Abstract
The first confirmed outbreak of highly pathogenic avian influenza (HPAI) virus infections in North America was caused by A/turkey/Ontario/7732/1966 (H5N9); however, the phylogeny of this virus is largely unknown. This study performed genomic sequence analysis of 11 avian influenza isolates from 1956 to 1979 for comparison with A/turkey/Ontario/7732/1966 (H5N9). Phylogenetic and genetic analyses included these viruses in combination with all known full-genome sequences of avian viruses isolated before 1981. It was shown that a low-pathogenic avian influenza virus, A/turkey/Ontario/6213/1966 (H5N1), that had been isolated 3 months previously, was the closest known genetic relative with six genome segments of common lineage encoding the polymerase subunits PB2, PB1 and PA, nucleoprotein (NP), haemagglutinin (HA) and non-structural (NS) proteins. The lineages of these genome segments included reassortment with other North American turkey viruses that were all rooted in North American wild waterfowl with the HA gene originating from the H5N2 serotype. The phylogenies demonstrated adaptation from North American wild birds to turkeys with the possible involvement of domestic waterfowl. The turkey isolate, A/turkey/Wisconsin/1968 (H5N9), was the second most closely related poultry isolate to A/turkey/Ontario/7732/1966 (H5N9), possessing five common lineage genome segments (PB2, PB1, PA, HA and neuraminidase). The A/turkey/Ontario/6213/1966 (H5N1) virus was more virulent than A/turkey/Wisconsin/68 (H5N9) for chicken embryos and mice, indicating a greater biological similarity to A/turkey/Ontario/7732/1966 (H5N9). Thus, A/turkey/Ontario/6213/1966 (H5N1) was identified as the closest known ancestral relative of HPAI A/turkey/Ontario/7732/1966 (H5N9), which will serve as a useful reference virus for characterizing the early genetic and biological properties associated with the emergence of pathogenic avian influenza strains.
Introduction
The first unconfirmed outbreaks of highly pathogenic avian influenza (HPAI) virus infections in North America occurred in the 1920s and were described as European ‘fowl-plague’ viruses on the basis of characteristic pathology; however, these virus isolates were not maintained and their identification remains speculative (reviewed by Lupiani & Reddy, 2009; Swayne, 2009). Except for an outbreak of neurological disease in ducklings in Manitoba in 1953 [A/duck/Manitoba/1953 (H10N7)] (Walker & Bannister, 1953), pathogenic influenza viruses were not isolated from North American poultry until the 1960s. Beginning with this time period, numerous influenza serotypes were isolated from turkeys (Lang & Ferguson, 1981), including the first confirmed North American isolate of HPAI, A/turkey/Ontario/7732/1966 (H5N9) [tk/ON/7732/66 (H5N9)], from an outbreak in a turkey breeder flock (Lang et al., 1968a). Several laboratories have confirmed the diagnosis of HPAI for tk/ON/7732/66 (H5N9) on infection of turkeys and chickens (Alexander et al., 1986; Lang et al., 1968a; Philpott et al., 1990), and the haemagglutinin (HA) sequence of this virus showed that it had acquired a multibasic cleavage site, which is characteristic of HPAI (GenBank accession no. AAA43205). The 1966 Ontario outbreak of HPAI was preceded, 3 months earlier, by an outbreak of low-pathogenic avian influenza virus A/turkey/Ontario/6613/1966 (H5N1) [tk/ON/6213/66 (H5N1)] in an unrelated turkey farm in Ontario (Lang et al., 1968b), which has been speculated to be a possible precursor virus (Swayne, 2009). The isolating laboratory did not report the neuraminidase (NA) serotype of this virus at the time (Lang et al., 1968b) but later identified it as H5N1 (Lang & Ferguson, 1981). Although tk/ON/6213/66 (H5N1) is classified as low pathogenic, it caused mortality in turkey and chicken embryos as well as young turkeys, which developed pancreatitis (Lang et al., 1968b; Rouse et al., 1968), which was a clinical feature that it shared with HPAI tk/ON/7732/66 (H5N9) for adult turkeys (Narayan et al., 1969a, b).
Host switching by influenza A virus from the wild aquatic bird reservoir often involves adaptation in domestic aquatic and terrestrial poultry with subsequent evolution to HPAI as seen for some H5 and H7 serotypes (Webster et al., 1992). HPAI was first observed in Europe in 1878 and during the early 1900s for H7 and later for H5 subtype viruses (Lupiani & Reddy, 2009). The current epizootic of HPAI H5N1 in Eurasia is caused by a lineage of viruses that first emerged in China as the A/goose/Guangdong/1/1996 (H5N1) strain, which continued to reassort with other avian strains to become endemic in terrestrial and aquatic poultry and later in wild birds (Guan et al., 2003; Mukhtar et al., 2007; Shaw et al., 2002; Zhao et al., 2008). The current H5N1 lineages continue to evolve and spread in wild and domestic birds with the continuing possibility of human adaptation (Brown et al., 2008; Li et al., 2004). The genesis of this lineage is complex, having derived genome segments from multiple progenitor viruses of low pathogenicity (Mukhtar et al., 2007). Little is known of the genetic features that are responsible for the evolution of virulence in this lineage of HPAI viruses, although roles have been demonstrated for the multibasic cleavage site in the HA receptor and RNA polymerase subunits (Basler & Aguilar, 2008). Genetic analysis has been hampered by the lack of a low-pathogenic H5N1 precursor for comparative analysis. This situation emphasizes the need to further understand host switching and virulence in influenza viruses so that emerging viruses in animals and humans can be predicted and eradicated.
To address the possibility that tk/ON/6213/66 (H5N1) was an evolutionary precursor of HPAI tk/ON/7732/66 (H5N9), we performed genome sequencing of tk/ON/6213/66 (H5N1) and 11 other early poultry and avian influenza isolates with collection dates spanning 1956–1979. We showed that tk/ON/6213/66 (H5N1) was the closest known potential genetic relative and donor of six of the eight genome segments of HPAI tk/ON/7732/66 (H5N9). Future studies of the role of individual genes of both of these viruses will provide insights into the evolution of virulence for tk/ON/7732/66 (H5N9).
Results
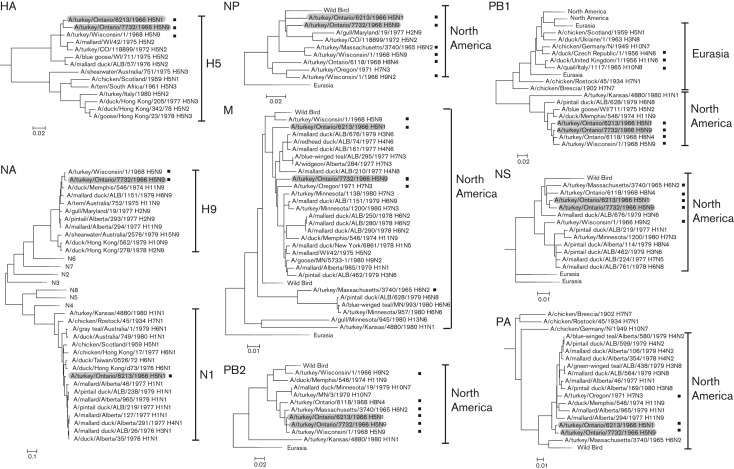
The genomic sequence of tk/ON/7732/66 (H5N9) was available; however, only two other North American avian influenza virus genome sequences were available from this early time period for comparative sequence and phylogenetic analysis before this report. We therefore sequenced the genomes of 11 early poultry and waterfowl virus isolates that were collected between 1956 and 1979, which are listed in Table 1. Viruses were grown in the allantoic cavity of embryonated chicken eggs with extraction of viral RNA for sequencing of each genome segment. The GenBank accession numbers for each genome segment are listed alphabetically in Table S1 (available in JGV Online). The tk/ON/6213/66 (H5N1) sequence analysis identified it as a mixture, with H3 and H4 genes from other viruses in the same sequencing project, indicating cross-contamination at some point in the analysis. We therefore purified tk/ON/6213/66 (H5N1) by two cycles of plaque isolation with partial sequencing of each genome segment to confirm that the genomic sequence was correct (data not shown). The serotypes of all other isolates were confirmed to be correct by blast analysis of their respective HA and NA genes. We then determined the phylogenetic relationship of each genomic segment of tk/ON/7732/66 (H5N9) compared with those of all available full-genome sequences from early avian virus isolates collected up to the end of 1980. Phylogenetic analysis of the HA genes showed that all the newly sequenced viruses mapped to branches that corresponded with their expected HA and NA serotypes (Fig. S1). The HPAI tk/ON/7732/66 (H5N9) H5 gene was most closely related to the turkey tk/ON/6213/66 (H5N1) and tk/WI/68 (H5N9) viruses on a branch associated with turkey and waterfowl viruses of the H5N2 serotype (Fig. 1 and Fig. S1). The NA phylogeny showed that the HPAI tk/ON/7732/66 (H5N9) was most closely related to the tk/WI/68 (H5N9) virus that was linked to a waterfowl isolate [A/duck/Memphis/546/1974 (H11N9)] embedded in a North American wild waterfowl lineage (Figs 1 and S1). This showed that, with respect to surface proteins, tk/ON/7732/66 (H5N9) derived its HA from a tk/ON/6213/66 (H5N1)-like turkey virus lineage emanating from H5N2 North American waterfowl, and that the NA was derived by reassortment with a North American waterfowl N9 virus of unknown HA serotype to form a common lineage with tk/WI/68 (H5N9)-like viruses. Alignment of the protein sequences of the HA cleavage-site region of the most closely related H5 viruses with the preceding HPAI viruses from Scotland [A/chicken/Scotland/1/1959 (H5N1)] and South Africa [A/tern/South Africa/1961 (H5N3)] demonstrated the close relationship of tk/ON/7732/66 (H5N9) to tk/ON/6213/66 (H5N1), as well as the presence of three additional basic amino acids, RKK, formed by nucleotide substitutions and a 3 nt insertion preceding the arginine residue at the normal cleavage-site motif of low-pathogenic avian influenza viruses (Fig. 2). The tk/ON/7732/66 (H5N9) cleavage site therefore possessed a sequence of five basic amino acids at the cleavage site with the sequence RRKKR/G (the three additional basic amino acids shown in bold).
Table 1. Viruses sequenced for phylogenetic analyses.
| Virus name | Abbreviation |
| A/duck/Czechoslovakia/1956 (H4N6) | dk/CZ/56 (H4N6) |
| A/turkey/Wisconsin/1968 (H5N9) | tk/WI/68 (H5N9) |
| A/turkey/Ontario/6213/1966 (H5N1)* | tk/ON/6213/66 (H5N1) |
| A/turkey/Ontario/7732/1966 (H5N9) | tk/ON/7732/66 (H5N9) |
| A/turkey/Massachusetts/3740/1965 (H6N2) | tk/MA/65 (H6N2) |
| A/turkey/Oregon/1971 (H7N3) | tk/OR/71 (H7N3) |
| A/turkey/Ontario/6118/1968 (H8N4) | tk/ON/68 (H8N4) |
| A/turkey/Wisconsin/1/1966 (H9N2) | tk/WI/66 (H9N2) |
| A/quail /Italy/1117/1965 (H10N8) | ql/IT/65 (H10N8) |
| A/duck/United Kingdom/1/1956 (H11N6) | dk/UK/56 (H11N6) |
| A/duck/Wisconsin/480/1979 (H12N6) | dk/WI/79 (H12N6) |
| A/gull/Maryland/704/1977 (H13N6) | gl/MD/77 (H13N6) |
Originally labelled as serotype H5N9.
Fig. 1.
Phylogenetic trees of the HA, NA, NP, M, PB2, PB1, NS and PA genome segments of avian influenza A viruses isolated before 1981. The tk/ON/6213/66 and tk/ON/7732/66 viruses are shaded, and sequenced viruses from this study (Table 1) are indicated by ▪. Full trees and a list of GenBank accession numbers of the viruses used are provided in Fig. S1 and Table S1. Bars, nucleotide substitutions per site.
Fig. 2.
Alignment of HA sequences of subtype H5 from different avian influenza A viruses over aa 281–350. The NCBI Protein numbers of the viruses used in the alignment are: AAC58994, ACZ48553, ACZ48585, AEA04387, AAA43205, AAC58998 and AAD37782, respectively. Amino acid substitutions within the alignment are shown in black.
The phylogenetic tree of the polymerase subunits PB2 and PB1 showed that tk/ON/7732/66 (H5N9) was most closely related to tk/ON/6213/66 (H5N1) and clustered among other closely related turkey viruses of various subtypes from the USA and Ontario including tk/ON/68 (H8N4) and tk/WI/68 (H5N9), with both genes originating from North American waterfowl (Figs 1 and S1). The polymerase subunit PA gene phylogeny showed that the tk/ON/7732/66 (H5N9) genome segment was most closely related to tk/ON/6213/66 (H5N1) among related North American aquatic waterfowl viruses that also included some turkey viruses (Figs 1 and S1). The nucleoprotein (NP) phylogenetic tree showed that tk/ON/7732/66 (H5N9) was on a common branch with a shorebird and other poultry viruses, with preceding branch points with linkage to Ontario and North American turkey viruses but with longer horizontal branch lengths indicating greater sequence divergence and thus evolutionary distances separating these viruses (Figs 1 and S1).
The non-structural (NS) gene phylogenies associated tk/ON/7732/66 (H5N9) and tk/ON/6213/66 (H5N1) on the same branch with other poultry viruses that were embedded among waterfowl branches and other turkey viruses (Figs 1 and S1). The phylogenetic tree of matrix (M) genes showed a distinct type of pattern, with tk/ON/7732/66 (H5N9) on its own branch embedded among a mixed lineage of turkey and wild aquatic bird viruses, with more distant linkages to later turkey isolates including tk/OR/71 (H7N3). The tk/ON/6213/66 (H5N1) M gene segment shared a common but more distant predecessor with tk/ON/7732/66 (H5N9) (Figs 1 and S1). Phylogenetic analysis of all tk/ON/7732/66 (H5N9) genome segments with all pre-1981 avian influenza virus genomes showed that six of the genome segments (PB2, PB1, PA, NP, HA and NS) were most closely related to tk/ON/6213/66 (H5N1); the NA gene was most closely related to tk/WI/68 (H5N9) and the M genes were most closely related to tk/OR/71(H7N3).
Comparison of tk/ON/7732/66 (H5N9) genome segments by nucleotide identity
To quantify further the extent of sequence similarity among avian influenza virus genes, we performed global blast analysis of each tk/ON/7732/66 (H5N9) genome segment against all influenza genes in GenBank. We found that the genes with the closest phylogenetic similarities to tk/ON/7732/66 (H5N9) in general also possessed the highest levels of nucleotide identity, with the highest identity levels observed for the tk/ON/6213/66 (H5N1) PB2 of 99.3 %, PB1 of 98.5 % [and 98.9 % for ON/68(H8N4)], PA of 99.2 %, HA of 97.8 %, NP of 96.2 % and NS of 99.1 % (Table 2). The NA gene was most closely related to tk/WI/68 (H5N9) (98.5 %) and the M genes with A/turkey/California/189/66 (H9N2) [99.2 %; also closely related to ON/68 (H8N4) with 97.1 % identity] (Table 2). These nucleotide identities represented the greatest similarities seen on blast except for the NP gene, which was most closely related to the early Canadian aquatic poultry virus isolate A/duck/Manitoba/1953 (H10N7) (96.9 % identity; Table 2). The genetic similarity by phylogenetic assessment of published full genomes was in general consistent with, and thus supported by, the extent of similarity assessed by phylogenetic comparison with all published influenza genome sequences.
Table 2. Extent of sequence identity of tk/ON/7732/66 genome segments to 1965–1968 North American turkey isolates.
The highest levels of nucleotide identity for each gene are indicated in bold. nd, Not determined.
| Gene(s) | Virus | |||||
| ON66 (H5N9) | ON66 (H5N1) | WI68 (H5N9) | ON68 (H8N4) | MA65 (H6N2) | WI66 (H9N2) | |
| PB2 | 100.0 | 99.3 | 98.8 | 96.5 | 98.2 | 91.0 |
| PB1 | 100.0 | 98.5 | 98.4 | 98.9 | <90 | <90 |
| PA | 100.0 | 99.2 | 93.7 | <91 | 94.9 | <91 |
| HA | 100.0 | 97.8 | 97.4 | nd | nd | nd |
| NP* | 100.0 | 96.2 | <95 | <95 | 95.0 | <95 |
| NA | 100.0 | nd | 98.5 | nd | nd | nd |
| M1/2† | 100.0 | <96 | <96 | 97.1 | <96 | <96 |
| NS1/2 | 100.0 | 99.1 | <96 | 98.2 | 97.9 | 96.8 |
A/duck/Manitoba/1/1953 (H10N7) possessed the highest identity (96.9 %) by blast analysis.
A/turkey/California/189/66 (H9N2) possessed the highest identity (99.2 %) by blast analysis.
Genotypic analysis demonstrates the origin of tk/ON/7732/66 (H5N9) genome segments from turkeys and aquatic birds
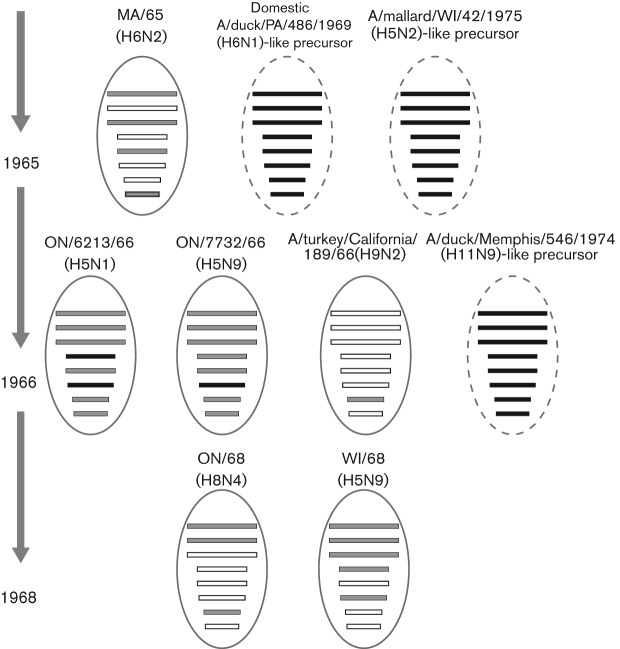
The shared lineage of genome segments among turkey viruses determined by phylogenetic and sequence relationships was used to map the genotypes of the tk/ON/7732/66 (H5N9) virus relative to other avian influenza virus genome segments. Genome segment lineages found in multiple poultry isolates are indicated in grey in Fig. 3, while turkey virus genome segments that were most closely related to aquatic bird segments are shown in black. The HA gene was probably derived by tk/ON/6213/66 (H5N1) from a North American A/mallard/WI/42/1975 (H5N2)-like wild waterfowl virus (Fig. 1) before it was transferred to tk/ON/7732/66 (H5N9) by a series of reassortment events in turkeys (shown in black for both turkey viruses; Fig. 3). The tk/ON/6213/66 (H5N1) virus had acquired its PB2, PA, NP and NS from a MA/65 (H6N2)-like virus and its N1 gene from a domestic aquatic poultry A/duck/Pennsylvania/486/1969 (H6N1)-like virus (Hwang et al., 1970) (highest similarity by global blast analysis; 97.6 % identity; Fig. 3). The tk/ON/7732/66 (H5N9) virus had acquired six of the tk/ON/6213/66 (H5N1)-like genes along with an NA gene from an A/duck/Memphis/546/1974 (H11N9)-like virus, which possessed the greatest genetic similarity among all avian influenza viruses (95.1 % nucleotide identity). tk/ON/7732/66 (H5N9) derived its M genome segments from an A/turkey/California/189/1966(H9N2)-like virus (with 99.2 % nucleotide identity), which was also similar to aquatic bird virus isolates. Two turkey viruses that were isolated 2 years later, tk/ON/68 (H8N4) and tk/WI/69 (H5N9), had acquired tk/ON/7732/66 (H5N9)-like PB2, PB1 and M versus PB2, PB1, PA, HA and NA genes respectively (Fig. 3). Thus, the tk/ON/7732/66 (H5N9) phylogenetic and gene similarity data indicated that the tk/ON/6213/66 (H5N1) virus that was isolated 3 months earlier (Lang & Ferguson, 1981) shared the largest number of related genome segments by possessing six in common, followed by the 1968 tk/WI/68 (H5N9) virus that possessed five genomes segments of common lineage. The phylogenetic data indicated exchange of genome segments by reassortment among several viruses to generate tk/ON/6213/66 (H5N1) (among H6N2, H5N2 and H6N1), followed by further reassortment to generate tk/ON/7732/66 (H5N9) (among H5N1, H11N9 and H9N2).
Fig. 3.
Lineage of the tk/ON/7732/66 virus. The eight genome segments are shown as horizontal bars in virus particles (ovals), in the order of segments one to eight: PB2, PB1, PA, HA, NP, NA, M and NS. Different lineages were determined by maximum similarity by blastn analysis to all tk/ON/7732/66 (H5N9) segments and tk/ON/6213/66 (H5N1) HA and NA. Genome segments are indicated by shaded bars: black indicates an origin from aquatic birds and grey indicates an origin from turkey. Open bars represent genome segments that were not assessed by blastn analysis.
tk/ON/6213/66 (H5N1) is more virulent for chicken embryos and mice than tk/WI/68 (H5N9)
To assess the biological similarities of the two viruses with the greatest similarity to tk/ON/7732/66 (H5N9), we assessed the relative virulence of tk/ON/6213/66 (H5N1) and tk/WI/68 (H5N9) viruses in mice and chicken embryos. We could not analyse tk/ON/7732/66 (H5N9) virus infection in eggs and mice because this virus was not available for analysis. Inoculation of the allantoic cavities of groups of four embryos with dosages of virus from 102 to 106 p.f.u. showed that all dosages of tk/ON/6213/66 (H5N1) resulted in 100 % mortality for the embryo by 2 days post-infection (p.i.) (LD50<101.5 p.f.u.), whereas all embryos remained viable for all dosages of tk/WI/68 (H5N9) (LD50>106.5 p.f.u.) (Table 3). Similarly intranasal infection of mouse lungs with 106 and 107 p.f.u. tk/ON/6213/66 (H5N1) resulted in 25 and 75 % mortality, respectively, indicating an LD50 of 106.5 p.f.u., whereas no mortality was observed on infection of mice with 106 p.f.u. tk/WI/68 (H5N9) (which was the highest achievable dose for this virus) for an LD50>106.5 p.f.u. (Table 3). Comparison of the disease severity by body weight loss for groups of mice infected with 106 p.f.u. for each virus showed that tk/WI/68 (H5N9) infection induced maximal weight loss at 3 days p.i. (5 %) followed by recovery, whereas tk/ON/6213/66 (H5N1)-infected mice continued to lose weight until day 6 p.i. with 13 % weight loss (P<0.01 by Student’s t-test; Fig. S2) with associated mortality (Table 3). Thus, tk/ON/6213/66 (H5N1) was more virulent than WI/68 (H5N9) for chicken embryos as well as the mammalian mouse host, indicating that it not only shared the greatest number of related genome segments with tk/ON/7732/66 (H5N9) but also shared virulence properties.
Table 3. tk/ON/6213/66 (H5N1) is more virulent than tk/WI/68 (H5N9) by assay of survival of infected chicken embryos and mice.
nd, Not done.
| Virus | Host | Dose (p.f.u.)* | LD50 | |||||
| 102 | 103 | 104 | 105 | 106 | 107 | |||
| tk/ON/6213/66 (H5N1) | Chicken embryo | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | nd | <101.5 |
| tk/WI/68 (H5N9) | Chicken embryo | nd | 4/4 | 4/4 | 4/4 | 4/4 | nd | >106.5 |
| tk/ON/6213/66 (H5N1) | Mouse | nd | nd | nd | nd | 3/4 | 1/4 | 106.5 |
| tk/WI/68 (H5N9) | Mouse | nd | nd | nd | nd | 4/4 | nd | >106.5 |
Survivors are indicated as the fraction of total mice or embryos infected with each dose.
Discussion
In order to determine the genealogical relationship of the first confirmed isolate of HPAI in North America, we performed genomic sequence analysis of a number of North American poultry isolates from this time period and compared them with tk/ON/7732/66 (H5N9). We identified tk/ON/6213/66 (H5N1) as the closest genetic relative and thus the precursor of the HPAI A/turkey/Ontario/7732/1966 (H5N9) isolate. This has expanded our understanding of the early evolutionary events in North American poultry that were associated with the genesis of the first confirmed outbreaks of HPAI in North America. We also confirmed that the tk/ON/6213/66 (H5N1) virus is the first H5N1 serotype in North America and that this virus is virulent for both chicken embryos and mice, indicating that the virus is biologically similar to tk/ON/7732/66 (H5N9).
We showed that tk/ON/6213/66 (H5N1) is the closest known genetic relative of tk/ON/7732/66 (H5N9) and thus is the probable precursor that donated six genome segments to A/turkey/Ontario/7732/1966 (H5N9) including the ribonucleocapsid components (polymerase subunits PB2, PB1 and PA plus NP), HA and NS. This set of genes parallels the findings of studies of experimental adaptive evolution of human influenza virus into mouse, which identified the ribonucleoprotein and HA genes as the most adaptive genetic aspects of host switching (Keleta et al., 2008; Ping et al., 2011), suggesting that these genes may also be important for adaptation of aquatic bird influenza to turkeys. Mouse adaptation has also identified several mutations in NS1 that are involved in host switching (Forbes et al., 2012). The N9 gene was most closely related to WI/68 (H5N9); however, this virus was the genetic acceptor of this genome segment because it was isolated 2 years later. The N9 gene originated from wild aquatic birds with the closest relative being A/duck/Memphis/546/1974 (H11N9), as determined by phylogenetic analysis and sequence identity.
In general, all the tk/ON/7732/66 (H5N9) genome segments were in shared lineages with other turkey isolates spanning 1965–1968 that were all rooted in phylogenetic trees emanating from the North American wild aquatic bird reservoir, indicating a flow of genes into poultry from the wild aquatic bird reservoir of influenza A viruses. The turkey H5 lineage originated from North American wild aquatic birds of the H5N2 serotype (Figs 1 and S1), which were also the precursors of subsequent HPAI viruses in the USA [A/chicken/Pennsylvania/1370/1983 (H5N2)] and Mexico [A/chicken/Puebla/8624-604/1994 (H5N2)] (Swayne, 2009), indicating a propensity for this lineage to generate highly pathogenic poultry viruses. The M genome segment was most closely related to A/turkey/California/189/66 (H9N2), which also possessed a close genetic linkage with the wild aquatic bird reservoir (data not shown). The NP gene was most closely related to the prior domestic aquatic H10N7 poultry virus isolate in Manitoba in 1953, linking transmission to terrestrial poultry from wild waterfowl via domestic aquatic poultry and suggesting that related viruses had been maintained in domestic or wild aquatic birds in the Mississippi flyway, which courses through Manitoba and Ontario. The evolutionary pattern is consistent with the adaptation of aquatic bird viruses to turkeys with continued evolution involving reassortment with other viruses introduced from wild aquatic birds (directly or indirectly from domestic aquatic birds). This is a repeated pattern of evolution of terrestrial poultry viruses that has been seen in Europe and Asia (Jadhao et al., 2009; Webster et al., 1992). Canadian aquatic poultry were characterized as endemically infected during this time period with avian influenza viruses (Lang & Ferguson, 1981), and surveys of North American aquatic poultry indicate endemic infection with a large number of serotypes of avian influenza but without associated disease (Sandhu & Hinshaw 2003). The most closely related N1 gene of tk/ON/6213/66 (H5N1) was a domestic duck isolate, A/duck/Pennsylvania/486/1969 (H6N1) (Fig. 3), which was associated with disease in ducks (Hwang et al., 1970), suggesting that prior adaptation to increased virulence in domestic ducks may have contributed to entry of this NA gene into turkey viruses. The same turkey farm that generated tk/ON/6213/66 (H5N1) virus was subject to an outbreak of A/turkey/Ontario/4689/1967 (H6N1) 1 year later (Lang & Ferguson, 1981), raising the possibility that a predecessor of this H6N1 virus could have been the source of the tk/ON/6213/66 (H5N1) NA genome segment (however, this gene sequence was unavailable for analysis). The poultry PB2 and NS1 gene lineages included tk/MA/65 (H6N2) virus, indicating movement of avian influenza among turkeys in North America through movement of turkey stock or possibly biological materials such as semen, which was associated with the outbreak of low-pathogenic tk/WI/68 (H5N9) virus of turkey flocks in Wisconsin in 1968 (Smithies et al., 1969).
Turkey flocks in Ontario were harbouring multiple subtypes of influenza in the 1960s where 65 outbreaks were recorded between 1962 and 1972 with isolation of viruses of multiple serotypes including H4N6, H5N1, H5N2, H5N9, H6N1, H6N2, H6N8, H8N4 and H9N2 (Lang & Ferguson, 1981), with a similar pattern of infections in this time period due to multiple serotypes seen in turkey flocks in the USA (Lupiani & Reddy, 2009). Ontario turkey farms are situated among wetlands, which provide an ideal habitat for waterfowl with the attendant opportunities for transmission of avian influenza viruses into turkeys. The increased occurrence of avian influenza outbreaks in turkeys in North America resulted in changes of husbandry practices from range rearing to housing in order to improve biosecurity and thus prevent infection from wild birds in the environment (Lang & Ferguson, 1981).
Although the phylogenetic and sequence similarity patterns are consistent with transmission from wild to domestic aquatic birds and then to turkeys, there is a need for further sequence data from wild and domestic poultry prior to 1966 in order to obtain a more complete understanding of the history of adaptation and reassortment events leading to the genesis of the pathogenic turkey tk/ON/7732/66 (H5N9) virus.
Although the tk/WI/68 (H5N9) and tk/ON/6213/66 (H5N1) viruses have five and six genome segments of shared lineage with tk/ON/7732/66 (H5N9), respectively, they differed markedly in their virulence for chicken embryos and mice (Table 3 and Fig. S2). The ability of tk/ON/6213/66 (H5N1) to cause lethal infections in mice at high dosage and in embryos at low dosage is consistent with the previously reported lethality in chicken embryos and young turkeys (Lang et al., 1968b; Rouse et al., 1968), whereas tk/WI/68 (H5N9) was not virulent in chicken embryos or mice, which is consistent with earlier reports that this virus was not pathogenic for turkeys and had a low ability to kill chicken embryos infected via the allantoic cavity (Smithies et al., 1969).
Although tk/ON/7732/66 (H5N9) followed the first report of the HPAI H5N1 infection of chicken in Scotland in 1959 [A/chicken/Scotland/1959 (H5N1)], there was no evidence of the introduction of genome segments from this lineage. The genealogies of all genome segments in tk/ON/7732/66 (H5N9) were distinct from those of the Eurasian lineage of viruses, indicating that there was no evidence of genetic exchange with earlier European HPAI viruses that had been reported to infect poultry in the 1920s in North America.
In conclusion, we demonstrated that tk/ON/6213/66 (H5N1) is the genetic precursor to the first confirmed HPAI virus in North America and thus that this virus can be used to analyse and determine the roles of individual genes and mutations in the pathogenicity of tk/ON/7732/66 (H5N9).
Methods
Viruses.
The viruses used in this study are listed in Table 1. Viruses were originally obtained from the D. A. MacLeod Repository, Health Canada (Ottawa) in 1983 and subsequently maintained at the University of Ottawa (E. G. Brown Repository). The viruses sequenced in this study have been deposited in the BioDefense and Emerging Infections Research Resources Repository (http://www.beiresources.org/). Viruses were cultivated in the allantoic cavity of 9-day-old specific-pathogen-free embryonated chicken eggs (Canadian Food Inspection Agency) for two passages using 0.001 ml inoculum in PBS, as described previously (Brown et al., 2001). The passage histories before receipt were unknown. tk/ON/6213/66 (H5N1) was also purified clonally by two cycles of plaque isolation in Madin–Darby canine kidney (MDCK) cells, as described previously (Brown et al., 2001), before amplification by two passages in eggs to generate seed and working stocks. Viruses were titrated by plaque assay in MDCK cells, as described previously (Brown et al., 2001).
Virulence assessment in chicken embryos and mice.
Survival of chicken embryos was assessed following infection of the allantoic cavity with dosages that were diluted in PBS to range from 102 to 106 p.f.u. in 0.1 ml in tenfold increments. Survival was monitored at 2 days p.i. Survival of groups of five 19–21 g female CD-1 mice was monitored for 14 days following intranasal infection with 106 or 107 p.f.u. of defined viruses, as described previously (Ping et al., 2010).
Ethics statement.
All procedures with animals were performed under the supervision of the University of Ottawa Animal Care and Veterinary Services. The animal study protocol was approved by the University of Ottawa Animal Care Committee. All in vivo research was performed in accordance with the guidelines of the Canadian Council on Animal Care (1993). All efforts were made to minimize suffering: mice were euthanized humanely at the experimental end point when infection resulted in >30 % body weight loss accompanied by respiratory distress and euthanized by CO2 narcosis.
Genomic sequencing.
Virus stocks were grown in embryonated eggs, and 140 µl stock was used for RNA isolation using an RNAeasy kit (Qiagen). cDNA was synthesized and amplified by PCR, as described previously (Ghedin et al., 2005). The avian influenza genomes sequenced in this study were deposited in GenBank under the accession numbers shown in Table S1.
Phylogenetic analysis.
A total of 127 complete reference avian influenza A virus genome sequences were included in the analysis (Table S1), representing all of the unique genome sets available from the Influenza Virus Resource (NCBI) isolated from poultry and waterfowl prior to and including 1980. Maximum-likelihood analysis was performed with mega5 (Tamura et al., 2011) using 500 bootstrap replicates. GenBank accession numbers used for phylogenetic reconstruction can be found in the Table S1. The protein sequences of seven H5 proteins were aligned using muscle version 3.7, spanning aa 281–350 (numbering includes the leader sequence). A/mallard/WI/169/1975 (H5N3) was used as the reference, and amino acid substitutions within the alignment are shown in black in Fig. 3.
blast analysis of extent of similarity.
blast (Altschul et al., 1990) was used on the NCBI website (http://blast.ncbi.nlm.nih.gov/) for comparison of all influenza A genome segments with the nucleotide sequence of each genome segment of A/Turkey/Ontario/7732/1966 (H5N9) (GenBank accession numbers are given in Table S1).
Acknowledgements
This project was funded in part through the Influenza Genome Sequencing Project with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract numbers N01-AI-30071 and HHSN272200900007C; CIHR Pandemic Preparedness Team grant TPA-90188 to the CIHR Canadian Influenza Pathogenesis Team (E. G. B.), and CIHR operating grant MOP-74526 (E. G. B.). N. E. F. was funded from an Ontario Graduate Scholarship in Science and Technology. Technical assistance was provided by Shuzhi Wang at the University of Ottawa, Ontario, Canada.
Footnotes
Two supplementary figures and a supplementary table are available with the online version of this paper.
References
- Alexander D. J., Parsons G., Manvell R. J. (1986). Experimental assessment of the pathogenicity of eight avian influenza A viruses of H5 subtype for chickens, turkeys, ducks and quail. Avian Pathol 15, 647–662 10.1080/03079458608436328 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Basler C. F., Aguilar P. V. (2008). Progress in identifying virulence determinants of the 1918 H1N1 and the Southeast Asian H5N1 influenza A viruses. Antiviral Res 79, 166–178 10.1016/j.antiviral.2008.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. G., Liu H., Kit L. C., Baird S., Nesrallah M. (2001). Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc Natl Acad Sci U S A 98, 6883–6888 10.1073/pnas.111165798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. G., Sattar S. A., Tetro J. A., Liu J. (2008). Trends in influenza A virus genetics: can we predict the natural evolution of a H5N1 Z? Curr Top Virol 7, 99–113 [Google Scholar]
- Canadian Council on Animal Care (1993). Guide to Care and Use of Experimental Animals, 2nd edn, vol. 1 Ottowa, ON: Canadian Council on Animal Care [Google Scholar]
- Forbes N. E., Ping J., Dankar S. K., Jia J. J., Selman M., Keleta L., Zhou Y., Brown E. G. (2012). Multifunctional adaptive NS1 mutations are selected upon human influenza virus evolution in the mouse. PLoS ONE 7, e31839 10.1371/journal.pone.0031839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E., Sengamalay N. A., Shumway M., Zaborsky J., Feldblyum T., Subbu V., Spiro D. J., Sitz J., Koo H. & other authors (2005). Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437, 1162–1166 10.1038/nature04239 [DOI] [PubMed] [Google Scholar]
- Guan Y., Peiris J. S., Poon L. L., Dyrting K. C., Ellis T. M., Sims L., Webster R. G., Shortridge K. F. (2003). Reassortants of H5N1 influenza viruses recently isolated from aquatic poultry in Hong Kong SAR. Avian Dis 47 (Suppl.), 911–913 10.1637/0005-2086-47.s3.911 [DOI] [PubMed] [Google Scholar]
- Hwang J., Lief F. S., Miller C. W., Mallinson E. T. (1970). An epornitic of type A influenza virus infection in ducks. J Am Vet Med Assoc 157, 2106–2108 [PubMed] [Google Scholar]
- Jadhao S. J., Nguyen D. C., Uyeki T. M., Shaw M., Maines T., Rowe T., Smith C., Huynh L. P., Nghiem H. K. & other authors (2009). Genetic analysis of avian influenza A viruses isolated from domestic waterfowl in live-bird markets of Hanoi, Vietnam, preceding fatal H5N1 human infections in 2004. Arch Virol 154, 1249–1261 10.1007/s00705-009-0429-2 [DOI] [PubMed] [Google Scholar]
- Keleta L., Ibricevic A., Bovin N. V., Brody S. L., Brown E. G. (2008). Experimental evolution of human influenza virus H3 hemagglutinin in the mouse lung identifies adaptive regions in HA1 and HA2. J Virol 82, 11599–11608 10.1128/JVI.01393-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G., Ferguson A. E. (1981). The extent and control of avian influenza in Canada. Can Vet J 22, 377–381 [PMC free article] [PubMed] [Google Scholar]
- Lang G., Narayan O., Rouse B. T., Ferguson A. E., Connell M. C. (1968a). A new influenza A virus infection in turkeys II. A highly pathogenic variant, A/turkey/Ontario 772/66. Can Vet J 9, 151–160 [PMC free article] [PubMed] [Google Scholar]
- Lang G., Rouse B. T., Narayan O., Ferguson A. E., Connell M. C. (1968b). A new influenza virus infection in turkeys. I. Isolation and characterization of virus 6213. Can Vet J 9, 22–29 [PMC free article] [PubMed] [Google Scholar]
- Li K. S., Guan Y., Wang J., Smith G. J., Xu K. M., Duan L., Rahardjo A. P., Puthavathana P., Buranathai C. & other authors (2004). Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430, 209–213 10.1038/nature02746 [DOI] [PubMed] [Google Scholar]
- Lupiani B., Reddy S. M. (2009). The history of avian influenza. Comp Immunol Microbiol Infect Dis 32, 311–323 10.1016/j.cimid.2008.01.004 [DOI] [PubMed] [Google Scholar]
- Mukhtar M. M., Rasool S. T., Song D., Zhu C., Hao Q., Zhu Y., Wu J. (2007). Origin of highly pathogenic H5N1 avian influenza virus in China and genetic characterization of donor and recipient viruses. J Gen Virol 88, 3094–3099 10.1099/vir.0.83129-0 [DOI] [PubMed] [Google Scholar]
- Narayan O., Lang G., Rouse B. T. (1969a). A new influenza A virus infection in turkeys. IV. Experimental susceptibility of domestic birds to virus strain turkey/Ontario 7732/1966. Arch Gesamte Virusforsch 26, 149–165 10.1007/BF01241184 [DOI] [PubMed] [Google Scholar]
- Narayan O., Lang G., Rouse B. T. (1969b). A new influenza A virus infection in turkeys. V. Pathology of the experimental disease by strain turkey/Ontario 7732/66. Arch Gesamte Virusforsch 26, 166–182 10.1007/BF01241185 [DOI] [PubMed] [Google Scholar]
- Philpott M., Hioe C., Sheerar M., Hinshaw V. S. (1990). Hemagglutinin mutations related to attenuation and altered cell tropism of a virulent avian influenza A virus. J Virol 64, 2941–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping J., Dankar S. K., Forbes N. E., Keleta L., Zhou Y., Tyler S., Brown E. G. (2010). PB2 and hemagglutinin mutations are major determinants of host range and virulence in mouse-adapted influenza A virus. J Virol 84, 10606–10618 10.1128/JVI.01187-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping J., Keleta L., Forbes N. E., Dankar S., Stecho W., Tyler S., Zhou Y., Babiuk L., Weingartl H. & other authors (2011). Genomic and protein structural maps of adaptive evolution of human influenza A virus to increased virulence in the mouse. PLoS ONE 6, e21740 10.1371/journal.pone.0021740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse B. T., Lang G., Narayan O. (1968). A new influenza A virus infection in turkeys. 3. Pathology of the experimental disease by virus strain turkey/Ontario/6213/66. J Comp Pathol 78, 525–533 10.1016/0021-9975(68)90053-4 [DOI] [PubMed] [Google Scholar]
- Sandhu T. H., Hinshaw V. (2003). Influenza A virus infection of domestic ducks. Avian Dis 47, 93–99 [Google Scholar]
- Shaw M., Cooper L., Xu X., Thompson W., Krauss S., Guan Y., Zhou N., Klimov A., Cox N. & other authors (2002). Molecular changes associated with the transmission of avian influenza a H5N1 and H9N2 viruses to humans. J Med Virol 66, 107–114 10.1002/jmv.2118 [DOI] [PubMed] [Google Scholar]
- Smithies L. K., Emerson F. G., Robertson S. M., Ruedy D. D. (1969). Two different type A influenza virus infections in turkeys in Wisconsin. II. 1968 outbreak. Avian Dis 13, 606–610 10.2307/1588535 [DOI] [PubMed] [Google Scholar]
- Swayne D. E. (2009). High pathogenicity avian influenza in the Americas. In Avian Influenza, pp. 191–216 Edited by Swayne D. E. Oxford, UK: Blackwell; 10.1002/9780813818634.ch8 [DOI] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R. V., Bannister G. L. (1953). A filterable agent in ducks. Can J Comp Med Vet Sci 17, 248–250 [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. (1992). Evolution and ecology of influenza A viruses. Microbiol Rev 56, 152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.-M., Shortridge K. F., Garcia M., Guan Y., Wan X.-F. (2008). Genotypic diversity of H5N1 highly pathogenic avian influenza viruses. J Gen Virol 89, 2182–2193 10.1099/vir.0.2008/001875-0 [DOI] [PubMed] [Google Scholar]