and implications for malaria treatment policies. Drug Resist Updat. 2004;7:279–288. doi: 10.1016/j.drup.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Bwijo B, Kaneko A, Takechi M, Zungu IL, Moriyama Y, Lum JK, Tsukahara T, Mita T, Takahashi N, Bergqvist Y, Bjorkman A, Kobayakawa T. High prevalence of quintuple mutant dhfr/dhps genes in Plasmodium infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003;85:363–373. doi: 10.1016/s0001-706x(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 16.Wellems T, Plowe C. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 17.White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 18.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 19.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayton K, Su XZ. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr Drug Targets Infect Disord. 2004;4:1–10. doi: 10.2174/1568005043480925. [DOI] [PubMed] [Google Scholar]
- 21.Warhurst DC. Drug resistance in Plasmodium falciparum malaria. Infection. 1999;27:S55–S58. doi: 10.1007/BF02561674. [DOI] [PubMed] [Google Scholar]
- 22.Ouellette M. Biochemical and molecular mechanisms of drug resistance in parasites. Trop Med Int Health. 2001;6:874–876. doi: 10.1046/j.1365-3156.2001.00777.x. [DOI] [PubMed] [Google Scholar]
- 23.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L. Artemisinin resistance: current status and scenarios for containment. Nat Rev Microbiol. 2010;8:272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- 25.Bonizzoni M, Afrane Y, Baliraine FN, Amenya DA, Githeko AK, Yan G. Genetic structure of Plasmodium falciparum populations between lowland and highland sites and antimalarial drug resistance in western Kenya. Infect Genet Evol. 2009;9:806–812. doi: 10.1016/j.meegid.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ndenga B, Githeko A, Omukunda E, Munyekenye G, Atieli H, Wamai P, Mbogo C, Minakawa N, Zhou G, Yan G. Population dynamics of malaria vectors in western Kenya highlands. J Med Entomol. 2006;43:200–206. doi: 10.1603/0022-2585(2006)043[0200:pdomvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 27.Wooden J, Kyes S, Sibley CH. PCR and strain identification in Plasmodium falciparum. Parasitol Today. 1993;9:303–305. doi: 10.1016/0169-4758(93)90131-x. [DOI] [PubMed] [Google Scholar]
- 28.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 29.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- 30.Oetting WS, Lee HK, Flanders DJ, Wiesner GL, Sellers TA, King RA. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics. 1995;30:450–458. doi: 10.1006/geno.1995.1264. [DOI] [PubMed] [Google Scholar]
- 31.Zhong D, Afrane Y, Githeko A, Yang Z, Cui L, Menge DM, Temu EA, Yan G. Plasmodium falciparum genetic diversity in western Kenya highlands. Am J Trop Med Hyg. 2007;77:1043–1050. [PubMed] [Google Scholar]
- 32.Anderson TJ, Haubold B, Williams JT, Estrada-Franco J, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 33.Vardo AM, Schall JJ. Clonal diversity of a lizard malaria parasite, Plasmodium mexicanum, in its vertebrate host, the western fence lizard: role of variation in transmission intensity over time and space. Mol Ecol. 2007;16:2712–2720. doi: 10.1111/j.1365-294X.2007.03355.x. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira MU, Nair S, Hyunh TV, Kawamoto F, Anderson TJ. Microsatellite characterization of Plasmodium falciparum from cerebral and uncomplicated malaria patients in southern Vietnam. J Clin Microbiol. 2002;40:1854–1857. doi: 10.1128/JCM.40.5.1854-1857.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong D, Afrane Y, Githeko A, Cui L, Menge D, Yan G. Molecular epidemiology of drug-resistant malaria in western Kenya highlands. BMC Infect Dis. 2008;8:105. doi: 10.1186/1471-2334-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco J, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 37.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- 38.Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis PO, Zaykin D. Genetic Data Analysis: Computer Program for the Analysis of Allelic Data. Version 1.1. 2002. http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php Available at.
- 40.Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered. 1999;90:502–503. [Google Scholar]
- 41.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haubold B, Hudson RR. LIAN 3.0: detecting linkage disequilibrium in multilocus data linkage analysis. Bioinformatics. 2000;9:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 43.Durand P, Michalakis Y, Cestier S, Oury B, Leclerc MC, Tibayrenc M, Renaud F. Significant linkage disequilibrium and high genetic diversity in a population of Plasmodium falciparum from an area (Republic of The Congo) highly endemic for malaria. Am J Trop Med Hyg. 2003;68:345–349. [PubMed] [Google Scholar]
- 44.Abdel-Wahab A, Abdel-Muhsin AA, Ali E, Suleiman S, Ahmed S, Walliker D, Babiker HA. Dynamics of gametocytes among Plasmodium falciparum clones in natural infections in an area of highly seasonal transmission. J Infect Dis. 2002;185:1838–1842. doi: 10.1086/340638. [DOI] [PubMed] [Google Scholar]
- 45.Malisa A, Pearce R, Abdulla S, Mshinda H, Kachur P, Bloland P, Roper C. Drug coverage in treatment of malaria and the consequences for resistance evolution: evidence from the use of sulphadoxine/pyrimethamine. Malar J. 2010;9:190. doi: 10.1186/1475-2875-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zalis MG, Pang L, Silveira MS, Milhous WK, Wirth DF. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am J Trop Med Hyg. 1998;58:630–637. doi: 10.4269/ajtmh.1998.58.630. [DOI] [PubMed] [Google Scholar]
- 47.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 49.Ogbonna A, Uneke CJ. Artemisinin-based combination therapy for uncomplicated malaria in sub-Saharan Africa: the efficacy, safety, resistance and policy implementation since Abuja 2000. Trans R Soc Trop Med Hyg. 2008;102:621–627. doi: 10.1016/j.trstmh.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 50.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Happi CT, Gbotosho GO, Folarin OA, Bolaji OM, Sowunmi A, Kyle DE, Milhous W, Wirth DF, Oduola AM. Association between mutations in Plasmodium falciparum chloroquine resistance transporter and P. falciparum multidrug resistance 1 genes and in vivo amodiaquine resistance in P. falciparum malaria-infected children in Nigeria. Am J Trop Med Hyg. 2006;75:155–161. [PubMed] [Google Scholar]
- 52.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 53.Kangwana BB, Njogu J, Wasunna B, Kedenge SV, Memusi DN, Goodman CA, Zurovac D, Snow RW. Malaria drug shortages in Kenya: a major failure to provide access to effective treatment. Am J Trop Med Hyg. 2009;80:737–738. [PMC free article] [PubMed] [Google Scholar]
- 54.Mokuolu OA, Okoro EO, Ayetoro SO, Adewara AA. Effect of artemisinin-based treatment policy on consumption pattern of antimalarials. Am J Trop Med Hyg. 2007;76:7–11. [PubMed] [Google Scholar]
- 55.Watsierah CA, Jura WG, Oyugi H, Abong'o B, Ouma C. Factors determining anti-malarial drug use in a peri-urban population from malaria holoendemic region of western Kenya. Malar J. 2010;9:295. doi: 10.1186/1475-2875-9-295. [DOI] [PMC free article] [PubMed] [Google Scholar]