Abstract
Recent studies indicate that human papillomaviruses (HPVs) from the genera Betapapillomavirus and Gammapapillomavirus are abundant in the human oral cavity. We report the cloning and characterization of a 7304 bp HPV120 genome from the oral cavity that is related most closely to HPV23 (L1 ORF, 83.7 % similarity), clustering it in the genus Betapapillomavirus (β-PV). HPV120 contains five early and two late genes, but no E5 ORF. HPV120 was detected from heterogeneous human biological niches, including the oral cavity, eyebrow hairs, anal canal and penile, vulvar and perianal warts. Characterization of the clinical spectrum of HPV120 infections indicates a broader spectrum of epithelial tropism than appreciated previously for HPV types from the genus β-PV.
Papillomaviruses are a family of heterogeneous, circular dsDNA viruses approximately 8 kb in size that can cause a variety of epithelial hyperplasias (reviewed by Bernard et al., 2010). Papillomaviruses isolated from humans (HPVs) can lead to benign or premalignant lesions and a subset are associated with invasive carcinomas (Li et al., 2011). Currently, over 150 HPV types have been characterized fully and the majority are classified into three major genera: Alphapapillomavirus (α-PV), typically isolated from genital lesions, and Betapapillomavirus (β-PV) and Gammapapillomavirus (γ-PV), predominantly found in skin lesions (Bernard et al., 2010).
Increasing evidence supports an association between a subset of oral and/or oropharyngeal cancers with persistent infection of certain high-risk human α-PV types (Marur et al., 2010). However, a recent report indicates that a wide spectrum of HPV types, predominantly from the genera β-PV and γ-PV, can be detected in the oral cavity (Bottalico et al., 2011). In addition, β-PVs have also been detected in head, neck and oesophageal tissues (de Villiers & Gunst, 2009). These observations could have significant implications for the epidemiological association of human α-PVs and β-PVs with oral and skin neoplasias, particularly if exposure is measured by serology.
A distinct papillomavirus type is established when the nucleotide sequence of the L1 gene of a cloned virus differs from that of any other characterized types by ≥10 % (Bernard et al., 2010); nucleotide differences of approximately 1.0–10.0 % and 0.5–1.0 % of the complete genomes have been used to define variant lineages and sublineages, respectively (Chen et al., 2011). This report describes the isolation and characterization of a novel β-PV, HPV120, which is related most closely to HPV23.
HPV120 was isolated from an oral-rinse sample collected from the Cancer, Longevity, Ancestry and Lifestyle (CL) study that was obtained as a source of genomic DNA for a case–control study of prostate cancer, as described previously (Agalliu et al., 2009). Additional oral-rinse samples were collected as part of other ongoing studies that obtained oral-rinse samples as a source of genomic DNA. All participants of this analysis gave written informed consent and the studies had Institutional Review Board approval.
HPV120 partial sequences were initially detected by PCR amplification with FAP primers (Bottalico et al., 2011). The respective 480 bp FAP fragment within the L1 ORF was sequenced directly. A blast search against GenBank indicated that the amplified fragment was identical to a partially FAP-sequenced isolate, FA16.1 (GenBank accession no. AF217658), but was <90 % similar to any other characterized PVs. Therefore, type-specific primer sets based on the FAP sequences were used to amplify and clone the complete genome in two overlapping fragments (1759 and 5922 bp) into the TOPO TA pCR2.1 vector (Invitrogen). Primers are available from the corresponding author upon request. DNA clones were sent to the Human Papillomavirus Reference Laboratory in Heidelberg, Germany, for official designation. The complete genome sequence of HPV120 from sample CL3857 was submitted to GenBank with accession no. GQ845442.
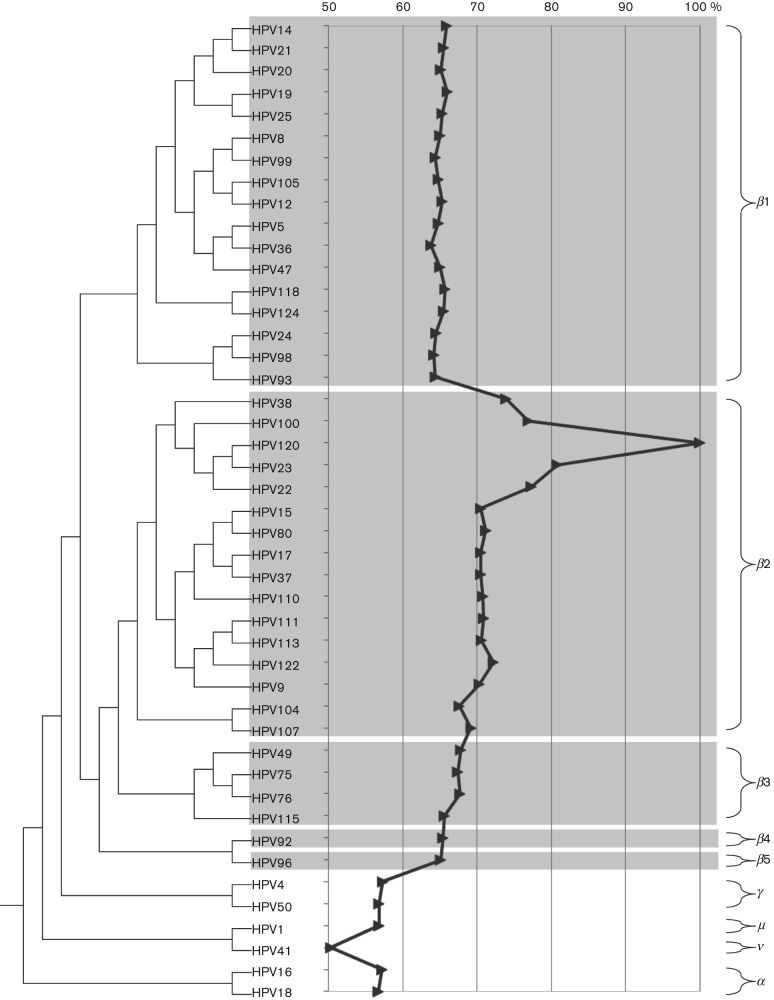
The complete genome of HPV120, 7304 bp in length (G+C content of 39.8 mol%), contains five early genes (E6, E7, E1, E2 and E4) and two late genes (L2 and L1), but no E5 ORF (Fig. S1a, available in JGV Online). A global alignment of the L1 nucleotide sequence revealed that HPV120 is related most closely to HPV23, with a pairwise identity of 83.7 %. A representative maximum-likelihood tree was inferred from the concatenated nucleotide sequences of six ORFs (E6, E7, E1, E2, L2 and L1) using RAxML MPI v7.2.8 (Stamatakis, 2006). The amino acid sequences of each ORF from representative HPVs were aligned using muscle v3.7 within the SeaView v4.1 program (Edgar, 2004); the nucleotide sequences of each codon region were aligned using the corresponding amino acid sequence alignments. The GTR+Γ model was set for among-site rate variation and allowed substitution rates of aligned sequences to be different. The prototype reference sequences of representative HPVs were accessed from GenBank. The phylogenetic tree (Fig. 1) clustered HPV120 with HPV22/23/38/100. Taken together, HPV120 fulfils the criteria of a novel HPV type within β-PV species group 2 (β2).
Fig. 1.
Phylogenetic position of HPV120. A maximum-likelihood tree was inferred from the concatenated nucleotide sequences of six ORFs (E6, E7, E1, E2, L2 and L2) from representative HPV genomes of all β-PVs and selected types from the other genera (indicated on the right of the figure) using RAxML MPI v7.2.8 (Stamatakis, 2006). Complete L1 nucleotide sequence identities of HPV120 compared with other types are shown in the centre panel. Areas in grey highlight HPV types within the β-PV species groups.
The first position of the complete genome of HPV120 was set as the first ATG of the E6 ORF. The putative E6 ORF contains two zinc-binding domains [CxxC(x)29CxxC], separated by 36 aa (reviewed by Lehoux et al., 2009). The putative E7 ORF contains one conserved zinc-binding domain, CxxC(x)29CxxC, and a motif (LxCxE) for binding to the pRB protein (reviewed by Lehoux et al., 2009). The ATP-binding site of the ATP-dependent helicase (GPPDTGKS) is conserved in the carboxy-terminal region of E1 (reviewed by McBride, 2008). The E4 ORF contains a start codon and overlaps the E2 ORF. A polyadenylation consensus sequence (AATAAA) for processing of early viral mRNA transcripts is present at the beginning of the L2 gene (reviewed by Zheng & Baker, 2006). The major (L1) and minor (L2) capsid proteins show a nuclear-localization signal at their 3′ end. The upstream regulatory region (URR) located between the stop codon of L1 and the first start codon of the E6 ORF consists of 390 bp and contains many cis-acting regulatory sequences and DNA–protein-binding motifs involved in viral transcription and replication (Fig. S1b).
To assess the genomic diversity and clinical importance of HPV120, this study also utilized specimens collected in Slovenia. Samples from the following sites/lesions – anal canal, eyebrow hair, penile, anal and perianal warts, vulvar and vaginal lesions, and cervical cancer – were collected prospectively during previous or ongoing studies of HPV distribution in Slovenia (Jancar et al., 2009; Kocjan et al., 2005, 2010; Milosevic et al., 2010; Potocnik et al., 2007). These studies were approved by the Ethics Committee of the Ministry of Health (Slovenia) and written informed consent was obtained from each patient. Samples of laryngeal papillomas, inverted sinonasal papillomas, oral papillomas and oral squamous cell carcinomas were retrieved from the tissue collection of paraffin-embedded samples of the Institute of Pathology, Faculty of Medicine, University of Ljubljana. Approval from the Institutional Review Board of the Institute of Pathology, Faculty of Medicine, University of Ljubljana, was obtained prior to starting work on samples included in this study. Sample collection, DNA extraction and PCR amplifications were performed as described previously (Bottalico et al., 2011; Jancar et al., 2009; Kocjan et al., 2005, 2010; Milosevic et al., 2010; Potocnik et al., 2007). A type-specific real-time PCR assay was developed to evaluate the presence of HPV120 in various tissues and lesions. Primers specific for HPV120 were selected within the viral L1 gene using ProbeFinder software v2.45 (http://qpcr.probefinder.com/roche3.html), resulting in a PCR product of approximately 90 bp. Primers RT-HPV120L1-F (5′-CATGTTGAGGAGTATCAGTTGTCTTT-3′, nt 6507–6532) and RT-HPV120L1-R (5′-AGAGTTCATTGCATTGATTTGTG-3′, nt 6599–6577) were used in combination with the QuantiTect SYBR Green PCR kit (Qiagen) on a LightCycler 2.0 Instrument (Roche Diagnostics), as recommended by the manufacturer. The sensitivity of the assay, estimated by testing serial HPV120 (SIBX-3a) plasmid dilutions, was established at approximately five viral copies per reaction. A semi-nested HPV120 type-specific PCR assay was developed to amplify the complete L1 gene (1521 bp) for 15 samples. The PCR products were purified and submitted for sequencing, as described previously (Kocjan et al., 2005).
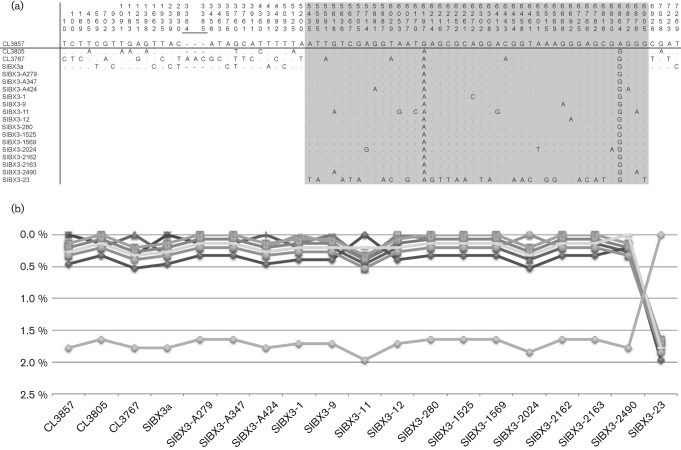
The genomic diversity of HPV120, a novel human β-PV, was determined from the sequences of four complete HPV120 genomes and the L1 ORFs of an additional 15 samples, described above. Cloned HPV120 genome and PCR fragments were sequenced on both strands. We calculated pairwise differences comparing HPV120 with all other types (Fig. 1) or HPV120 prototype with all other variants (Fig. 2) based on the aligned nucleotide sequences using the p-distance method in mega5 (Tamura et al., 2011). The same program was used to determine the single-nucleotide polymorphisms (SNPs) from the above alignments. Positions of SNPs are based on the prototype reference sequence (CL3857; GenBank accession no. GQ845442). In total, 72 nucleotide changes among 7304 bp (a 3 bp indel within isolate CL3767 at nt 3384 was counted as one event) were observed (Fig. 2a). The complete genomes displayed <0.5 % pairwise difference in nucleotide sequences and, therefore, these four variants cluster within a single lineage (Burk et al., 2011) (Fig. 2a, b). This level of diversity is also reflected through comparison of predicted amino acids in these genomes that differ by a maximum of 0.3 %. Interestingly, isolate SIBX3-23 was distinctly separated from other variants, with 25 nt changes within the L1 ORF (pairwise differences of 1.6–2.0 %), suggesting that it represents a second variant lineage (Fig. 2a, b). However, as variant lineages are based on comparison of complete PV genomes (Bernard et al., 2010; Chen et al., 2011), it is premature to assign variant lineages to HPV120.
Fig. 2.
Genomic diversity of HPV120 variants. (a) SNPs of HPV120 variant isolates. Positions of SNPs are based on the prototype reference sequence CL3857 (GenBank accession no. GQ845442). The L1 region is highlighted in grey. (b) Percentage nucleotide sequence differences within the L1 ORF nucleotide region were calculated for each isolate compared with all other isolates. Values for each comparison of a given isolate are connected by lines and the comparison to self is indicated by the 0 % difference point. Each isolate has a unique symbol and line to facilitate visual comparisons. Except for isolate SIBX3-23, all other isolates are related closely, as detailed in (a).
The prevalence of HPV120 was 0.9 % (four of 446) in a set of oral-rinse samples without information on pathological conditions in the oral cavity (Table 1). However, in more detailed studies, no HPV120 DNA was detected in oral squamous cell carcinomas and/or oral papillomas (Table 1). Additional studies of tissue specimens collected from a variety of studies indicated that HPV120 was identified in 14 of 63 (22.2 %) plucked eyebrow hair samples from immunocompetent patients. Over 3 % of anal and perianal samples, some from warts, were HPV120-positive. Real-time PCR analyses with an HPV120-specific assay indicated equivalent low viral load within these tissues. Several samples from penile warts, laryngeal papillomas and vulvar/vaginal lesions also contained HPV120 DNA. Thus, HPV120 was detected in various human bio-niches; however, the pathological consequence of these infections requires further investigation. It is unclear whether the development of warts in some subjects was the result of infection by HPV120 or another HPV type, as approximately two-thirds of the HPV120 DNA-positive samples, particularly those from wart tissues, contained multiple HPV types (Table S1).
Table 1. Prevalence of HPV120 infection from different anatomical sites and lesions.
| Type of specimen | Total no. of samples* | No. of HPV120-positive samples | Prevalence of HPV120 (%) |
| Oral cavity (rinse specimens) | 446 | 4 | 0.9 |
| Oral papillomas | 65 | 0 | 0 |
| Oral squamous cell carcinomas | 65 | 0 | 0 |
| Eyebrow hairs (immunocompetent patients) | 63 | 14 | 22.2 |
| Swab of the anal canal | 210 | 7 | 3.3 |
| Anal and perianal warts | 144 | 5 | 3.5 |
| Penile warts | 56 | 2 | 3.6 |
| Laryngeal papillomas | 58 | 1 | 1.7 |
| Inverted sinonasal papillomas | 60 | 0 | 0 |
| Vulvar/vaginal lesions (warts, VIN1-3, VaIN1-3) | 80 | 1 | 1.3 |
| Cervical squamous cell carcinomas | 61 | 0 | 0 |
| Total | 1308 | 34 | 2.6 |
Each sample is from a separate subject, thus no. of samples = no. of subjects.
The characterization and classification of HPV120 add it to the repertoire of human β-PVs. Detailed analysis of multiple anatomical sites and lesions containing HPV120 DNA suggests that this HPV has expanded into multiple human epithelial niches (Grice et al., 2009). The role of symbiotic HPV infections, their relationship with the host (e.g. commensal) and the emergence of their pathogenic potential remains to be better understood.
Acknowledgements
We thank Anja Kovanda for her help in the initial HPV120 (SIBX-3a) cloning experiments and for reviewing the annotation of HPV120 (SIBX-3a) ORFs. This work was supported in part by the National Cancer Institute (CA78527) (R. D. B.), the Einstein–Montefiore Center for AIDS funded by the National Institutes of Health (AI-51519) and the Einstein Cancer Research Center funded by the National Cancer Institute (P30CA013330).
Footnotes
A supplementary table and figure are available with the online version of this paper.
References
- Agalliu I., Gern R., Leanza S., Burk R. D. (2009). Associations of high-grade prostate cancer with BRCA1 and BRCA2 founder mutations. Clin Cancer Res 15, 1112–1120 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H. U., Burk R. D., Chen Z., van Doorslaer K., Hausen H., de Villiers E. M. (2010). Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401, 70–79 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottalico D., Chen Z., Dunne A., Ostoloza J., McKinney S., Sun C., Schlecht N. F., Fatahzadeh M., Herrero R. & other authors (2011). The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J Infect Dis 204, 787–792 10.1093/infdis/jir383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk R. D., Chen Z., Harari A., Smith B. C., Kocjan B. J., Maver P. J., Poljak M. (2011). Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages. Acta Dermatovenerol Alp Panonica Adriat 20, 113–123 [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Schiffman M., Herrero R., Desalle R., Anastos K., Segondy M., Sahasrabuddhe V. V., Gravitt P. E., Hsing A. W., Burk R. D. (2011). Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS ONE 6, e20183 10.1371/journal.pone.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers E. M., Gunst K. (2009). Characterization of seven novel human papillomavirus types isolated from cutaneous tissue, but also present in mucosal lesions. J Gen Virol 90, 1999–2004 10.1099/vir.0.011478-0 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A., Kong H. H., Conlan S., Deming C. B., Davis J., Young A. C., Bouffard G. G., Blakesley R. W., Murray P. R. & other authors (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancar N., Kocjan B. J., Poljak M., Lunar M. M., Bokal E. V. (2009). Distribution of human papillomavirus genotypes in women with cervical cancer in Slovenia. Eur J Obstet Gynecol Reprod Biol 145, 184–188 10.1016/j.ejogrb.2009.04.030 [DOI] [PubMed] [Google Scholar]
- Kocjan B. J., Poljak M., Seme K., Potocnik M., Fujs K., Babic D. Z. (2005). Distribution of human papillomavirus genotypes in plucked eyebrow hairs from Slovenian males with genital warts. Infect Genet Evol 5, 255–259 10.1016/j.meegid.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Kocjan B. J., Poljak M., Seme K. (2010). Universal ProbeLibrary based real-time PCR assay for detection and confirmation of human papillomavirus genotype 52 infections. J Virol Methods 163, 492–494 10.1016/j.jviromet.2009.10.024 [DOI] [PubMed] [Google Scholar]
- Lehoux M., D’Abramo C. M., Archambault J. (2009). Molecular mechanisms of human papillomavirus-induced carcinogenesis. Public Health Genomics 12, 268–280 10.1159/000214918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Franceschi S., Howell-Jones R., Snijders P. J., Clifford G. M. (2011). Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int J Cancer 128, 927–935 10.1002/ijc.25396 [DOI] [PubMed] [Google Scholar]
- Marur S., D’Souza G., Westra W. H., Forastiere A. A. (2010). HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol 11, 781–789 10.1016/S1470-2045(10)70017-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A. A. (2008). Replication and partitioning of papillomavirus genomes. Adv Virus Res 72, 155–205 10.1016/S0065-3527(08)00404-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic M., Poljak M., Mlakar B. (2010). Anal HPV infection in Slovenian men who have sex with men. Cent Eur J Med 5, 698–703 10.2478/s11536-010-0019-4 [DOI] [Google Scholar]
- Potocnik M., Kocjan B. J., Seme K., Poljak M. (2007). Distribution of human papillomavirus (HPV) genotypes in genital warts from males in Slovenia. Acta Dermatovenerol Alp Panonica Adriat 16, 91–96, 98 [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z. M., Baker C. C. (2006). Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 11, 2286–2302 10.2741/1971 [DOI] [PMC free article] [PubMed] [Google Scholar]