Abstract
Helicobacter pullorum is an enterohepatic Helicobacter species (EHS) that was recently reported as a naturally acquired infection in mice. Faecal samples from 18 out of 20 Brown Norway (BN) rats, housed in the same barrier as the H. pullorum-infected mice, were positive for H. pullorum using species-specific PCR. In addition, we determined whether H. pullorum was able to persistently colonize the gastrointestinal tract and/or biliary tree and elicit tissue inflammation as well as a serum IgG response in BN rats. Six (four male, two female) 6-week-old, H. pullorum-negative BN rats were orally dosed with 4×108 c.f.u. of H. pullorum every other day for a total of three doses. At 2 weeks post-infection, all rats were H. pullorum-positive by faecal PCR. Five out of the six BN rats remained H. pullorum-positive for the entire 30 week study. PCR analysis of tissue collected at necropsy confirmed that the colon and caecum were the primary sites of H. pullorum colonization. Rats that were persistently colonized by H. pullorum had a sustained H. pullorum-specific IgG response measured by ELISA. Intestinal or hepatic pathology associated with H. pullorum infection was not noted. To our knowledge, this is the first report documenting that rats can be persistently colonized with an EHS that also infects humans.
Introduction
Helicobacter species are a group of taxonomically related Gram-negative, microaerophilic bacteria, some of which are pathogenic and known to colonize the gastrointestinal and biliary tracts of many animal species. These pathogens are generally separated into two groups, gastric and enterohepatic, based on their preferred site of colonization. During the last decade, enterohepatic Helicobacter species (EHS) have gained recognition in the field of emerging infectious pathogens (Fox, 2002). Infection by this group of micro-organisms is generally characterized by colonization of the distal gastrointestinal tract and, in select cases, the biliary tree. As reported for the gastric pathogen H. pylori, gastrointestinal colonization by EHS can be associated with chronic inflammation and neoplasia (Fox et al., 1996; García et al., 2008; Ward et al., 1994).
Helicobacter pullorum, an EHS, was first isolated by Stanley et al. (1994) from the faeces of diarrhoeic humans and the intestinal contents and livers of chickens. The organism is suspected to cause vibrionic hepatitis in chickens. Infection with this organism is most often associated with farm-raised birds, including chickens, turkeys and guinea fowl (Nebbia et al., 2007; Stanley et al., 1994). In one report, H. pullorum was isolated from human faeces 3 months following the patients’ initial presentation with diarrhoea (Steinbrueckner et al., 1997). In another case, H. pullorum was isolated from the faeces of a male with diarrhoea and elevated liver enzymes (Burnens et al., 1994). H. pullorum has also been identified by PCR in humans with inflammatory bowel disease, hepatitis, cholecystitis and hepatocelluar carcinoma (Castéra et al., 2006; Fox et al., 1998; Rocha et al., 2005; Veijola et al., 2007). A recent report identified an association between EHS and Crohn’s disease, with H. pullorum being one of the most prevalent EHS identified (Laharie et al., 2009).
In July 2009, routine surveillance testing detected a Helicobacter species in C57BL/6NTac mice and BN/MolTac Brown Norway (BN) rats at Taconic Farms, Germantown, New York. This Helicobacter species was later confirmed to be H. pullorum (Boutin et al., 2010). To our knowledge, prior to this event, H. pullorum had not previously been reported to infect rodents either naturally or experimentally. The purpose of this report is to describe H. pullorum infection in naturally and experimentally infected BN rats and demonstrate that this rat strain can be persistently infected with an EHS know to infect humans.
Methods
Natural infection of Brown Norway rats.
Faecal DNA samples from 20 BN rats were shipped to our laboratory from Taconic Farms after routine testing discovered Helicobacter in mice where the BN rats were co-housed in the same barrier. Samples were screened for Helicobacter species by PCR using 16S rRNA gene genus-specific primers and H. pullorum cytolethal distending toxin B (cdtB) gene species-specific primers (Boutin et al., 2010; Fox et al., 1998; Rocha et al., 2005). Samples confirmed as H. pullorum were subjected to restriction fragment length polymorphism (RFLP) analysis following amplification using Helicobacter genus-specific primers.
Experimental infection of Brown Norway rats.
All rats were maintained in an AAALAC accredited facility and housed singly in filter topped polycarbonate cages. Rats were fed a standard pelleted rodent diet and provided water ad libitum. BN rats were free from the following murine agents certified by the vendor: Kilham’s rat virus, rat coronavirus, rat minute virus, rat parvovirus, sialodacryoadenitis virus, Toolan’s H-1 parvovirus, encephalomyelitis virus, pneumonia virus of mice, Sendai virus, hantaan virus, mouse adenovirus (MAV-1, MAV-2), respiratory enteric virus III, β-haemolytic streptococcus, Bordetella bronchiseptica, Clostridium piliforme, Corynebacterium kutscheri, Klebsiella oxytoca, Klebsiella pneumoniae, Mycoplasma, Pasteurella, Salmonella, Streptobacillus moniliformis, Streptococcus pneumoniae, CAR bacillus, Helicobacter, Pneumocystis carinii, endoparasites and ectoparasites. All experimental procedures were reviewed and approved by the MIT Institutional Animal Care and Use Committee.
Six Helicobacter-free BN weanling rats (two female, four male) were obtained from Taconic Farms. After acclimation, each rat was inoculated by oro-gastric gavage with 400 µl of OD600 (4×108 c.f.u.) of an H. pullorum isolate (MIT 09-6635) that was cultured from the caeca of the H. pullorum-infected mice identified in the initial outbreak (Boutin et al., 2010). Each rat was gavaged once every other day for a total of three doses. Animals were housed singly and tested every 2 weeks [except for 12 weeks post-infection (w.p.i.)] for H. pullorum colonization by a species-specific faecal PCR and serum ELISA, respectively. Rats were monitored for weight loss and diarrhoea. At 30 w.p.i., animals were euthanized.
PCR.
Faecal DNA extraction was performed using the High Pure PCR Template Preparation kit (Roche) or the QIAamp DNA stool mini kit (Qiagen). Pre-infection Helicobacter-free status was confirmed using Helicobacter genus-specific primers C97 (5′-GCTATGACGGGTATCC-3′) and C05 (5′-ACTTCACCCCAGTCGCTG-3′). The amplified 1200 bp PCR product was also used for 16S rRNA gene sequencing and RFLP analysis (see below). The H. pullorum cdtB-gene-specific primers (F1, 5′-GTCTTTTGAGTGGATTGGATTCT-3′) and (R2, 5′-CACTCCGGGTGCTTGTGTAT-3′) were used to determine infection status at all other time points (Laharie et al., 2009).
Real-time quantitative PCR.
The SyBr-based real-time quantitative PCR assay (qPCR) used for quantifying levels of H. pullorum is described elsewhere (Turk et al., 2012). Briefly, a standard curve containing a serial 10-fold dilution of known genomic copies (106 to 101) of H. pullorum MIT 98-5489 was used to enumerate H. pullorum in tissue and faecal DNA samples from rats using a 7500 Fast sequence detection system (Applied Biosystems). qPCR primers were derived from the cdtB gene as described previously (Boutin et al., 2010). Mucosal and faecal H. pullorum numbers calculated from standard curves were then normalized to micrograms of rat chromosomal DNA and measured by qPCR using the 18S rRNA gene-based primers and probe mixture (Applied Biosystems).
Restriction fragment length polymorphism.
RFLP was performed using previously described methods (Shen et al., 2000). Briefly, the amplified 1200 bp H. pullorum, H. bilis, H. muridarum and H. trogontum DNA samples were digested using AluI and HhaI in a 37 °C water bath for 3 h. Digested samples were subsequently electrophoresed on a 3 % agarose gel and stained with ethidium bromide.
Culture.
One faecal pellet was suspended in 1.5 ml of freeze medium (20 % glycerol in Brucella broth); 0.3 ml to 0.5 ml of suspended faeces was collected and filtered using a 0.45 µm syringe filter (Pall Corporation) and streaked onto sheep blood agar plates (Remel). An unfiltered faecal suspension was streaked on CVA plates containing cefoperazone, vancomycin and amphotericin B (Becton Dickinson). Plates were incubated under microaerobic conditions (10 % CO2, 10 % H2, 80 % N2) and observed for growth every 2–4 days. Single colonies with visual and physical characteristics of H. pullorum were replated to obtain a pure culture. Cultures were harvested and confirmed to be H. pullorum using an H. pullorum-specific PCR. Faecal cultures from BN rats were performed at both the pre-infection and final end points as well as intermittently throughout the 30-week study.
ELISA.
H. pullorum outer-membrane protein (OMP) antigen was prepared by methods described previously (Whary et al., 1998). The OMP concentration was measured using a BCA Protein Assay kit (Pierce). Immunlon 2HB 96-well plates (Thermo Electron Corporation) were coated with 100 µl OMP in carbonate buffer (pH 9.6) at a concentration of 1 µg ml−1 per well and incubated overnight at 4 °C. All wells were blocked by incubating with 200 µl 2 % BSA (BSA) for 1 h. Serum was assayed at a dilution of 1 : 100 with 1 % BSA-PBS. Biotinylated goat anti-rat secondary antibodies (Southern Biotech) at a concentration of 1 : 1000 were used to detect H. pullorum-specific IgG. Plates were then incubated with extravidin peroxidase (Sigma) followed by 2-2′-azino-di-(3-ethyl-benzthiazoline-6-sulfonate) (ABTS) (Kirkegaard & Perry Laboratories) for colour development. The optical density (OD) of each sample was measured and recorded at both 405 and 590 nm at 30 min by a Powerwave X ELISA plate reader (Bio-Tek Instruments). All samples were run in triplicate. Seroconversion, as determined by the presence of H. pullorum-specific IgG, was defined as OD values exceeding the mean plus 2×sd of the OD of the samples at the pre-infection time point. The Student’s t-test was used to determine significance at P<0.05.
Pathology.
All rats were euthanized by CO2 inhalation and blood was collected to assess specific IgG response to H. pullorum. At necropsy, the liver and entire large intestine were fixed in 10 % formalin, paraffin-embedded, sectioned at 5 µm and stained with haematoxylin and eosin.
Tissue sections were evaluated by a board-certified veterinary pathologist who was blinded to sample identity. The severity of lesions in the liver and large intestine was scored, using an ascending scale from 0 to 4, based on the degree of lesion severity: 0, absent; 1, mild; 2, moderate; 3, marked; and 4, severe.
For the liver, a hepatitis index was calculated by combining individual scores for lobular, portal and interface hepatitis, as well as the number of lobes (out of a total of four) that contained five or more inflammatory lesions. Hepatitis was defined by a hepatitis index equal to or greater than 4 (Rogers et al., 2007).
For the large intestine, lesions or inflammation, oedema, epithelial defects, crypt atrophy, hyperplasia and dysplasia were scored at separate locations (caecum at the ileocecocolic junction, proximal colon and distal colon) (Turk et al., 2012).
Results
Molecular analyses indicated a high prevalence of naturally acquired H. pullorum infection in Brown Norway rats
Eighteen out of the 20 faecal DNA samples collected from BN rats housed in the barrier where the H. pullorum outbreak was first detected in mice were positive by PCR using both Helicobacter genus-specific and H. pullorum-specific primers. Ten of the 1200 bp helicobacter PCR products were subjected to RFLP analysis using AluI and HhaI restriction enzymes and yielded patterns identical to H. pullorum MIT 09-6635. Using HhaI digestion, H. pullorum had the same pattern as H. bilis, but a different pattern from those of H. muridarum and H. trogontum. When analysing AluI restriction digestion patterns, H. bilis and H. trogontum had similar patterns, but all three rat Helicobacter species (H. bilis, H. muridarum, H. trogontum) were different from the restriction pattern of H. pullorum (Figs 1 and 2). 16S rRNA analysis of the 1200 bp products from faeces of two rats and the H. pullorum mouse isolate (MIT 09-6635) indicated 99 % sequence identity with H. pullorum ATCC 51801 isolated from humans.
Fig. 1.
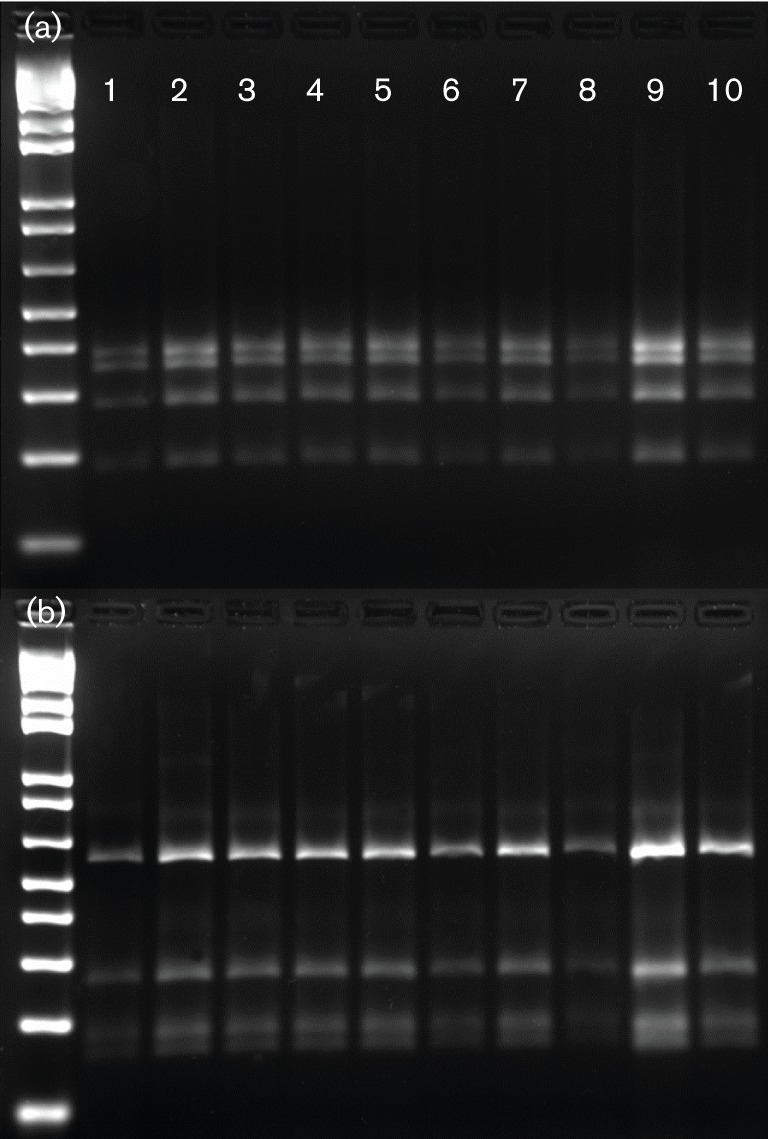
RFLP using HhaI (a) and AluI (b) digestion of 1200 bp PCR products from 10 rat caecal DNA samples from the initial outbreak.
Fig. 2.
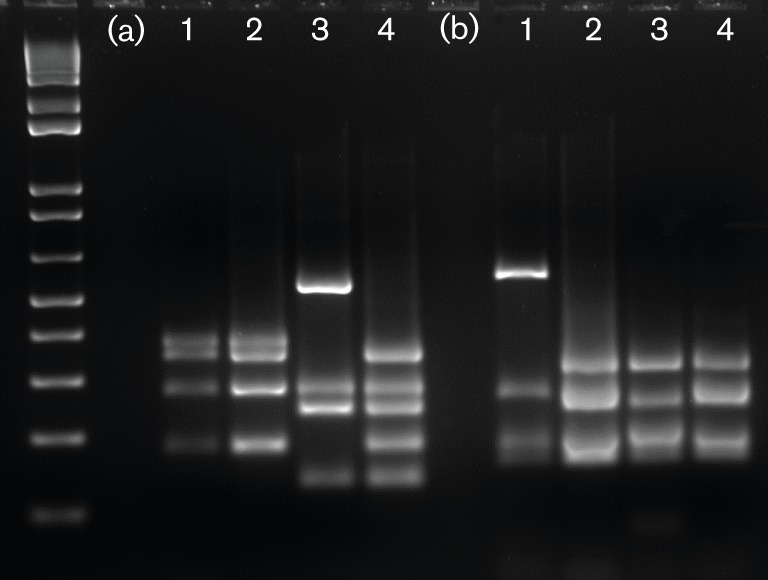
RFLP using HhaI (a) and AluI (b) digestion of 1200 bp PCR products from H. pullorum (MIT 09-6635) from a BN rat. For comparison, lanes 2, 3 and 4 are PCR products from H. bilis, H. muridarum and H. trogontum, respectively.
Experimentally infected brown Norway rats were persistently infected with H. pullorum and developed an H. pullorum IgG immune response
Pre-inoculation faecal samples analysed by PCR were negative for Helicobacter genus-specific and H. pullorum-specific DNA. All rats were confirmed as H. pullorum-positive at 2 w.p.i. by both species-specific faecal PCR and faecal culture. Positive faecal cultures were confirmed by H. pullorum-specific PCR. At 4 w.p.i. and continuing through 30 w.p.i., BN rats #2–6 tested positive (except at 6 w.p.i. and 20 w.p.i.) by either species-specific faecal PCR or faecal culture. BN rat #1 was positive for H. pullorum by species-specific PCR and faecal culture at 2 weeks and PCR positive at 4 w.p.i. but negative at all subsequent time points (Table 1). H. pullorum was detected at necropsy in 4/6 caecal samples and 0/6 liver samples by PCR and in 5/6 caecal samples and 1/6 liver samples by qPCR. qPCR detected quantities of H. pullorum ranging from 11 to 558 684 bacteria mg−1 host DNA in faecal samples collected at necropsy. IgG seroconversion to H. pullorum was apparent in 4/6 BN rats by 2 w.p.i. and in all five colonized rats by 12 w.p.i. and was maintained through 24 w.p.i. (See Fig. 3). During the 30-week experiment, all rats were clinically normal with no signs of diarrhoea.
Table 1. H. pullorum PCR and culture results in experimentally infected BN rats over the 30 w.p.i.
Values are presented as PCR/culture results, respectively. Single values represent PCR results where cultures were not taken. +, Positive; –, negative.
| W.p.i. | BN1 | BN2 | BN3 | BN4 | BN5 | BN6 |
| 0 (pre-dosing) | –/– | –/– | –/– | –/– | –/– | –/– |
| 2 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| 4 | +/– | +/+ | +/+ | +/+ | +/+ | +/+ |
| 6 | – | – | – | + | + | + |
| 8 | – | + | + | + | + | + |
| 10 | – | + | + | + | – | + |
| 14 | –/– | +/+ | +/+ | +/+ | –/+ | +/+ |
| 16 | – | + | + | + | + | + |
| 18 | – | + | + | + | + | + |
| 20 | – | + | – | + | – | + |
| 22 | – | + | + | + | + | + |
| 24 | –/– | +/+ | +/+ | +/+ | +/+ | +/+ |
| 26 | –/– | +/+ | +/+ | +/+ | +/+ | +/+ |
| 28 | – | + | + | + | + | + |
| 30 | –/– | +/+ | +/+ | +/+ | +/+ | +/+ |
Fig. 3.
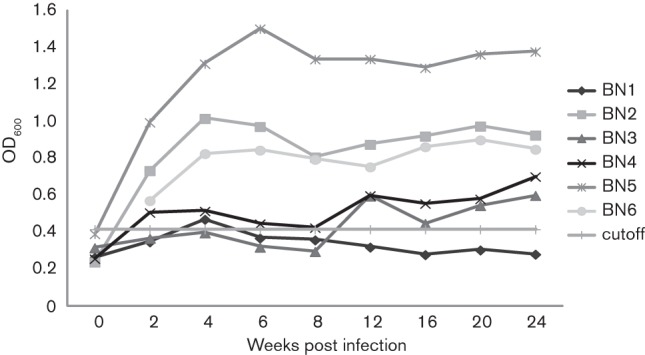
IgG response to experimental infection with H. pullorum. Rats 2–6 were persistently infected with H. pullorum and developed moderate to robust H. pullorum IgG responses. Rat 1 did not colonize with H. pullorum and did not seroconvert.
Pathology
No significant lesions in the intestine were observed in the experimentally infected group. All six BN rats had minimal portal liver inflammation and one rat had minimal lobular inflammation.
Discussion
Eighteen of 20 BN rats surveyed from the natural outbreak were infected with H. pullorum based on H. pullorum-specific PCR and RFLP analyses, which was confirmed by 16S rRNA gene sequencing. In experimentally infected BN rats, 5/6 rats became persistently colonized for the 30 week duration of the experiment. The remaining rat was negative by PCR and culture after 4 w.p.i. PCR results for BN2 and BN3 at 6 w.p.i. and BN3 and BN5 at 20 w.p.i. were negative using our detection methods, which most likely indicates intermittent shedding of H. pullorum. ELISA data measuring IgG response to H. pullorum in BN rats indicated seroconversion in four out of the six rats by 2 w.p.i. and five out of the six rats by 12 w.p.i. These data are most likely representative of the persistent H. pullorum infection we noted in the natural H. pullorum outbreak in BN rats and indicate that serological testing may be a useful screening tool to determine natural exposure or infection with H. pullorum. Similar to EHS infections in mice, and H. pylori infections in humans and mice, seroconversion denotes a persistent infection and indicates that serum antibodies to these helicobacters are not protective to the host.
Gastrointestinal colonization with H. pullorum has been confirmed in humans, poultry and, more recently, mice (Boutin et al., 2010; Stanley et al., 1994; Veijola et al., 2007). Our results confirmed the ability of H. pullorum to successfully colonize the gastrointestinal tract of BN rats. However, H. pullorum infection did not elicit clinical signs or cause gastrointestinal pathology in rats. This finding is consistent with the lack of gastrointestinal lesions in C57BL/6NTac mice persistently infected with H. pullorum (Boutin et al., 2010; Turk et al., 2012).
Faeces from wild rodents and geese around the exterior of the barrier structure where the H. pullorum outbreak occurred were collected and tested for the presence of Helicobacter species by genus-specific PCR. Although H. pullorum was not identified, another avian Helicobacter species was identified; H. brantae naturally colonizes geese and is 97 % similar to H. pullorum by 16S rRNA gene analysis (Boutin et al., 2010; Fox et al., 2006). The source of the H. pullorum outbreak in the commercially received mice and rats remains unknown; however, the H. pullorum strain isolated (MIT 09-6635) was 99 % similar to human H. pullorum isolate NCTC 12826 by 16S rRNA gene analysis, indicating that an infected human may have been responsible for initiating the H. pullorum outbreak in the rodent barrier (Boutin et al., 2010). In humans, H. pullorum has been cultured from diarrhoeic patients and identified by PCR in patients with inflammatory bowel disease and hepatobiliary disease (Castéra et al., 2006; Fox, 2002; Fox et al., 1998). EHS including H. pullorum have also been identified by PCR in patients with hepatobiliary tumours (Rocha et al., 2005).
Prior to this study, only three EHS, H. bilis, H. trogontum and H. muridarum, were known to naturally colonize rats. Of the aforementioned, only H. bilis has been shown to cause clinical disease in rats, causing colitis in naturally and experimentally infected nu/nu rats (Haines et al., 1998; Lee et al., 1992; Mendes et al., 1996). Our finding of natural H. pullorum infection in rats and the ability of H. pullorum to experimentally infect rats warrants further studies to determine whether wild rats are infected with H. pullorum and serve as reservoirs capable of contaminating food stuffs used for poultry or human consumption.
Isolation of H. pullorum from naturally infected rats and mice also supports the rationale that this bacterium should be included in the differential diagnosis when Helicobacter genus-specific PCR results are positive. Additionally, the ability of H. pullorum to colonize multiple species, including humans, and its demonstrated persistent shedding in the faeces of rats and mice pose a zoonotic risk to personnel exposed to H. pullorum-infected animals (Turk et al., 2012). To our knowledge, this is the first report confirming that rats, like mice, can be persistently colonized by an EHS known to colonize humans. Further studies on the epidemiology and pathogenesis of H. pullorum will be facilitated by the availability of the H. pullorum genome (http://www.ncbi.nlm.nih.gov/genome/?term=helicobacter%20pullorum).
Acknowledgements
Grant support was provided by grants from the National Institutes of Health (nos R01RR032307, R01AI50952, T32RR070036 and P30-ES02109). Partial funding for this project was also provided by Taconic Farms Inc., Germantown, NY.
Abbreviations:
- BN
Brown Norway
- EHS
enterohepatic Helicobacter species
- qPCR
quantitative PCR
- RFLP
restriction fragment length polymorphism
- w.p.i.
weeks post-infection
References
- Boutin S. R., Shen Z., Roesch P. L., Stiefel S. M., Sanderson A. E., Multari H. M., Pridhoko E. A., Smith J. C., Taylor N. S. & other authors (2010). Helicobacter pullorum outbreak in C57BL/6NTac and C3H/HeNTac barrier-maintained mice. J Clin Microbiol 48, 1908–1910 10.1128/JCM.02531-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnens A. P., Stanley J., Morgenstern R., Nicolet J. (1994). Gastroenteritis associated with Helicobacter pullorum. Lancet 344, 1569–1570 10.1016/S0140-6736(94)90376-X [DOI] [PubMed] [Google Scholar]
- Castéra L., Pedeboscq A., Rocha M., Le Bail B., Asencio C., de Lédinghen V., Bernard P. H., Laurent C., Lafon M. E. & other authors (2006). Relationship between the severity of hepatitis C virus-related liver disease and the presence of Helicobacter species in the liver: a prospective study. World J Gastroenterol 12, 7278–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. G. (2002). The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50, 273–283 10.1136/gut.50.2.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. G., Yan L., Shames B., Campbell J., Murphy J. C., Li X. (1996). Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect Immun 64, 3673–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. G., Dewhirst F. E., Shen Z., Feng Y., Taylor N. S., Paster B. J., Ericson R. L., Lau C. N., Correa P. & other authors (1998). Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114, 755–763 10.1016/S0016-5085(98)70589-X [DOI] [PubMed] [Google Scholar]
- Fox J. G., Taylor N. S., Howe S., Tidd M., Xu S., Paster B. J., Dewhirst F. E. (2006). Helicobacter anseris sp. nov. and Helicobacter brantae sp. nov., isolated from feces of resident Canada geese in the greater Boston area. Appl Environ Microbiol 72, 4633–4637 10.1128/AEM.02876-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A., Ihrig M. M., Fry R. C., Feng Y., Xu S., Boutin S. R., Rogers A. B., Muthupalani S., Samson L. D., Fox J. G. (2008). Genetic susceptibility to chronic hepatitis is inherited codominantly in Helicobacter hepaticus-infected AB6F1 and B6AF1 hybrid male mice, and progression to hepatocellular carcinoma is linked to hepatic expression of lipogenic genes and immune function-associated networks. Infect Immun 76, 1866–1876 10.1128/IAI.01044-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines D. C., Gorelick P. L., Battles J. K., Pike K. M., Anderson R. J., Fox J. G., Taylor N. S., Shen Z., Dewhirst F. E. & other authors (1998). Inflammatory large bowel disease in immunodeficient rats naturally and experimentally infected with Helicobacter bilis. Vet Pathol 35, 202–208 10.1177/030098589803500305 [DOI] [PubMed] [Google Scholar]
- Laharie D., Asencio C., Asselineau J., Bulois P., Bourreille A., Moreau J., Bonjean P., Lamarque D., Pariente A. & other authors (2009). Association between entero-hepatic Helicobacter species and Crohn’s disease: a prospective cross-sectional study. Aliment Pharmacol Ther 30, 283–293 10.1111/j.1365-2036.2009.04034.x [DOI] [PubMed] [Google Scholar]
- Lee A., Phillips M. W., O’Rourke J. L., Paster B. J., Dewhirst F. E., Fraser G. J., Fox J. G., Sly L. I., Romaniuk P. J. & other authors (1992). Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int J Syst Bacteriol 42, 27–36 10.1099/00207713-42-1-27 [DOI] [PubMed] [Google Scholar]
- Mendes E. N., Queiroz D. M., Dewhirst F. E., Paster B. J., Moura S. B., Fox J. G. (1996). Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol 46, 916–921 10.1099/00207713-46-4-916 [DOI] [PubMed] [Google Scholar]
- Nebbia P., Tramuta C., Ortoffi M., Bert E., Cerruti Sola S., Robino P. (2007). Identification of enteric Helicobacter in avian species. Schweiz Arch Tierheilkd 149, 403–407 10.1024/0036-7281.149.9.403 [DOI] [PubMed] [Google Scholar]
- Rocha M., Avenaud P., Ménard A., Le Bail B., Balabaud C., Bioulac-Sage P., de Magalhães Queiroz D. M., Mégraud F. (2005). Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut 54, 396–401 10.1136/gut.2004.042168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A. B., Theve E. J., Feng Y., Fry R. C., Taghizadeh K., Clapp K. M., Boussahmain C., Cormier K. S., Fox J. G. (2007). Hepatocellular carcinoma associated with liver-gender disruption in male mice. Cancer Res 67, 11536–11546 10.1158/0008-5472.CAN-07-1479 [DOI] [PubMed] [Google Scholar]
- Shen Z., Feng Y., Fox J. G. (2000). Identification of enterohepatic Helicobacter species by restriction fragment-length polymorphism analysis of the 16S rRNA gene. Helicobacter 5, 121–128 10.1046/j.1523-5378.2000.00019.x [DOI] [PubMed] [Google Scholar]
- Stanley J., Linton D., Burnens A. P., Dewhirst F. E., On S. L., Porter A., Owen R. J., Costas M. (1994). Helicobacter pullorum sp. nov.-genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology 140, 3441–3449 10.1099/13500872-140-12-3441 [DOI] [PubMed] [Google Scholar]
- Steinbrueckner B., Haerter G., Pelz K., Weiner S., Rump J. A., Deissler W., Bereswill S., Kist M. (1997). Isolation of Helicobacter pullorum from patients with enteritis. Scand J Infect Dis 29, 315–318 10.3109/00365549709019053 [DOI] [PubMed] [Google Scholar]
- Turk M. L., Cacioppo L. D., Ge Z., Shen Z., Whary M. T., Parry N., Boutin S. R., Klein H. J., Fox J. G. (2012). Persistent Helicobacter pullorum colonization in C57BL/6NTac mice: a new mouse model for an emerging zoonosis. J Med Microbiol 61, 720–728 10.1099/jmm.0.040055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veijola L., Nilsson I., Halme L., Al-Soud W. A., Mäkinen J., Ljungh A., Rautelin H. (2007). Detection of Helicobacter species in chronic liver disease and chronic inflammatory bowel disease. Ann Med 39, 554–560 10.1080/07853890701545714 [DOI] [PubMed] [Google Scholar]
- Ward J. M., Fox J. G., Anver M. R., Haines D. C., George C. V., Collins M. J., Jr, Gorelick P. L., Nagashima K., Gonda M. A. & other authors (1994). Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst 86, 1222–1227 10.1093/jnci/86.16.1222 [DOI] [PubMed] [Google Scholar]
- Whary M. T., Morgan T. J., Dangler C. A., Gaudes K. J., Taylor N. S., Fox J. G. (1998). Chronic active hepatitis induced by Helicobacter hepaticus in the A/JCr mouse is associated with a Th1 cell-mediated immune response. Infect Immun 66, 3142–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]


