Abstract
To date, 11 studies conducted in different countries to test the association between Plasmodium falciparum Na+/H+ exchanger gene (pfnhe-1; PF13_0019) polymorphisms and in vitro susceptibility to quinine have generated conflicting data. In this context and to extend our knowledge of the genetic polymorphism of Pfnhe gene, we have sequenced the ms4760 locus from 595 isolates collected in the Comoros (N = 250; an area with a high prevalence of chloroquine and sulfadoxine-pyrimethamine resistance) and Madagascar (N = 345; a low drug-resistance area). Among them, 29 different alleles were observed, including 8 (27%) alleles not previously described. Isolates from the Comoros showed more repeats in block II (DNNND), which some studies have found to be positively associated with in vitro resistance to quinine, compared with isolates from Madagascar. Additional studies are required to better define the mechanisms underlying quinine resistance, which involve multiple gene interactions.
Quinine (QN), a natural quinoline derivative compound found in Cinchona bark, has been used for centuries in malaria-endemic regions.1 To date, resistance to QN remains particularly patchy and rare,2–12 and only few cases of clinical failure have been reported in Asia and South America. The mechanism underlying QN resistance is not well-understood, and it is probably complex and multigenic. Since the seminal work by Ferdig and others13 in 2004, 11 studies have been conducted in different countries to evaluate implication of ms4760 polymorphisms in QN resistance,14–24 and conflicting data have been reported, likely because of the different geographical origin of parasites (implying different genetic backgrounds), the type of parasites used (fresh isolates, culture-adapted strains, and reference lines), and the method used to assess in vitro QN susceptibility.21,25
In this context and to extend our previous work regarding ms4760 polymorphisms in Plasmodium falciparum parasites circulating in malaria-endemic areas in the Indian Ocean,14 we have analyzed ms4760 sequences from 595 isolates (Madagascar, N = 345; Comoros, N = 250).
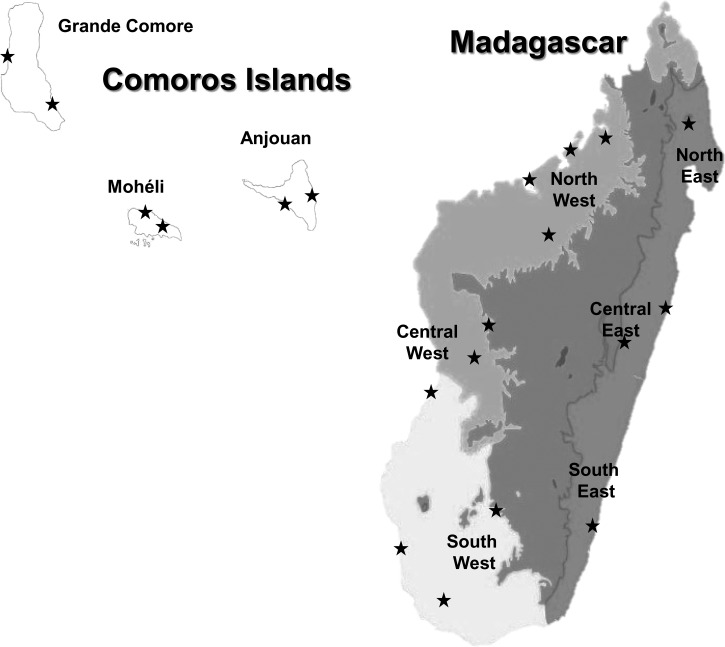
P. falciparum isolates from Madagascar were collected in 2006 and 2007 as part of the surveillance of antimalarial drug resistance from symptomatic malaria-infected patients before treatment in 14 health centers (northwest: Antohihy, Analalava, Mahajanga, and Maevatanana; central west: Tsiroanomandidy, Miandrivazo, and Morondava; southwest: Ihosy, Ejeda, and Toliara; northeast: Andapa; central east: Toamasina and Moramanga; southeast: Faranfagana). Comorian isolates were collected from finger prick onto filter paper in 2006 in six different sites (Grande Comore: Moroni and Foumbouni; Mohéli: Fomboni and Wanani; Anjouan: Pomoni and Domoni) (Figure 1). Informed written consent was provided by all patients or their parents/guardians before inclusion in the study, and blood collections were conducted in accordance to the Ethics Committee of the Ministries of Health of Madagascar and the Comoros (N°007/SANPF/2007; registration number ISRCTN36517335).
Figure 1.
Samples collection sites in the Comoros Islands and Madagascar from 2006 to 2007.
Parasite DNA was extracted from blood spots with Instagene matrix (Bio-Rad, Marnes la Coquette, France) according to the manufacturer's instructions or directly from 100 μL infected blood by the phenol-chloroform method.26 The parasite species was confirmed by real-time polymerase chain reaction (PCR) as described in the work by Mangold and others.27 Amplification and sequencing of the ms4760 locus in the P. falciparum Na+/H+ exchanger gene (pfnhe-1) was performed in accordance with the protocol described earlier.14 pfnhe-1 ms4760 alleles were constructed from a full sequence presenting an unambiguous single-allele signal at all positions and used P. falciparum 3D7 (sodium/hydrogen exchanger, Na+, H+ antiporter, PF13_0019, XM_001349726) as the reference.
Genetic diversity was assessed by Nei's unbiased expected heterozygosity (He) from haploid data and calculated as He = [n/(n – 1)][1 − pi] (n = the number of isolates sampled; pi = the frequency of the ith allele).28 Population genetic differentiation was measured using Wright's F statistics (Fst).29 A P value < 0.05 was considered statistically significant.
Nucleotide sequences of new ms4760 haplotypes were deposited in the GenBank database under accession numbers from JX472441 to JX472448.
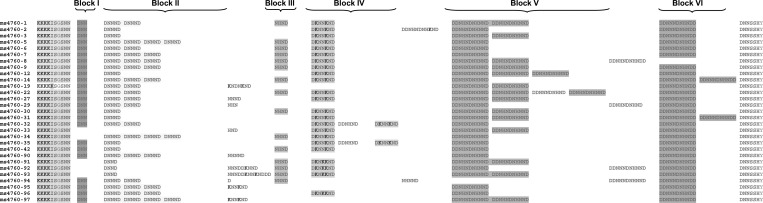
Among the 595 P. falciparum isolates (Madagascar, N = 345; Comoros, N = 250), 29 different alleles were observed, including 8 alleles not previously described (ms4760-90 to ms4760-97) (Table 1). ms4760-1 was the most prevalent (180/595; 30.3%) followed by ms4760-3 (96/595; 16.1%), ms4760-7 (77/595; 12.9%), and ms4760-6 or ms4760-9 (43/595; 7.2%); 15 ms4760 alleles (ms4760-1, ms4760-2, ms4760-3, ms4760-6, ms4760-7, ms4760-8, ms4760-9, ms4760-12, ms4760-27, ms4760-29, ms4760-30, ms4760-35, ms4760-91, ms4760-95, and ms4760-96) were distributed in both countries, whereas others were exclusively found in Madagascar (N = 10; ms4760-19, ms4760-22, ms4760-31, ms4760-32, ms4760-33, ms4760-42, ms4760-92, ms4760-93, ms4760-94, and ms4760-97) or the Comoros (N = 4; ms4760-5, ms4760-14, ms4760-34, and ms4760-90). Details are given in Table 1, and multiple amino acid sequence alignments are shown in Figure 2.
Table 1.
Distribution of ms4760 alleles among Indian Ocean isolates collected in 2006–2007
| Allele ms4760 | No. | No. of isolates | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNNND repeats | NHNDNHNNDDD repeats | Comoros Islands | Madagascar | Indian Ocean | GenBank accession numbers | ||||||||||||||||||||||
| Anjouan | Grande Comore | Mohéli | Total | Northwest | Northeast | Central west | Central east | Southwest | Southeast | Total | |||||||||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||||
| ms4760-1 | 2 | 2 | 19 | 28 | 43 | 40 | 17 | 23 | 79 | 31.6 | 23 | 41 | 4 | 25 | 44 | 28 | 11 | 25 | 10 | 20 | 9 | 41 | 101 | 29.3 | 180 | 30.3 | |
| ms4760-2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0.8 | 3 | 5 | 1 | 6 | 4 | 3 | 4 | 9 | 1 | 2 | 13 | 3.8 | 15 | 2.5 | |||||
| ms4760-3 | 1 | 2 | 9 | 13 | 7 | 6 | 11 | 15 | 27 | 10.8 | 6 | 11 | 5 | 31 | 30 | 19 | 10 | 23 | 14 | 28 | 4 | 18 | 69 | 20.0 | 96 | 16.1 | |
| ms4760-5 | 4 | 1 | 5 | 5 | 1 | 1 | 6 | 2.4 | 6 | 1.0 | |||||||||||||||||
| ms4760-6 | 2 | 1 | 5 | 7 | 6 | 6 | 5 | 7 | 16 | 6.4 | 4 | 7 | 13 | 8 | 2 | 5 | 7 | 14 | 1 | 5 | 27 | 7.8 | 43 | 7.2 | |||
| ms4760-7 | 3 | 1 | 8 | 12 | 13 | 12 | 13 | 17 | 34 | 13.6 | 6 | 11 | 2 | 13 | 19 | 12 | 8 | 18 | 6 | 12 | 2 | 9 | 43 | 12.5 | 77 | 12.9 | |
| ms4760-8 | 3 | 2 | 1 | 1 | 1 | 0.4 | 5 | 3 | 1 | 5 | 6 | 1.7 | 7 | 1.2 | |||||||||||||
| ms4760-9 | 3 | 2 | 4 | 6 | 3 | 3 | 6 | 8 | 13 | 5.2 | 6 | 11 | 1 | 6 | 13 | 8 | 1 | 2 | 5 | 10 | 4 | 18 | 30 | 8.7 | 43 | 7.2 | |
| ms4760-12 | 1 | 3 | 8 | 7 | 1 | 1 | 9 | 3.6 | 1 | 6 | 5 | 3 | 1 | 2 | 1 | 2 | 8 | 2.3 | 17 | 2.9 | |||||||
| ms4760-14 | 3 | 1 | 2 | 2 | 2 | 0.8 | 2 | 0.3 | |||||||||||||||||||
| ms4760-19 | 2 | 2 | 2 | 1 | 2 | 0.6 | 2 | 0.3 | |||||||||||||||||||
| ms4760-22 | 2 | 3 | 2 | 5 | 2 | 0.6 | 2 | 0.3 | |||||||||||||||||||
| ms4760-27 | 2 | 2 | 2 | 3 | 3 | 3 | 1 | 1 | 6 | 2.4 | 3 | 5 | 1 | 2 | 4 | 1.2 | 10 | 1.7 | |||||||||
| ms4760-29 | 2 | 1 | 9 | 8 | 3 | 4 | 12 | 4.8 | 2 | 1 | 2 | 0.6 | 14 | 2.4 | |||||||||||||
| ms4760-30 | 1 | 2 | 2 | 3 | 1 | 1 | 3 | 1.2 | 1 | 2 | 6 | 4 | 2 | 4 | 9 | 2.6 | 12 | 2.0 | |||||||||
| ms4760-31 | 1 | 2 | 4 | 3 | 1 | 2 | 5 | 1.4 | 5 | 0.8 | |||||||||||||||||
| ms4760-32 | 2 | 1 | 1 | 6 | 1 | 1 | 2 | 0.6 | 2 | 0.3 | |||||||||||||||||
| ms4760-33 | 0 | 2 | 1 | 1 | 1 | 0.3 | 1 | 0.2 | |||||||||||||||||||
| ms4760-34 | 4 | 1 | 1 | 1 | 1 | 0.4 | 1 | 0.2 | |||||||||||||||||||
| ms4760-35 | 1 | 1 | 1 | 1 | 1 | 0.4 | 1 | 1 | 1 | 2 | 2 | 0.6 | 3 | 0.5 | |||||||||||||
| ms4760-42 | 1 | 1 | 1 | 5 | 1 | 0.3 | 1 | 0.2 | |||||||||||||||||||
| ms4760-90 | 3 | 1 | 3 | 4 | 1 | 1 | 1 | 1 | 5 | 2.0 | 5 | 0.8 | JX472441 | ||||||||||||||
| ms4760-91 | 1 | 2 | 1 | 1 | 3 | 4 | 4 | 1.6 | 1 | 6 | 2 | 1 | 1 | 2 | 4 | 1.2 | 8 | 1.3 | JX472442 | ||||||||
| ms4760-92 | 1 | 1 | 1 | 1 | 1 | 0.3 | 1 | 0.2 | JX472443 | ||||||||||||||||||
| ms4760-93 | 1 | 2 | 1 | 2 | 1 | 0.3 | 1 | 0.2 | JX472444 | ||||||||||||||||||
| ms4760-94 | 2 | 0 | 2 | 4 | 2 | 0.6 | 2 | 0.3 | JX472445 | ||||||||||||||||||
| ms4760-95 | 3 | 1 | 7 | 10 | 4 | 4 | 9 | 12 | 20 | 8.0 | 2 | 1 | 1 | 2 | 3 | 0.9 | 23 | 3.9 | JX472446 | ||||||||
| ms4760-96 | 3 | 1 | 6 | 9 | 2 | 2 | 1 | 1 | 9 | 3.6 | 3 | 5 | 1 | 1 | 1 | 2 | 5 | 1.4 | 14 | 2.4 | JX472447 | ||||||
| ms4760-97 | 4 | 2 | 1 | 2 | 1 | 1 | 2 | 0.6 | 2 | 0.3 | JX472448 | ||||||||||||||||
| Total | 67 | 108 | 75 | 250 | 56 | 16 | 157 | 44 | 50 | 22 | 345 | 595 | |||||||||||||||
Figure 2.
Amino acid alignment of 29 ms4760 haplotypes found in the Comoros Islands and Madagascar from 2006 to 2007.
The distribution and prevalence of the alleles were significantly different between countries (P < 0.0001) and within country only for isolates from the Comoros (between Anjouan/Mohéli and Grande Comore, P = 0.003 and P = 0.02, respectively).
According to the number of repeats in block II (DNNND) and block V (NHNDNHNNDDD), which have been associated with modulation of in vitro susceptibility to QN,13 ms4760 alleles were grouped in 12 different profiles (from ms4760-A to ms4760-L) as presented in Table 2. The number of repeats in block II (DNNND) varied from zero (ms-4760-A) to four (ms4760-K and ms4760-L), whereas the number of repeats in block V (NHNDNHNNDDD) varied from zero (ms4760-E) to three (ms4760-D and ms4760-H); 73% of isolates were grouped in three profiles: ms-4760-C ([DNNND]1; [NHNDNHNNDDD]2; 20.5%), ms-4760-G ([DNNND]2; [NHNDNHNNDDD]2; 32.3%), and ms-4760-I ([DNNND]3; [NHNDNHNNDDD]1; 20.3%). Seven profiles (ms-4760-B, -C, -D, -F, -G, -I, and -J) were found in both countries, four profiles (ms-4760-A, -E, -H, and -L) were found only in Madagascar, and one profile (ms-4760-H) was found only in the Comoros. The mean number of DNNND repeats was significantly higher in the Comoros (2.21) compared with Madagascar (1.93; P < 0.001). Inversely, the number of NHNDNHNNDDD repeats was significantly lower in the Comoros (1.60) compared with Madagascar (1.73; P = 0.005). Consequently, the mean ratio of DNNND/NHNDNHNNDDD repeats was significantly higher in the Comoros (1.77 versus 1.47; P < 0.001). The prevalence of the pfnhe-1 ms4760 profiles according to the geographical location of the isolates (country and region) significantly differed between the two countries (P < 0.0001). In both countries, three profiles were predominant (ms-4760-G, 32.3%; ms-4760-I, 20.3%; ms-4760-C, 20.5%). Two profiles (ms-4760-I, P < 0.0001; ms-4760-K, P = 0.006) were significantly more frequent in the Comoros, and one profile (ms-4760-C, P = 0.02) was significantly more frequent in Madagascar.
Table 2.
Pfhne-1 ms4760 profile groups according to the number of repeats in block II (DNNND) and block V (DDNHNDNHNND) and the geographical location of the isolates
| ms4760 alleles number | ms4760 profiles | No. of allele | Block II (DNND) | Block V (DDNHNDNHNND) | Frequency (%) | |
|---|---|---|---|---|---|---|
| Comoros Islands | Madagascar | |||||
| 33 | ms4760-A | 1 | 0 | 2 | 0 | 0.3 |
| 2, 35, 42, 92 | ms4760-B | 4 | 1 | 1 | 1.2 | 4.9 |
| 3, 30, 31, 91, 93 | ms4760-C | 5 | 1 | 2 | 13.6 | 25.5 |
| 12 | ms4760-D | 1 | 1 | 3 | 3.6 | 2.3 |
| 94 | ms4760-E | 1 | 2 | 0 | 0 | 0.6 |
| 6, 29, 32 | ms4760-F | 3 | 2 | 1 | 11.2 | 9.0 |
| 1, 19, 27 | ms4760-G | 3 | 2 | 2 | 34.0 | 31.0 |
| 22 | ms4760-H | 1 | 2 | 3 | 0 | 0.6 |
| 7, 14, 90, 95, 96 | ms4760-I | 5 | 3 | 1 | 28 | 14.8 |
| 8, 9 | ms4760-J | 2 | 3 | 2 | 5.6 | 10.4 |
| 5, 34 | ms4760-K | 2 | 4 | 1 | 2.8 | 0 |
| 97 | ms4760-L | 1 | 4 | 2 | 0 | 0.6 |
Genetic diversity, assessed by Nei's unbiased expected heterozygosity (He), was similar between countries (Madagascar = 0.84, ranging from 0.75 for the southeast area to 0.85 for the central west area; Comoros = 0.85, ranging from 0.80 for Grande Comore to 0.87 for Mohéli). However, the degree of genetic differentiation of the ms4760 profiles within parasite populations, estimated by Fst values, indicated a large divergence between Grande Comore populations and Malagasy populations from the northwest, central east, west, and southwest areas (Table 3).
Table 3.
Pairwise population genetic distances (Fst according to Weir and Cockerham)29
| Comoros Islands | Madagascar | |||||||
|---|---|---|---|---|---|---|---|---|
| Mohéli | Anjouan | Northeast | Northwest | Central east | Central west | Southeast | Southwest | |
| Comoros Islands | ||||||||
| Grande Comore | 0.037 | 0.003 | 0.033 | 0.0014* | 0.0014* | 0.0014* | 0.020 | 0.0014* |
| Mohéli | 0.346 | 0.490 | 0.018 | 0.114 | 0.0389 | 0.232 | 0.109 | |
| Anjouan | 0.58 | 0.119 | 0.019 | 0.003 | 0.125 | 0.018 | ||
| Madagascar | ||||||||
| Northeast | 0.115 | 0.843 | 0.785 | 0.479 | 0.540 | |||
| Northwest | 0.051 | 0.037 | 0.380 | 0.061 | ||||
| Central east | 0.183 | 0.193 | 0.115 | |||||
| Central west | 0.680 | 0.443 | ||||||
| Southwest | 0.305 | |||||||
| Southeast | ||||||||
P > 0.05.
The data represented here are an extension of our previous study performed in 2010.14 By using a large number of P. falciparum isolates from Indian Ocean malaria-endemic areas (Comoros and Madagascar), we confirm the extended polymorphisms of ms4760 allele in pfnhe-1 gene in this region. Among the 595 studied sequences, we have observed 29 different alleles, including 8 new alleles (27%). By compiling our data with previous published sequences available in GenBank,14–24 we estimate that 101 different ms4760 alleles have been described to date. However, in most publications, the numbering of the ms4760 alleles did not always taking into account the previously described alleles, making data comparison difficult. This finding raises the need to establish a standard nomenclature for ms4760 alleles.
As expected and found in previous studies, four alleles (ms4760-1, ms4760-3, ms4760-6, and ms4760-7) were predominant in both countries.14,15,17–22 However, significant differences in the distribution and prevalence of allele were observed both between countries and within sites in the Comoros (Table 1). The genetic diversity of the ms4760 allele observed in our study was similar between both countries (0.84 and 0.85), and it was comparable with the genetic diversity previously described in African (Congo = 0.76, Uganda = 0.79, and Kenya = 0.66)15,17,20 and Indian isolates (0.68)23 and significantly higher than the diversity found in Asian isolates (China/Myanmar = 0.68, P = 0.04; Vietnam = 0.49, P < 0.0001).19,22 This situation is likely reflecting the level of malaria transmission, but it also could be related to the prevalence of resistant parasites to quinoline antimalarial drugs. This latter hypothesis is strengthened by our data, which show that isolates from the Comoros (an area with a high prevalence of antimalarial drugs resistance, although specific data about QN resistance are lacking) had significantly more repeats in block II (DNNND) than those isolates from Madagascar (a low drug-resistance area); these findings are consistent with some previous findings observed in culture-adapted parasites from Asia,13,18,19,21,22 India,23 and East Africa.20
In conclusion, current observations from molecular surveys that aimed to define an association between potential contributors to QN resistance, such as ms4760 allele polymorphism, have generated conflicting data and do not allow for proposing a simple molecular typing methodology of global application based on this molecular marker. The level of genetic diversity observed in the present study was comparable with the level found in African countries and not comparable with the level found in Asian countries, where the pfnhe-1 polymorphisms seemed more often usable as molecular markers of QN resistance.19,22 The higher mean number of DNNND repeats found in isolates from the Comoros compared with Madagascar underlined the importance of the geographical origin of parasites, even at this regional level. Additional studies are required to better define the mechanisms underlying QN resistance, which involve multiple gene interactions.
ACKNOWLEDGMENTS
The authors thank the patients and healthcare workers involved in the studies performed in Madagascar and Comoros. This work was supported by grants from Natixis Banques and the Genomics Platform, Pasteur Génopôle, Pasteur Institute, France. Sample collection was funded in Comoros by the FSP/RAI 2001-168 project (Fonds de Solidarité Prioritaire - Résistance aux Anti-Infectieux, French Ministry of Foreign Affairs) and sample collection in Madagascar was funded by Global Fund Project Round 3 Grant MDG-304-G05-M. B.W. is supported by a post-doctoral fellowship from the Division International, Institut Pasteur (2011–2013). C.B. is supported by a grant from the Fondation Pierre Ledoux, Jeunesse Internationale (2012). D.M. is supported by the French Ministry of Foreign Affairs.
Footnotes
Authors' addresses: Valérie Andriantsoanirina and Rémy Durand, Hôpital Avicenne, AP-HP, Laboratoire de Parasitologie-Mycologie, Bobigny, France, E-mails: landyvalerie@gmail.com and remy.durand@avc.aphp.fr. Nimol Khim, Benoit Witkowski, Lydie Canier, Christophe Benedet, and Didier Ménard, Institut Pasteur du Cambodge, Malaria Molecular Epidemiology Unit, Phnom Penh, Cambodia, E-mails: knimol@pasteur-kh.org, bwitkowski@pasteur-kh.org, lcanier@pasteur-kh.org, christophe.benedet@pasteur-kh.org, and dmenard@pasteur-kh.org. Arsene Ratsimbasoa, Ministère de la Santé, du Planning Familial et de la Protection Sociale—National Malaria Control Programme, Antananarivo, Madagascar, E-mail: arsene.ratsimbasoa@laposte.net. Christiane Bouchier and Magali Tichit, Institut Pasteur, Génopôle de l'Ile de France, Plate-Forme Génomique, Paris, France, E-mails: bouchier@pasteur.fr and mtichit@pasteur.fr.
References
- 1.Baird JK. Effectiveness of antimalarial drugs. N Engl J Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- 2.Achan J, Tibenderana JK, Kyabayinze D, Wabwire Mangen F, Kamya MR, Dorsey G, D'Alessandro U, Rosenthal PJ, Talisuna AO. Effectiveness of quinine versus artemether-lumefantrine for treating uncomplicated falciparum malaria in Ugandan children: randomised trial. BMJ. 2009;339:b2763. doi: 10.1136/bmj.b2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam I, Ali DM, Noureldien W, Elbashir MI. Quinine for the treatment of chloroquine-resistant Plasmodium falciparum malaria in pregnant and non-pregnant Sudanese women. Ann Trop Med Parasitol. 2005;99:427–429. doi: 10.1179/136485905X36217. [DOI] [PubMed] [Google Scholar]
- 4.Adegnika AA, Breitling LP, Agnandji ST, Chai SK, Schutte D, Oyakhirome S, Schwarz NG, Grobusch MP, Missinou MA, Ramharter M, Issifou S, Kremsner PG. Effectiveness of quinine monotherapy for the treatment of Plasmodium falciparum infection in pregnant women in Lambarene, Gabon. Am J Trop Med Hyg. 2005;73:263–266. [PubMed] [Google Scholar]
- 5.Chongsuphajaisiddhi T, Sabchareon A, Attanath P. Treatment of quinine resistant falciparum malaria in Thai children. Southeast Asian J Trop Med Public Health. 1983;14:357–362. [PubMed] [Google Scholar]
- 6.de Vries PJ, Bich NN, Van Thien H, Hung LN, Anh TK, Kager PA, Heisterkamp SH. Combinations of artemisinin and quinine for uncomplicated falciparum malaria: efficacy and pharmacodynamics. Antimicrob Agents Chemother. 2000;44:1302–1308. doi: 10.1128/aac.44.5.1302-1308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGready R, Ashley EA, Moo E, Cho T, Barends M, Hutagalung R, Looareesuwan S, White NJ, Nosten F. A randomized comparison of artesunate-atovaquone-proguanil versus quinine in treatment for uncomplicated falciparum malaria during pregnancy. J Infect Dis. 2005;192:846–853. doi: 10.1086/432551. [DOI] [PubMed] [Google Scholar]
- 8.McGready R, Brockman A, Cho T, Cho D, van Vugt M, Luxemburger C, Chongsuphajaisiddhi T, White NJ, Nosten F. Randomized comparison of mefloquine-artesunate versus quinine in the treatment of multidrug-resistant falciparum malaria in pregnancy. Trans R Soc Trop Med Hyg. 2000;94:689–693. doi: 10.1016/s0035-9203(00)90235-9. [DOI] [PubMed] [Google Scholar]
- 9.Pukrittayakamee S, Chantra A, Vanijanonta S, Clemens R, Looareesuwan S, White NJ. Therapeutic responses to quinine and clindamycin in multidrug-resistant falciparum malaria. Antimicrob Agents Chemother. 2000;44:2395–2398. doi: 10.1128/aac.44.9.2395-2398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pukrittayakamee S, Supanaranond W, Looareesuwan S, Vanijanonta S, White NJ. Quinine in severe falciparum malaria: evidence of declining efficacy in Thailand. Trans R Soc Trop Med Hyg. 1994;88:324–327. doi: 10.1016/0035-9203(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 11.Rahman MR, Paul DC, Rashid M, Ghosh A, Bangali AM, Jalil MA, Faiz MA. A randomized controlled trial on the efficacy of alternative treatment regimens for uncomplicated falciparum malaria in a multidrug-resistant falciparum area of Bangladesh–narrowing the options for the National Malaria Control Programme? Trans R Soc Trop Med Hyg. 2001;95:661–667. doi: 10.1016/s0035-9203(01)90108-7. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization . Global Report on Antimalarial Efficacy and Drug Resistance: 2000–2010. Geneva: World Health Organization; 2010. [Google Scholar]
- 13.Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 14.Andriantsoanirina V, Menard D, Rabearimanana S, Hubert V, Bouchier C, Tichit M, Bras JL, Durand R. Association of microsatellite variations of Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene with reduced in vitro susceptibility to quinine: lack of confirmation in clinical isolates from Africa. Am J Trop Med Hyg. 2010;82:782–787. doi: 10.4269/ajtmh.2010.09-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baliraine FN, Nsobya SL, Achan J, Tibenderana JK, Talisuna AO, Greenhouse B, Rosenthal PJ. Limited ability of Plasmodium falciparum pfcrt, pfmdr1, and pfnhe1 polymorphisms to predict quinine in vitro sensitivity or clinical effectiveness in Uganda. Antimicrob Agents Chemother. 2010;55:615–622. doi: 10.1128/AAC.00954-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briolant S, Henry M, Oeuvray C, Amalvict R, Baret E, Didillon E, Rogier C, Pradines B. Absence of association between piperaquine in vitro responses and polymorphisms in the pfcrt, pfmdr1, pfmrp, and pfnhe genes in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:3537–3544. doi: 10.1128/AAC.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briolant S, Pelleau S, Bogreau H, Hovette P, Zettor A, Castello J, Baret E, Amalvict R, Rogier C, Pradines B. In vitro susceptibility to quinine and microsatellite variations of the Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene: the absence of association in clinical isolates from the Republic of Congo. Malar J. 2011;10:37. doi: 10.1186/1475-2875-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry M, Briolant S, Zettor A, Pelleau S, Baragatti M, Baret E, Mosnier J, Amalvict R, Fusai T, Rogier C, Pradines B. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob Agents Chemother. 2009;53:1926–1930. doi: 10.1128/AAC.01243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng H, Zhang R, Yang H, Fan Q, Su X, Miao J, Cui L, Yang Z. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob Agents Chemother. 2010;54:4306–4313. doi: 10.1128/AAC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okombo J, Kiara SM, Rono J, Mwai L, Pole L, Ohuma E, Borrmann S, Ochola LI, Nzila A. In vitro activities of quinine and other antimalarials and pfnhe polymorphisms in Plasmodium isolates from Kenya. Antimicrob Agents Chemother. 2010;54:3302–3307. doi: 10.1128/AAC.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelleau S, Bertaux L, Briolant S, Ferdig MT, Sinou V, Pradines B, Parzy D, Jambou R. Differential association of Plasmodium falciparum Na+/H+ exchanger polymorphism and quinine responses in field- and culture-adapted isolates of Plasmodium falciparum. Antimicrob Agents Chemother. 2011;55:5834–5841. doi: 10.1128/AAC.00477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinou V, Quang le H, Pelleau S, Huong VN, Huong NT, Tai le M, Bertaux L, Desbordes M, Latour C, Long LQ, Thanh NX, Parzy D. Polymorphism of Plasmodium falciparum Na(+)/H(+) exchanger is indicative of a low in vitro quinine susceptibility in isolates from Viet Nam. Malar J. 2011;10:164. doi: 10.1186/1475-2875-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinayak S, Alam MT, Upadhyay M, Das MK, Dev V, Singh N, Dash AP, Sharma YD. Extensive genetic diversity in the Plasmodium falciparum Na+/H+ exchanger 1 transporter protein implicated in quinine resistance. Antimicrob Agents Chemother. 2007;51:4508–4511. doi: 10.1128/AAC.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyomtip T, Suwandittakul N, Sitthichot N, Khositnithikul R, Tan-ariya P, Mungthin M. Polymorphisms of the pfmdr1 but not the pfnhe-1 gene is associated with in vitro quinine sensitivity in Thai isolates of Plasmodium falciparum. Malar J. 2012;11:7. doi: 10.1186/1475-2875-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okombo J, Ohuma E, Picot S, Nzila A. Update on genetic markers of quinine resistance in Plasmodium falciparum. Mol Biochem Parasitol. 2011;177:77–82. doi: 10.1016/j.molbiopara.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Rakotonirina H, Barnadas C, Raherijafy R, Andrianantenaina H, Ratsimbasoa A, Randrianasolo L, Jahevitra M, Andriantsoanirina V, Menard D. Accuracy and reliability of malaria diagnostic techniques for guiding febrile outpatient treatment in malaria-endemic countries. Am J Trop Med Hyg. 2008;78:217–221. [PubMed] [Google Scholar]
- 27.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB, Jr, Peterson LR, Kaul KL. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005;43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]