Abstract
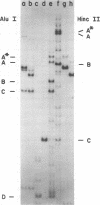
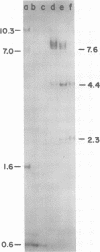
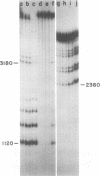
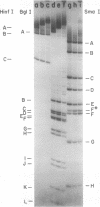
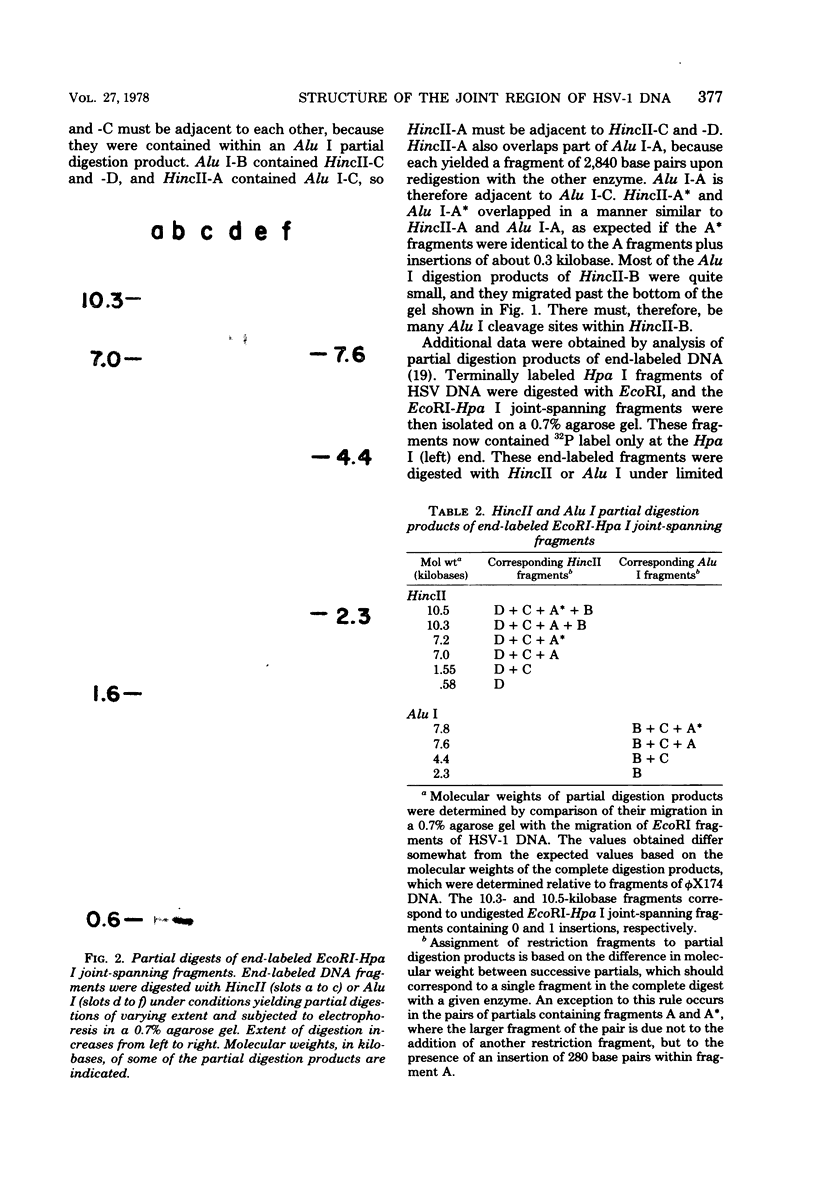
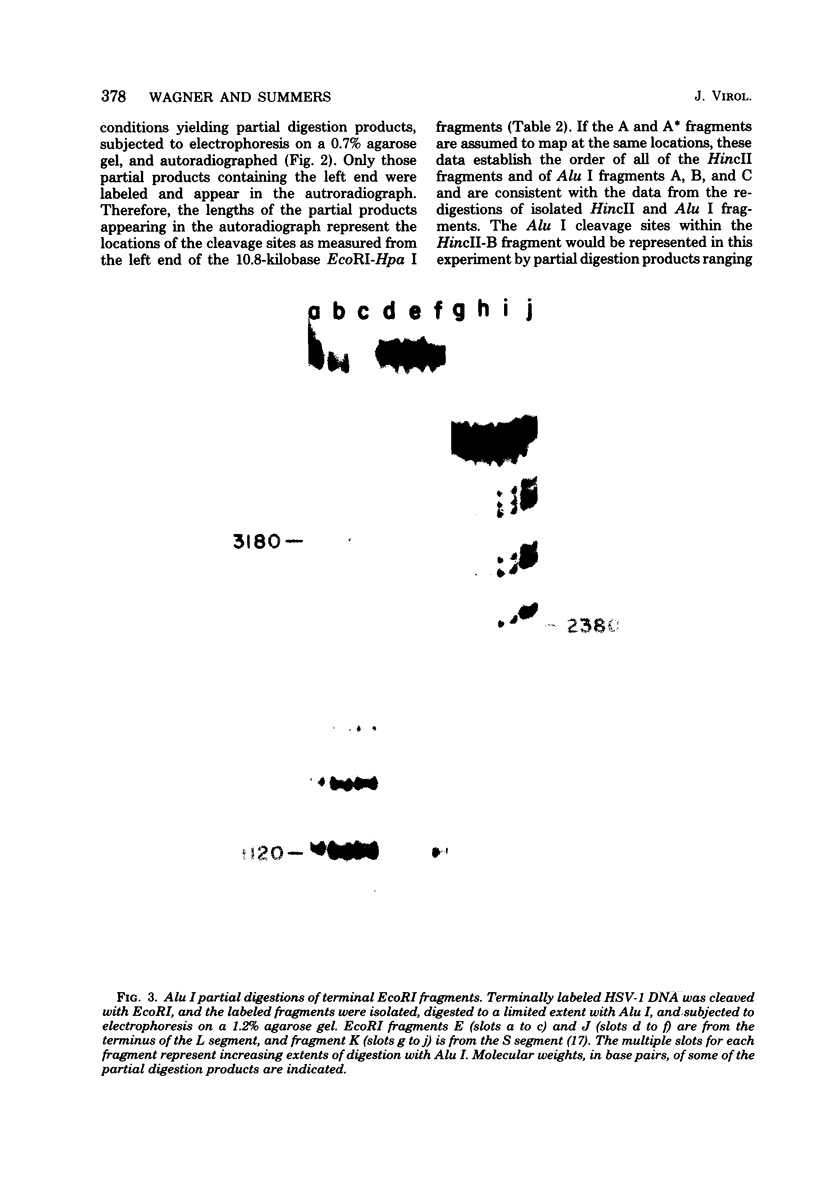
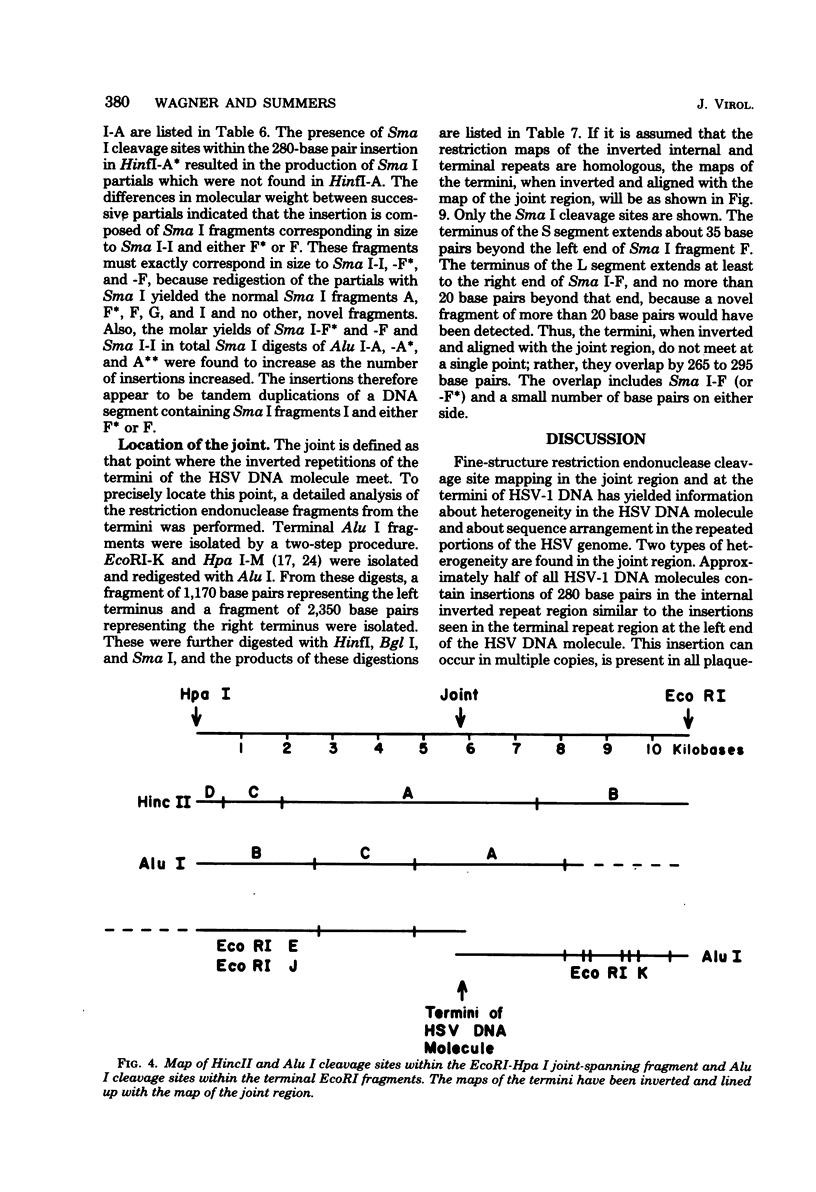
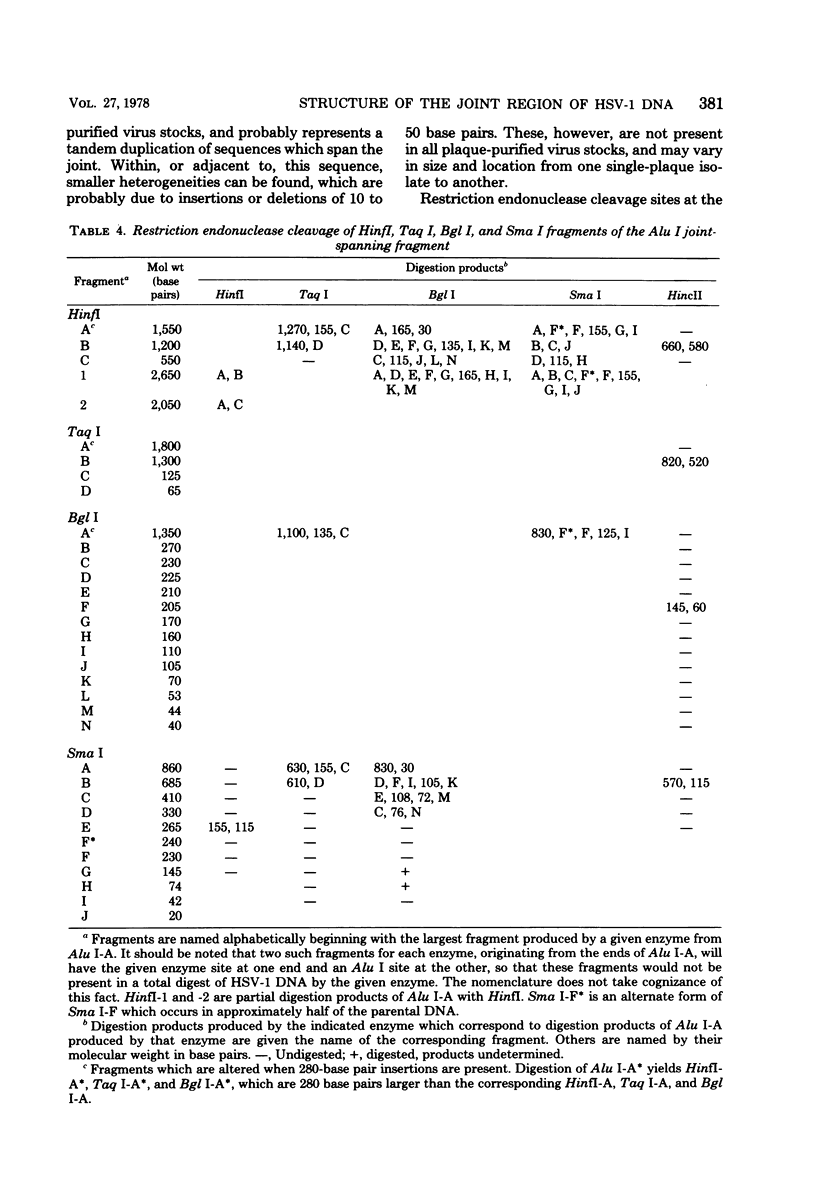
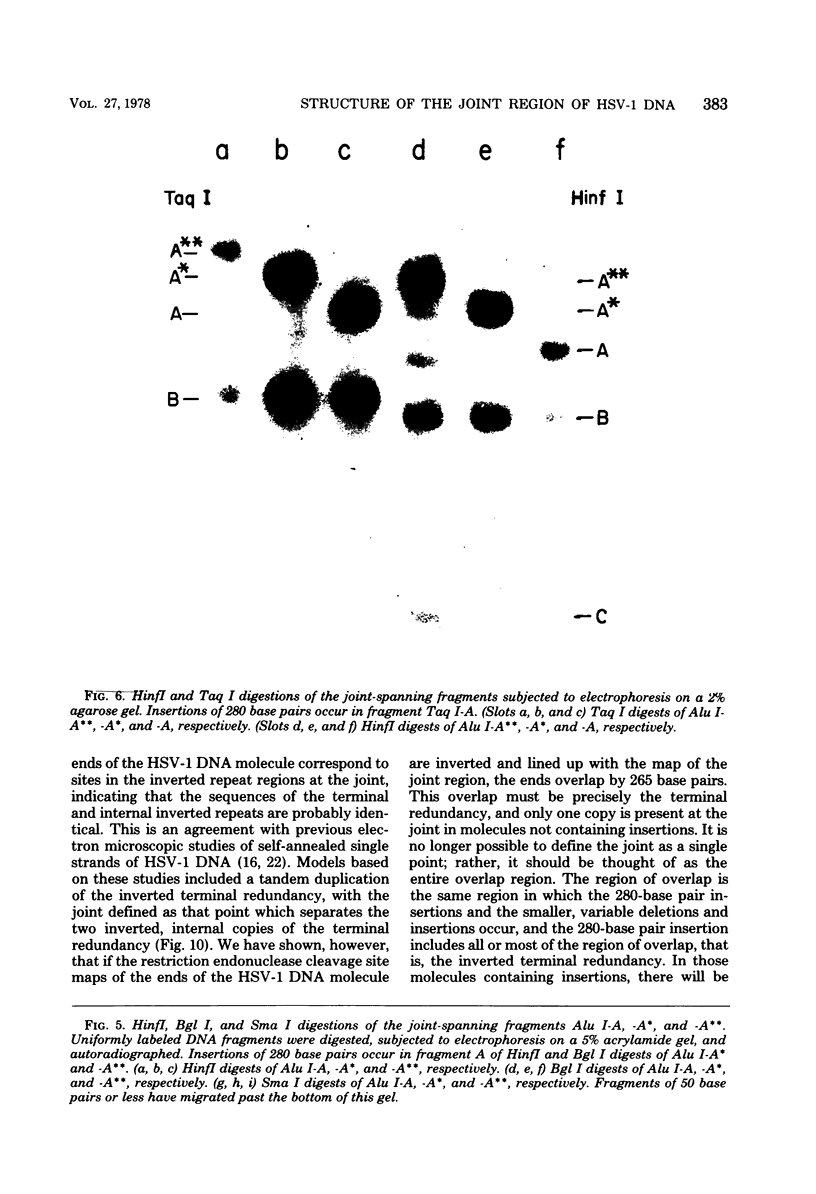
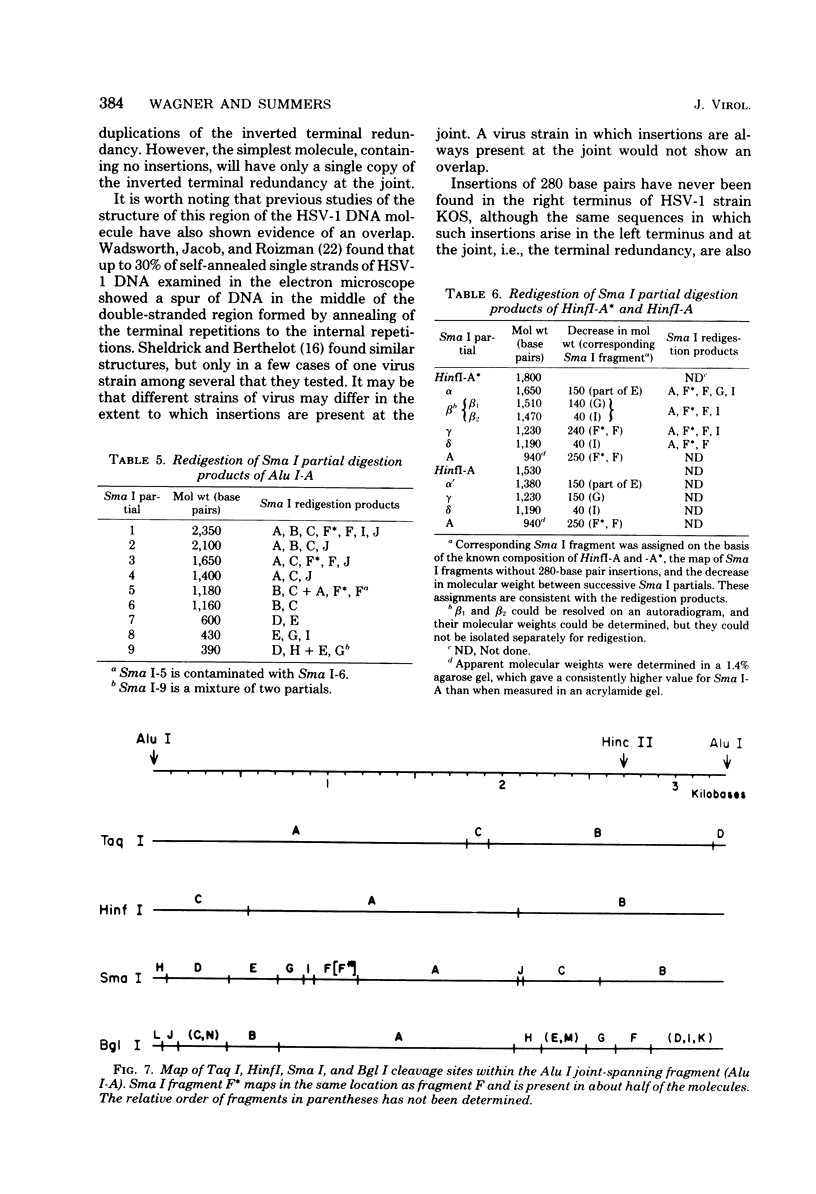
Analysis of restriction endonuclease cleavage sites within the inverted, repeated sequences in the joint region of the DNA of herpes simplex virus type 1 strain KOS revealed the presence of two types of sequence heterogeneity. The first was an insertion of 280 base pairs or multiples of 280 base pairs which was found in approximately half of all DNA molecules from every plaque-purified stock of virus. These insertions seemed to be tandem duplications of sequences which were present at the joint and correspond closely to the inverted terminal redunancy. The second type of heterogeneity was due to variable insertions and deletions which were present in some, but not all, plaque-purified virus stocks. Comparison of restriction fragments from the joint region with fragments from the termini indicated that in the simplest observed molecules of herpes simplex virus type 1 DNA, only one copy of the inverted terminal redundancy was present at the joint. A map of restriction endonuclease cleavage sites in the joint region is presented.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker Y., Dym H., Sarov I. Herpes simplex virus DNA. Virology. 1968 Oct;36(2):184–192. doi: 10.1016/0042-6822(68)90135-9. [DOI] [PubMed] [Google Scholar]
- Clements J. B., Cortini R., Wilkie N. M. Analysis of herpesvirus DNA substructure by means of restriction endonucleases. J Gen Virol. 1976 Feb;30(2):243–256. doi: 10.1099/0022-1317-30-2-243. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W. Terminal repetitions in herpes simplex virus type 1 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):679–681. doi: 10.1101/sqb.1974.039.01.081. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W., Burke S., Kudler L. A nearby inverted repeat of the terminal sequence of herpes simplex virus DNA. Biochem Biophys Res Commun. 1976 Jan 26;68(2):609–615. doi: 10.1016/0006-291x(76)91189-x. [DOI] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Arthrobacter luteus. J Mol Biol. 1976 Mar 25;102(1):157–165. doi: 10.1016/0022-2836(76)90079-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. P., Summers W. C. Structure and function of herpesvirus genomes. I. comparison of five HSV-1 and two HSV-2 strains by cleavage their DNA with eco R I restriction endonuclease. J Virol. 1975 Apr;15(4):726–732. doi: 10.1128/jvi.15.4.726-732.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J Virol. 1976 Feb;17(2):503–512. doi: 10.1128/jvi.17.2.503-512.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Skare J., Summers W. C. Analysis of DNA of defective herpes simplex virus type 1 by restriction endonuclease cleavage and nucleic acid hybridization. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):683–686. doi: 10.1101/sqb.1974.039.01.082. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]