Abstract
The regulation and control of gene expression in response to differing environmental stimuli is crucial for successful pathogen adaptation and persistence. The regulatory gene vru of Streptococcus uberis encodes a stand-alone response regulator with similarity to the Mga of group A Streptococcus. Mga controls expression of a number of important virulence determinants. Experimental intramammary challenge of dairy cattle with a mutant of S. uberis carrying an inactivating lesion in vru showed reduced ability to colonize the mammary gland and an inability to induce clinical signs of mastitis compared with the wild-type strain. Analysis of transcriptional differences of gene expression in the mutant, determined by microarray analysis, identified a number of coding sequences with altered expression in the absence of Vru. These consisted of known and putative virulence determinants, including Lbp (Sub0145), SclB (Sub1095), PauA (Sub1785) and hasA (Sub1696).
Introduction
Improvements in milking hygiene, routine dry cow therapy and culling of persistently infected animals have significantly reduced mastitis rates caused by Streptococcus agalactiae and Staphylococcus aureus (Hillerton et al., 1995); however, the same is not the case for infections caused by pathogens from environmental sources, namely Escherichia coli and Streptococcus uberis. S. uberis is an opportunistic pathogen, responsible for a significant proportion of clinical cases of mastitis in the UK, and remains one of the major causative agents of bovine mastitis worldwide (Botrel et al., 2010; Bradley et al., 2007; Hillerton et al., 1993; Olde Riekerink et al., 2008; Shum et al., 2009). S. uberis has been isolated from a range of environmental sources, including soil, pasture, faeces and bedding materials (Bramley, 1982; Field et al., 2003; Lopez-Benavides et al., 2007; Zadoks et al., 2005), and other bovine host sites including skin, genital tract, gastrointestinal tract and tonsils, all of which are colonized asymptomatically (Cruz Colque et al., 1993; Epperson et al., 1993; Kruze & Bramley, 1982).
The ability of S. uberis to adapt to such varied niches is likely to contribute to the organisms’ continued persistence within dairy systems. Such adaptation is likely to be influenced by regulatory genes, and the characterization of their role can be used to identify specific gene subsets of relevance to different conditions. Furthermore, interference with the activity of such regulatory elements may provide novel targets against which control strategies to prevent or eliminate bacterial infections may be mounted. Streptococcus pyogenes [group A Streptococcus (GAS)] is evolutionarily closely related to S. uberis (Ward et al., 2009), and is specifically a human pathogen with a remarkable ability to cause a wide range of infections, ranging from pharyngitis to necrotizing fasciitis and streptococcal toxic-shock syndrome (Cunningham, 2000; Johansson et al., 2010). Unlike many other prokaryotes, which co-ordinate the regulation of gene expression through alternative sigma factors, GAS favours control through transcriptional regulators, which can act as both activators and repressors, and can be categorized as either response regulators or two-component signal transduction systems (TCSs) (Kreikemeyer et al., 2003; Opdyke et al., 2001). Analysis of GAS genomes has revealed around 13 TCSs, many of which contribute to bacterial virulence. These contain a sensor kinase that responds to changes in signal by phosphorylation of a linked DNA-binding response regulator to alter patterns of gene expression (Hondorp & McIver, 2007). GAS also frequently use so called ‘stand-alone’ response regulators that have been shown to control multiple virulence genes. The positive transcriptional regulator Mga (Caparon & Scott, 1987) activates expression of a number of GAS virulence-related genes, including M protein (emm) (Perez-Casal et al., 1991), fibronectin-binding protein (fba) (Terao et al., 2001), collagen-like protein (sclA) (Rasmussen et al., 2000), a C5a peptidase (scpA) (Chen et al., 1993) and M-like immunoglobulin-binding proteins (including mrp and enn) (Podbielski et al., 1995, 1996).
Coding sequences (CDSs) homologous to the regulator Mga and some of the regulated CDSs (in particular M protein and C5a peptidase) have been identified in other streptococci of human and/or animal origin, including Streptococcus pneumoniae, Streptococcus dysgalactiae and Streptococcus iniae (Locke et al., 2008; Tettelin et al., 2001; Vasi et al., 2000). As such, these sequences can be postulated to represent a conserved regulon of products linked to early colonization and infection in a range of streptococcal species.
Analysis of the S. uberis genome has identified a number of potential regulatory elements, one of which, sub0144 (vru); shows some similarity to Mga of S. pyogenes. The translation product of the putative transcriptional regulator (Vru) displays 38 % sequence identity (57 % similarity) to Mgc, a transcriptional regulator common to group C streptococci (GCS) (Geyer & Schmidt, 2000), and 33 % identity (56 % similarity) to Mga, the counterpart in GAS. The S. uberis 0140J Vru CDS displays good homology to functional regions of these regulators extending over the fourth crucial helix–turn–helix DNA-binding domain identified in GAS Mga (McIver & Myles, 2002), supporting a putative DNA-binding mode of action in S. uberis. However unlike GAS and GCS, S. uberis lacks many of virulence determinants typically controlled via Mga/Mgc (Ward et al., 2009), although CDSs with limited similarity to both C5a peptidase (sub1154) and collagen-like protein (sub1095) have been identified (Ward et al., 2009). Both of these sequences, along with that of sub0145, shown previously to encode a lactoferrin-binding protein, have been demonstrated to play a role in virulence of S. uberis; in each case the absence of the gene product reduces the ability to colonize and/or cause disease in the target species (Leigh et al., 2010).
In this study, we investigated the role of Vru in vivo and the effect of its absence on transcriptional control of gene expression using a custom-designed genome-wide microarray.
Methods
Bacterial strains and media.
S. uberis strain 0140J (strain ATCC BAA-854/0140J), originally isolated from a clinical case of bovine mastitis in the UK, was used throughout this study. The sub0145, sub0826, sub0888 and sub1095 mutants were isolated following PCR screening of an S. uberis 0140J/pGh9:ISS1 mutant bank, as previously described (Egan et al., 2010; Leigh et al., 2010; Ward et al., 2001). The vru mutant, containing an insertional disruption in the reverse orientation to the CDS, was isolated from the same mutant bank using a phenotypic screening method by identification of zones of clearing on Todd–Hewitt agar containing skimmed milk (1 %, v/v) and bovine plasminogen, as previously described (Ward et al., 2003). Southern analysis and DNA sequencing were used to confirm the nature and location of mutation events (data not shown). Strains were routinely grown in Todd–Hewitt broth (THB) (Oxoid) and plated on sheep blood agar containing 1 % (w/v) aesculin (ABA) at 37 °C.
Isolation of bacterial RNA.
Aliquots of bacterial culture (15–20 ml) of S. uberis 0140J and vru mutant were harvested once cultures reached OD550 0.42 for early exponential or OD550 0.75 for late-exponential growth phase. Two volumes of RNAprotect Bacteria Reagent (Qiagen) were immediately added to each sample and the mixture was incubated for 5 min at room temperature. Samples were centrifuged (5000 g, 10 min), the supernatant was discarded and the cell pellets were stored at −80 °C until RNA extraction was performed.
Total RNA was extracted following resuspension of the cells in 1 ml RLT buffer (Qiagen) containing 1 % (v/v) β-mercaptoethanol (Sigma–Aldrich). Cells were mechanically disrupted in a Mini-Beadbeater-8 (Biospec) using acid-washed glass beads (Sigma). Following disruption, glass beads and cell debris were removed by centrifugation (17 000 g, 10 s). Supernatants were removed and mixed with an equal volume of 70 % ethanol. Total RNA was extracted using an RNeasy column (Qiagen) according to the manufacturer’s instructions, with RNA finally eluted in 50 µl nuclease-free water. Contaminating DNA was removed from the samples by DNase treatment using a DNA-free DNase Treatment kit (Ambion), and resulting samples were further purified by ethanol precipitation. The integrity and quality of the RNA were determined by gel electrophoresis and by spectrophotometry using an ND-1000 spectrophotometer (NanoDrop Technologies).
Design and hybridization to S. uberis 0140J gene expression microarray.
A custom-designed S. uberis 0140J DNA microarray was generated and probed by Oxford Gene Technology (OGT). The array encompassed the whole bacterial genome and was composed of 3780 features, with optimized 60-mer oligonucleotide probes printed in triplicate. Two separate probes were designed to match each individual gene determined from the S. uberis 0140J genome annotation (Ward et al., 2009), with regions of homology discounted. Arrays were printed on glass microscope slides, as described elsewhere (Hughes et al., 2001).
Fluorescently labelled probes for microarray hybridization were generated from 500 ng total RNA using the MessageAmp II-Bacteria RNA Amplification kit (Ambion) according to the manufacturer’s instructions. A total of 5 µg of the resulting amplified antisense RNA was purified using the RNeasy MinElute Cleanup kit (Qiagen) and hybridization was performed using a Gene Expression Hybridization kit (Agilent). Samples were labelled with either Cy3 or Cy5 N-hydroxysuccinimide ester reactive dyes (GE healthcare) according to the manufacturer’s instructions, with sample combinations generated following a loop design (Townsend, 2003). Three independent repetitions were performed of two growth-point comparisons, giving a total of six microarray hybridizations. Cy3- and Cy5-labelled probes were mixed together in the appropriate combinations, with the fragmented RNA and hybridization buffer applied to the microarray hybridization chamber (Agilent Technologies) and incubated at 65 °C for 17 h with 10 r.p.m. rotation. Following hybridization, microarrays were washed using Gene Expression Wash Buffer (Agilent Technologies). Arrays were scanned on an Agilent C scanner at 5 µm using the XDR function at 10 %. The images were then feature-extracted using Agilent feature extraction version 10.5.1.1 and the GE2_105_Jan09 extraction protocol.
Bioinformatic and statistical analyses.
The microarray data were loess- and scale-normalized (Smyth & Speed, 2003), and analysed for differential expression using limma (Smyth, 2004). Linear models were fitted to the data to compare the vru mutant with the wild-type 0140J strain at both the early and late exponential growth phase. The resultant P values were adjusted for the false discovery rate (Benjamini & Hochberg, 1995). Expression values were visualized using ProGenExpress (Watson, 2005). Genes were linked to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Kanehisa & Goto, 2000) and statistically enriched pathways identified using corna (Wu & Watson, 2009). Sequence analysis of the intergenic and promoter regions of the genes sub0144 (vru) and sub0145 (lbp) was performed using Artemis (Rutherford et al., 2000) and bprom from Softberry sequence analysis tools (http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb).
Challenge of lactating dairy cows with S. uberis and analysis of milk samples.
The requirement of a functional vru gene for S. uberis strain 0140J virulence was determined by intramammary challenge of Holstein–Friesian cows, 2–10 weeks into their first lactation, with either S. uberis 0140J or Vru mutant. Animal selection was based on previously well-established criteria (Leigh et al., 2010) with the same virulent wild-type strain 0140J previously used in a number of challenge experiments (Field et al., 2003; Leigh et al., 2010; Smith et al., 2003). Bacteria were grown overnight in THB at 37 °C, and cells were recovered by centrifugation (10 000 g, 10 min) and resuspended in pyrogen-free saline (Sigma). Cell suspensions were diluted to approximately 1000 c.f.u. ml−1 and held on ice, and the number of viable bacteria was determined from aliquots prior to and post challenge. Animals were challenged in mammary quarters by infusion of 1 ml of the suspension; one animal was challenged in two mammary quarters with 1×103 c.f.u. of strain 0140J, and two animals were challenged each in two mammary quarters with a similar dose of the Vru mutant.
Following challenge, animals were milked and inspected twice daily (07 : 00 h and 15 : 30 h) and were treated with appropriate antibiotics once clinical end points had been reached using criteria previously described (Field et al., 2003; Leigh et al., 2010), including the presence of clotted/discoloured milk and a swollen udder quarter. Milk samples were taken at each milking and analysed for the presence of bacteria and somatic cells. The number of viable bacteria present was estimated by direct plating of up to 1 ml of each milk sample onto ABA. Samples were also diluted in saline, and 50 µl of each dilution plated directly onto ABA. In each case, the presence and/or number of S. uberis was determined, and the genotype of the recovered isolates was determined by RFLP analysis (Hill & Leigh, 1989) and PCR amplification of the vru locus. The number of somatic cells present in milk samples was determined using a DeLaval portable cell counter in line with the manufacturer’s instructions.
Production of recombinant Sub0145, Sub0888 and Sub1095 and corresponding antisera.
The predicted mature CDS of Lbp/Sub0145 was amplified from genomic DNA (S. uberis 0140J) by PCR using the primer pair 5′-GAAGATTTATTTACTATAAATAATTCAG-3′ and 5′-TAGGAAAGTTTCTGGTACCCCTTTGTTA-3′ to generate an amplicon incorporating a KpnI restriction site for efficient directional cloning into a PvuII, KpnI and alkaline phosphatase (New England Biolabs) treated pQE1 vector (Qiagen). Similarly, the Sub1095 CDS was amplified using the primer pair 5′-CAAATGCCAAGTCATTCTCATC-3′ and 5′-CGCTTAATCACAAGGTACCTCTGA-3′ with an incorporated KpnI restriction site. Amplification of the predicted mature CDS of Sub0888 was performed using the primer pair 5′-GAAGAAGTAGTGTCTTCATTAGGG-3′ and 5′-CATCTTATTTGGTCGACTACTCTTC-3′, incorporating a SalI restriction site for efficient directional cloning into a PvuII, SalI and alkaline phosphatase-treated pQE1 vector. Ligated plasmid was generated, desalted and transformed into E. coli M15 pREP4 (Qiagen). Recombinant clones were selected on LB kanamycin (25 µg ml−1) ampicillin (50 µg ml−1) agar plates. Overnight cultures containing each recombinant (6×His-tagged) protein were subcultured by dilution (1 : 30) into LB broth containing 50 µg ampicillin ml−1 and 25 µg kanamycin ml−1 and grown at 37 °C with agitation (200 r.p.m.) for 2 h. Expression of the recombinant proteins, subsequent purification and generation of antisera were performed as described previously (Egan et al., 2010).
Preparation of bacterial extracts and immunoblot analysis.
Culture supernatant samples from S. uberis 0140J and vru mutant strains grown in THB to early exponential (OD550 0.42) and late-exponential (OD550 0.75) growth phases were harvested following centrifugation (16 000 g, 10 min). Complete Protease Inhibitor (Roche Diagnostics) was added to the supernatant, which was subsequently filter-sterilized through a 0.22 µm pore-size filter (Millipore), and proteins therein were concentrated by precipitation, as previously described (Egan et al., 2010). Concentrated supernatant proteins were resuspended in SDS-loading buffer and separated by SDS-PAGE using 15 % (w/v) polyacrylamide gels. Proteins were subsequently either stained with InstantBlue (Expedeon Protein Solutions) or transferred (200 mA for 1 h in a Transblot apparatus, Bio-Rad) in transfer buffer [25 mM Tris base, 192 mM glycine, 20 % (v/v) methanol, pH 8.1–8.4] to nitrocellulose membranes for immunoblotting.
Membranes were incubated in a blocking solution [0.5 % skimmed milk (Marvel) in PBS] at 4 °C overnight, washed three times for 5 min (PBS containing 0.1 % Tween-20; PBST) and then incubated for 1 h with anti-Lbp rabbit antisera (1 : 10 000 in blocking solution) or anti-PauA EC3 (Leigh, 1994) mouse monoclonal antibody (1 : 1000 in blocking solution). Membrane washes were repeated following incubation with either goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP; Southern Biotech) or rabbit anti-mouse IgG conjugated to peroxidase (Sigma), each diluted 1 : 1000 in blocking buffer, for 1 h. Membranes were washed (as above) and HRP conjugate was detected following incubation in 4-chloronaphthol (0.5 mg ml−1) containing methanol [16.7 % (v/v)] and H2O2 [0.00015 % (v/v)] under dark conditions for 1 h. Membranes were subsequently rinsed in PBS and allowed to dry, and were stored under light-tight conditions.
Detection of bacterial proteins on whole cells by ELISA.
S. uberis strains were grown until they reached OD550 0.42 or 0.75, and cultures were centrifuged (16 000 g, 5 min) and bacterial pellets washed in PBS, then recentrifuged, as before. Bacteria were resuspended in ELISA coating buffer (carbonate/bicarbonate buffer, pH 9.6; Sigma) and each suspension was adjusted to OD550 1. Triplicate wells were coated with 100 µl of each bacterial strain and incubated at 4 °C overnight. Wells were washed five times with PBST, and 300 µl blocking solution [0.5 % (w/v) casein/PBS] was added and plates were incubated at 4 °C overnight. Blocked wells were washed as above and individual antisera added at the following concentrations: anti-Sub0145 1 : 4000, anti-Sub0826 (Egan et al., 2010) and anti-Sub0888 1 : 2000, and anti-Sub1095 1 : 500 in blocking solution, and plates were incubated for 1 h at room temperature. After repeated washes with PBST, 100 µl anti-rabbit–HRP conjugate (Sigma) was added at a dilution of 1 : 5000 in blocking solution to each well and incubated at room temperature for 1 h. Wells were then emptied and reactions developed by adding 100 µl 3,3′,5,5′-Tetramethylbenzidine (TMB) Liquid Substrate (Sigma) and stopped by the addition of 100 µl Stop Reagent for TMB Substrate (Sigma). The colorimetric reaction was measured by A450, and differences between datasets were assessed using one-way ANOVA followed by Tukey’s multiple comparisons to determine statistically significant values between each mutant strain and test antigen.
Results
Comparison of bacterial virulence by experimental challenge with S. uberis 0140J and Vru mutant
The virulence of the S. uberis 0140J strain and that of the mutant with a defective virulence regulatory gene (vru, sub0144) was assessed using a well-established experimental model for bovine mastitis (Field et al., 2003; Leigh et al., 2010; Smith et al., 2003). The Vru mutant contained an insertional disruption (the 8 bp repeat signature of which spanned from −5 bp upstream of the ATG start codon to +3 bp), which was confirmed by PCR and DNA sequencing (data not shown).
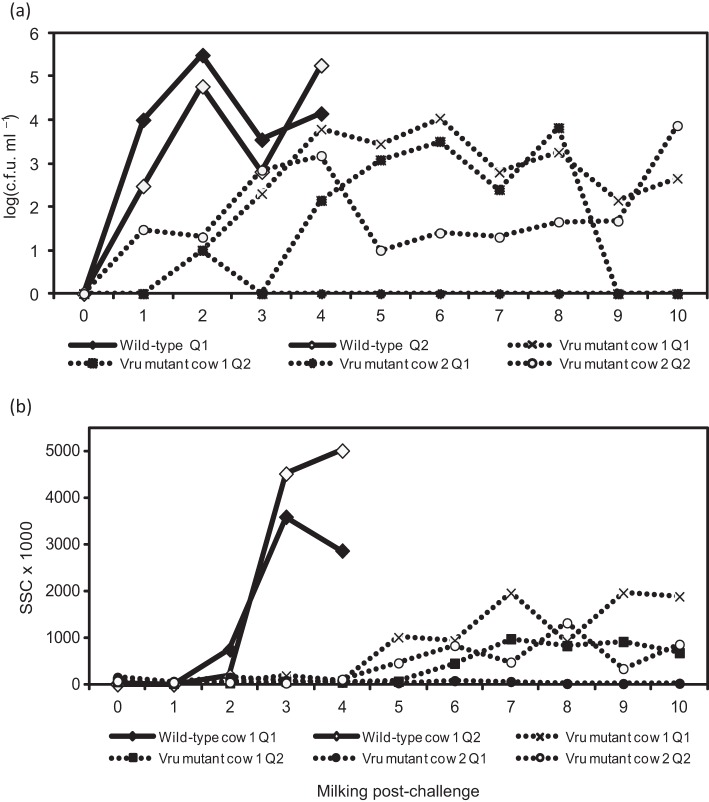
Both challenged quarters of the animal that received wild-type S. uberis became infected and shed bacteria at around 1×105 c.f.u. ml−1 by 48 h post-challenge (Fig. 1a). Animals challenged with the Vru mutant shed considerably fewer bacteria in their milk. This low level of infection, which remained consistent and detectable for the remainder of the experiment (5 days), peaked at approximately 103 c.f.u. ml−1 by milking 5 (Fig. 1a).
Fig. 1.
Bacterial isolation and somatic cell count following challenge with S. uberis 0140J and Vru mutant in dairy cattle. (a) Bacterial recovery of S. uberis, quantified after each milking following challenge. Data are represented as the number of bacteria obtained from the milk of animals challenged with either strain 0140J (n = 2) or the Vru mutant (n = 4) from each individual challenged quarter, as indicated above. (b) Cellular influx measured after each milking following challenge. Data are represented by the number of somatic cells (somatic cell count; SCC) obtained from the milk of animals challenged with either strain 0140J (n = 2) or the Vru mutant (n = 4) from each individual challenged quarter, as indicated above.
The cellular infiltration into the mammary gland in response to infection was markedly lower in quarters challenged with the Vru mutant (Fig. 1b), with no overt clinical signs of mastitis. In contrast, the animal challenged with the wild-type strain showed clinical signs of mastitis (severe changes in milk composition/colour and the inflammatory response of the mammary gland) and required antibiotic treatment after milking 4.
Comparison of differences in gene transcription by microarray analysis
Gene expression profiles of S. uberis 0140J and isogenic vru mutant were compared by microarray analysis using a custom-designed S. uberis 0140J array optimized using two different probes per gene to match individual genes as determined from the genome annotation (Ward et al., 2009). Two different growth time points were analysed (early and late-exponential phases of growth), and genes were considered to be differentially expressed in the wild-type and mutant if the difference between the two strains was greater than fourfold by analysis of the duplicate probes for each gene and the comparison of the datasets preferentially showed an adjusted P value of <0.001.
Using these criteria, a total of 19 genes were found to be expressed at lower levels by the Vru mutant, eight of which (sub0051, sub0052, sub0145, sub1480, sub1174, sub1696, sub1697 and sub1785) were common to both early and late-exponential growth phases (Table 1). Genes relating to capsule production (Ward et al., 2001), hasA and hasB1 (sub1697 and sub1696, respectively), were significantly and substantially downregulated in the Vru mutant compared with wild-type in both early (>250-fold) and late (>13-fold) exponential phases of growth. Also, both pauA (sub1785), previously shown to encode a plasminogen activator (Rosey et al., 1999), and lbp (sub0145), encoding a lactoferrin-binding protein (Moshynskyy et al., 2003), were dramatically downregulated in the Vru mutant cultures during early exponential growth (>100-fold) and to a substantial, but lesser, extent (>20-fold) during late growth periods. Pathway analysis showed differences in the exponential phase of growth, where two pathways were significantly downregulated in the Vru mutant compared with the wild-type 0140J strain, i.e. the valine, leucine and isoleucine biosynthesis and aminoacyl-tRNA biosynthesis pathways (Table 2).
Table 1. Differentially transcribed genes expressed at a higher level in early and late-exponential phase cultures of S. uberis 0140J compared with the S. uberis Vru mutant.
| Gene* | Protein annotation | Early exponential growth phase | Late-exponential growth phase | ||
| Fold change† | P value‡ | Fold change† | P value‡ | ||
| hasA/Sub1697 | Hyaluronan synthase | 380.0/250.2 | <0.001 | 17.9/13.8 | <0.001 |
| hasB1/Sub1696 | UDP-glucose 6-dehydrogenase 1 | 366.0/357.9 | <0.001 | 8.5/8.1 | 0.006/<0.001 |
| lbp/Sub0145 | Lactoferrin-binding protein | 255.1/153.8 | <0.001 | 41.2/27.8 | <0.001/0.1262 |
| pauA/Sub1785 | PauA protein precursor (streptokinase precursor) | 169.6/168.3 | <0.001 | 32.1/23.6 | <0.001 |
| Sub0051 | Hypothetical protein | 15.0/13.3 | <0.001 | 16.6/10.5 | <0.001/0.034 |
| Sub0052 | Membrane protein | 14.8/14.5 | <0.001 | 13.4/8.5 | <0.001 |
| Sub0177 | RNA polymerase sigma factor protein | – | – | 12.3/7.8 | <0.001 |
| Sub0176 | Exported protein | – | – | 9.4/6.6 | <0.001 |
| Sub1480 | Permease | 9.2/8.7 | <0.001 | 8.1/4.1 | <0.001 |
| Sub1174 | ABC transporter protein | 8.3/4.2 | 0.033/0.002 | 16.4/12.9 | <0.001 |
| Sub0543 | Dihydroxyacetone kinase subunit DhaK | – | – | 8.7/5.2 | <0.001 |
| Sub1173 | Membrane protein | – | – | 8.5/6.7 | 0.027/<0.001 |
| Sub0155 | Basic membrane protein | 8.1/7.7 | <0.001 | – | – |
| sclB/Sub1095 | Collagen-like surface-anchored protein | 6.9/6.5 | 0.012/<0.001 | – | – |
| pyrD/Sub1264 | Dihydroorotate dehydrogenase 1A | 6.0/5.3 | <0.001 | – | – |
| Sub0118 | Competence protein | – | – | 5.6/4.5 | <0.001 |
| pyrF/Sub1025 | Orotidine 5′-phosphate decarboxylase | 5.4/5.0 | <0.001 | – | – |
| Sub0544 | Dihydroxyacetone kinase family protein | – | – | 4.9/4.4 | <0.001 |
| Sub0137 | Lipoprotein | – | – | 4.1/4.0 | <0.001 |
Gene and protein annotation according to the genomic sequence of S. uberis 0140J (Ward et al., 2009).
Mean fold change represented by S. uberis 0140J versus the S. uberis vru mutant for each of the probes tested.
Where the P value for data from one of the two probes is not in excess of 0.001, the mean of both P values is provided.
Table 2. corna-based pathway analysis for early and late-exponential phase cultures of the S. uberis Vru mutant compared with S. uberis 0140J.
| Pathway regulation in Vru mutant compared with wild-type strain | Pathway | Total genes involved in pathway | Expectation | Observation | Fisher distribution P value |
| Downregulated in early exponential growth phase | Sub00290; valine, leucine and isoleucine biosynthesis | 7 | 2 | 7 | 0.013 |
| Downregulated in early exponential growth phase | Sub00970; aminoacyl-tRNA biosynthesis | 25 | 7 | 16 | 0.015 |
| Upregulated in late-exponential growth phase | Sub00230; purine metabolism | 47 | 13 | 28 | <0.0002 |
| Upregulated in late-exponential growth phase | Sub00061; fatty acid biosynthesis | 12 | 3 | 11 | <0.0002 |
A number of CDSs were only differentially expressed in the early exponential phase of growth; those downregulated in the Vru mutant included sub0155, sub1095, sub1264 and sub1025. Other CDSs (sub0176, sub0177, sub0543, sub0544, sub1173, sub0118 and sub0137) were found to be downregulated in the Vru mutant only in the late-exponential phase of growth.
A total of 24 genes were found to be expressed at higher levels by the Vru mutant, 12 of which were common to both early and late-exponential growth (Table 3). A number of these genes were involved in purine biosynthesis, including purH, purN, purF, purM, purC, purK, purE, purB and purD. Pathway analysis unsurprisingly indicated that the Sub00230 purine metabolism pathway was upregulated in the absence of functional vru at both time points, but also identified that the Sub00061 fatty acid biosynthesis pathway was also significantly upregulated in the absence of Vru at the stationary growth phase (Table 2).
Table 3. Differentially transcribed genes expressed at a higher level in early and late-exponential phase cultures of the S. uberis Vru mutant compared with S. uberis 0140J.
| Gene* | Protein annotation | Early exponential growth phase | Late-exponential growth phase | ||
| Mean fold change† | Mean P value‡ | Mean fold change† | Mean P value‡ | ||
| purH/Sub0030 | Bifunctional phosphoribosyl-aminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | 91.1/90.0 | <0.001 | 14.8/12.2 | <0.001/0.065 |
| Sub0865 | Guanosine 5′-monophosphate oxidoreductase | 83.1/75.9 | <0.001 | 8.2/7.1 | <0.001 |
| purN/Sub0029 | Phosphoribosylglycinamide formyltransferase | 72.8/63.1 | <0.001 | 10.9/9.1 | <0.001 |
| purF/Sub0027 | Amidophosphoribosyltransferase | 69.8/67.9 | <0.001 | 31.0/29.7 | <0.001 |
| purM/Sub0028 | Phosphoribosylaminoimidazole synthetase | 67.7/65.0 | <0.001 | 21.4/8.6 | 0.033/0.011 |
| Sub0026 | Phosphoribosylformylglycinamidine synthase protein | 60.0/21.5 | <0.001 | 50.7/40.8 | <0.001 |
| purC/Sub0025 | Phosphoribosylaminoimidazole-succinocarboxamide synthase | 20.0/18.0 | <0.001 | 61.7/55.3 | <0.001 |
| purA/Sub0152 | Adenylosuccinate synthetase | 18.9/15.5 | <0.001 | – | – |
| Sub0987 | Hypothetical protein | 14.8/13.6 | <0.001 | – | – |
| purK/Sub0055 | Phosphoribosylaminoimidazole carboxylase ATPase subunit | 12.0/11.3 | <0.001 | 21.2/13.2 | <0.001 |
| purE/Sub0054 | Phosphoribosylaminoimidazole carboxylase catalytic subunit | 8.2/7.6 | <0.001 | 23.4/20.4 | <0.001 |
| purB/Sub0056 | Adenylosuccinate lyase | 7.0/6.5 | <0.001 | 4.9/4.7 | <0.001 |
| purD/Sub0053 | Phosphoribosylamine-glycine ligase | 7.0/6.7 | <0.001 | 18.9/18.0 | <0.001 |
| Sub0410 | Membrane protein | 7.0/6.3 | <0.001 | 4.5/4.3 | <0.001 |
| Sub0411 | PAP2 superfamily protein | 6.1/5.9 | <0.001 | – | – |
| Sub0826 | Surface-anchored subtilase family protein | – | – | 6.6/6.1 | <0.001 |
| fhs1/Sub0942 | Formate-tetrahydrofolate ligase | – | – | 6.1/5.8 | <0.001 |
| spxA/Sub1777 | Transcriptional regulator Spx | 5.9/5.6 | <0.001 | – | – |
| Sub0746 | Membrane protein | 5.4/4.3 | <0.001 | – | – |
| Sub1260 | Formate/nitrite transporter family protein | 4.9/4.4 | <0.001 | – | – |
| fabD/Sub1497 | Malonyl-CoA-acyl carrier protein transacylase | – | – | 4.7/4.7 | <0.001 |
| fabK/Sub1498 | Enoyl-ACP reductase | – | – | 4.4/4.3 | <0.001 |
| Sub1233 | Major facilitator superfamily protein | 4.3/4.2 | <0.001 | – | – |
| rpmB/Sub0331 | 50S ribosomal protein L28 | 4.2/4.2 | 0.01/<0.001 | – | – |
Gene and protein annotation according to the genomic sequence of S. uberis 0140J (Ward et al., 2009).
Mean fold change represented by S. uberis 0140J versus the S. uberis vru mutant for each of the probes tested.
Where the P value for data from one of the two probes is not in excess of 0.001, the mean of both P values is provided.
Detection of proteins in concentrated culture supernatants of the wild-type and Vru-deficient mutant and whole-cell ELISA
Mouse monoclonal antibody directed towards PauA (Sub1785) was used to detect the presence of PauA in concentrated culture supernatants of S. uberis 0140J and the Vru mutant (Fig. 2a). Similarly, rabbit antisera directed against Lbp (Sub0145) were used to detect the presence of Lbp (Fig. 2b). In each case, proteins corresponding to the approximate sizes of the secreted versions of the PauA (30.6 kDa) and Lbp (54.6 kDa) proteins were detected in culture supernatant from the wild-type strain, although no cross-reactive proteins were evident in that from the Vru mutant strain.
Fig. 2.

Identification of PauA and Lbp from extracts of S. uberis 0140J and Vru mutant. Immunoblots of protein extracts from S. uberis 0140J and the Vru mutant probed with either mouse monoclonal antibody generated against PauA (Sub1785) (a) or rabbit antisera generated against Lbp (Sub0145) (b). Precipitated media extracts from the late-exponential growth phase (OD550 0.75) from the Vru mutant (lane 1) and 0140J (lane 2) were analysed; molecular mass standards are indicated.
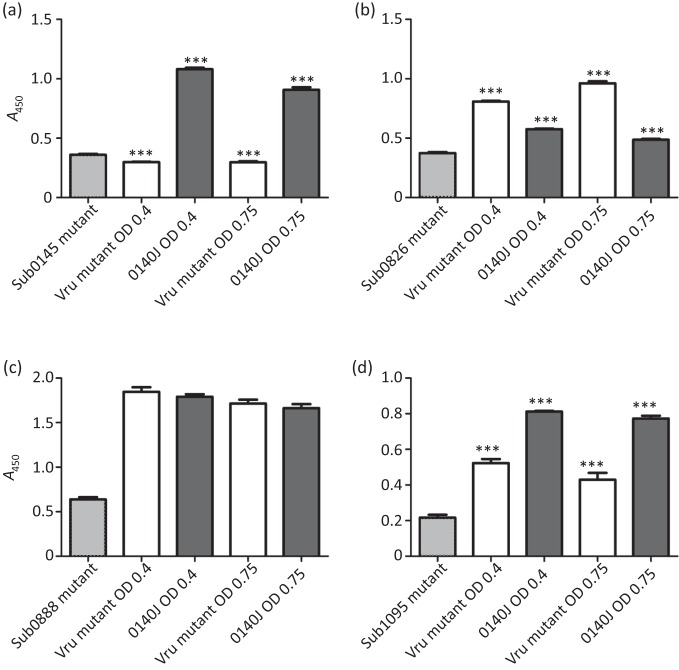
As a number of the differentially expressed gene products (Sub0145, Sub0826 and Sub1095) were known to be anchored to the cell wall by SrtA in S. uberis (Egan et al., 2010), the presence of these proteins on whole cells was determined by ELISA (Fig. 3). By way of comparison, a similarly anchored gene product (Sub0888) that appeared from the microarray analysis to be unaffected by the absence of Vru was also assayed by whole-cell ELISA. In each case the reactions of the wild-type and Vru mutant were compared with that of a relevant specific CDS mutant previously shown to lack the gene product of interest (Leigh et al., 2010).
Fig. 3.
Differential protein expression between S. uberis 0140J and Vru mutant. Whole-cell ELISA was used to detect protein expression by S uberis 0140J wild-type and Vru mutant strains for Sub0145 (a), Sub0826 (b), Sub0888 (c) and Sub1095 (d). Results are expressed as the means of triplicate values; error bars, sd. In each ELISA, statistical significance was determined by ANOVA and Tukey’s multiple comparison, with triple asterisks indicating a P value of <0.001. The analysis displayed is between strains at same growth point, with each strain statistically significantly different from the individual mutant strain (P<0.001) in each assay, except in (a), in which the significant difference between the Vru mutant and Sub0145 mutant was at P<0.01.
Sub0145 was only detected on the wild-type strain (Fig. 3a); the level of reaction for the Vru mutant being similar to that for the Sub0145-negative mutant. The Sub0826 protein (Fig. 3b) was detectable on both the wild-type and the Vru mutant, but at significantly higher levels on the Vru mutant compared with the wild-type. Conversely, the Sub1095 protein, which was also detected on both strains (Fig. 3d), was detected at a higher level on the wild-type than on the Vru mutant. The Sub0888 protein was detected on both strains at a similar level (Fig. 3c).
Discussion
The ability of S. uberis to survive and/or proliferate within a range of external environments and in various host niches is critical for the continued persistence of this opportunistic pathogen. Such adaptation requires co-ordinated regulation of gene expression in response to differing conditions (Ward et al., 2009). Survival and transmission of the pathogen between cattle may be further enhanced by the ability to control and co-ordinate virulence. In modern dairy settings, overtly infected/diseased cattle may be treated with antimicrobial compounds to eliminate the infection; however, from an evolutionary standpoint, the ability to colonize the mammary gland at high levels, coincidentally resulting in disease in the host, may favour transmission of the bacterium between individuals or niches.
The importance of the putative stand-alone gene regulator Vru for virulence of S. uberis was demonstrated in the current study. None of the bovine mammary quarters challenged with the Vru mutant showed high-level bacterial colonization or overt signs of mastitis; in contrast, those that received the genetically intact wild-type strain shed bacteria at high levels and displayed clinical signs similar to those previously reported following intramammary challenge with S. uberis (Field et al., 2003; Leigh et al., 2010).
The vru sequence exhibits a good level of homology to other stand-alone gene regulators of streptococci, mga and mgc (Caparon & Scott, 1987; Geyer & Schmidt, 2000), both of which positively influence expression of virulence-related sequences. Consistent with a role as a specific gene regulator, the absence of Vru in S. uberis resulted in altered levels of transcription from 42 CDSs. Of these, 19 CDSs, including five that corresponded to the production of putative virulence determinants of streptococci, were transcribed at lower levels in the absence of Vru. Conversely, 24 CDSs with higher levels of transcription were identified. It was not determined whether Vru directly or indirectly altered gene expression, but the presence of both positively and negatively altered transcription may be indicative of secondary regulatory events. To further substantiate this view, two CDSs with altered expression in the Vru mutant showed homology to other known regulatory elements. Transcription of a gene encoding an RNA polymerase sigma factor was suppressed, whereas that encoding a putative transcriptional regulator, spxA (Kajfasz et al., 2009), was enhanced.
In the context of the current investigation it is difficult to envisage how enhanced transcription would mediate a reduction in virulence, but it was of interest to note that 10 of the 23 CDSs showing upregulation in the absence of Vru were identifiably associated with purine biosynthesis. These fell into two gene clusters: sub0025–sub0030, carrying purCFMNH and the unassigned CDS sub0026; and sub0053–sub0056, carrying purDEKB. A further purine biosynthesis-related sequence, sub0152/purA, located at a distant locus was also upregulated, indicating co-ordinate regulation. Lesions within purine metabolism (purM, purC) resulting in gene disruption have been shown to affect growth in raw bovine milk (Smith, 2000), but unsurprisingly, no such effect was noted for the Vru mutant.
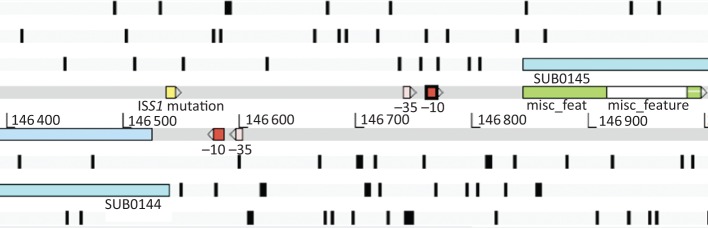
One highly downregulated sequence, lbp, was adjacent to vru within the S. uberis genome. We speculated whether mutation of vru may physically affect transcription from lbp by preventing read through of a single transcript. To investigate this, the 303 bp intergenic region (nucleotides 146 540–146 843 on the 0140J genome sequence) separating sub0144/vru and sub0145 was scanned in both directions for bacterial promoter sequences using bprom and displayed using the Artemis program (Fig. 4) to illustrate promoter sequence orientation with respect to the flanking CDSs. Sequence analysis of this intergenic region identified two divergent pairs of candidate promoter sequences. This further substantiated the assertion that disruption of the vru ORF in the vicinity of its start codon was likely to be influencing sub0145 expression through altered expression of its gene product, rather than abrogation of a possible shared transcriptional promoter. Furthermore, similarly altered expression of Sub0145 protein was seen in immunoblots of concentrated media proteins in a second sub0144 mutant background where the ISS1 insertion mapped to a position distant from candidate promoter sequences (within 100 bp of the stop codon of sub0144), which further excludes the possibility that both CDSs were transcribed from a shared divergent promoter (data not shown).
Fig. 4.
Highlighted promoter regions of the S. uberis 0140J, sub0144 and sub0145 genes. Snapshot of the Artemis program in database mode displaying the S. uberis 0140J chromosome, with the genes and σ70-type promoter regions of both sub0144 (vru) and sub0145 (lbp) with their orientation shown with respect to the flanking CDSs. The −10 box 5′-CATCATAAT-3′ and a −35 box, 5′-TTGAAG-3′, can be seen on the negative strand upstream of the vru start codon, whilst the corresponding promoter sequences are identified on the positive strand upstream of the sub0145 gene. The location of the mutation in the sub0144/vru gene is also highlighted.
In GAS, the Mga regulator controls expression of a number of major virulence determinants, including M protein, C5a peptidase, ScpA, and collagen-like binding protein SclA (Barnett & Scott, 2002; Caparon & Scott, 1987; Chen et al., 1993; Rasmussen et al., 2000). Analysis of the S. uberis genome did not reveal a gene encoding M protein but did indicate a C5a peptidase-like precursor (Sub1154) and a collagen-like binding protein (Sub1095) (Egan et al., 2010; Ward et al., 2009). Unlike the situation with Mga in GAS, Vru did not appear to influence the expression of Sub1154, but did appear to alter expression of the collagen-like protein, which was downregulated at the transcriptional level during early exponential growth.
Hondorp & McIver (2007) speculated that only those CDSs that show high-level activation/suppression in the presence/absence of Mga (respectively) are under its direct control. A similar consideration of the data from this study would suggest that genes encoding production of capsule, plasminogen activator and lactoferrin-binding protein are those most likely to be under the direct control of Vru.
The sequences downregulated in the absence of Vru responsible for production of capsule, hasA and hasB1 (Ward et al., 2001), plasminogen activator, pauA (Leigh, 1994; Rosey et al., 1999), lactoferrin-binding protein, lbp, and collagen-like protein, Sub1095 (Ward et al., 2009), have been subject to previous investigation to determine their individual roles in virulence (Field et al., 2003; Leigh et al., 2010; Ward et al., 2003). While Lbp and the collagen-like protein have been shown to be required for the full expression of the virulent phenotype of S. uberis (Leigh et al., 2010), the absence of PauA or capsule alone was not deemed to affect virulence in lactating cattle (Field et al., 2003; Ward et al., 2003).
The transcriptional data relating to lbp, pauA and Sub1095, the collagen-like protein gene, were validated with respect to their effect on protein expression. In the absence of Vru, neither PauA nor Lbp was detectable, and the collagen-like protein, which was detected on whole cells, was shown to be present at a significantly lower level. These data, along with that related to Sub0826 (upregulated in the absence of Vru) and Sub0888 (unaffected by Vru), were consistent with their comparative transcriptional levels in the presence/absence of Vru, thus indicating that transcriptional analysis was reflected in protein expression.
It is possible that the reduced virulence observed for the Vru mutant during this study was solely due to reduced expression of Lbp. A mutant strain in the same genetic background (strain 0140J) that lacks the ability to produce Lbp has been shown to be of reduced virulence. This mutant, like the Vru mutant, is less able to colonize the lactating bovine mammary gland, and induces a lesser inflammatory response (lower somatic cell count) and fewer clinical signs than the wild type (Leigh et al., 2010). However, any reduction in virulence of the Vru mutant due to the absence of Lbp may be augmented by the reduction in expression of the collagen-like protein (Sub1095), the absence of which has been shown to affect both colonization at high bacterial numbers and bacterial persistence within the bovine mammary gland (Leigh et al., 2010). Although the precise mechanism by which these two proteins exert their effects on pathogenesis is currently unknown, it is unlikely that they show redundancy in their mechanisms, as the absence of each alone measurably reduced virulence. Consequently, it is likely that the absence (or reduced expression) of both has a greater effect on virulence than the absence (or reduced expression) of either alone.
A possible role for either capsule and/or the plasminogen activator PauA is more speculative. Whilst attractive as virulence determinants, neither has been shown independently to alter the virulence of S. uberis (Field et al., 2003; Ward et al., 2003), implying that either these proteins do not have a function in pathogenesis or that they exert any effect in a highly redundant manner alongside other molecules.
The hyaluronic acid capsule of a number of streptococcal species has been implicated in disease pathogenesis, linked to invasion, adherence and evasion of phagocytic host defences. Capsule production in S. uberis has been shown to be dependent on hasA and hasC (Ward et al., 2001). This is distinct from the situation with GAS, in which capsule expression is dependent on hasA and hasB but not hasC. S. uberis carries a homologue of hasB (sub1027/hasB2) elsewhere within the genome (Ward et al., 2009), and unlike GAS does not appear to encode another means of producing UDP-glucose, the chemical product resulting from the action of HasC. Therefore, of the genes required for capsule production in S. uberis, only hasA (as part of the hasAB1 gene cluster) showed significantly reduced transcription in the Vru mutant; hasC, located around 3 kb downstream from the hasAB1 gene cluster and hasB2, showed no marked differences in expression in the absence of Vru. Mutation of hasA in the same genetic background (strain 0140J) as used in the present study has been shown experimentally not to alter virulence. The hasAB1 gene cluster is not universally present in S. uberis, although this operon has been shown to be over-represented in isolates from clinically apparent infections (Field et al., 2003). Together, these data imply some level of evolutionary significance with respect to virulence; however, the precise role of the capsule, and whether it acts a genetic marker of other, more functional, CDSs remain to be determined.
Streptococcal species have evolved, or acquired by horizontal gene transfer, a number of apparently distinct products that typically activate plasminogen in a species-specific manner that invariably includes that from the target species (Caballero et al., 1999; Kinnby et al., 2008; Leigh et al., 1998). Indeed, different strains of S. uberis have been shown to possess two different and mutually exclusive CDSs encoding distinct products, PauA and PauB (Rosey et al., 1999; Ward & Leigh, 2002). Parallel evolution and/or acquisition and retention of such CDSs imply the utility of the activity, a view that is further reinforced by the apparent lack of variation within a selection of PauA sequences analysed (Ward & Leigh, 2004). However disabling mutations of PauA in S. uberis have no effect on virulence in lactating dairy cattle, indicating that any role for this activity is not linked to virulence within the lactating bovine mammary gland (Ward et al., 2003). Again, the presence of PauA (or PauB) is not ubiquitous in S. uberis, although it appears to be present in more than 90 % of the isolates investigated (Ward & Leigh, 2002), a greater proportion than have acquired (and retained) the hasAB1 gene cluster.
Of the remaining CDSs that were downregulated in the absence of Vru, none has previously been shown to affect virulence or have any putative or speculated role regarding the host–pathogen interaction. Some gene products were putatively involved in solute transport via ABC type transporters (Sub0137, 1174 and 1173), while others were linked to metabolism of pyrimidines (Sub1264 and 1025). However, these did not affect growth of the Vru mutant in either THB or milk.
In conclusion, it would appear justified to speculate that the observed reduced virulence of an S. uberis Sub0144 (Vru) mutant was mediated largely by its effect on expression from the adjacent CDS (Sub0145), encoding a surface-anchored protein shown by others to bind lactoferrin (Moshynskyy et al., 2003). Such effects could be further enhanced by the concomitant reduced expression of a collagen-like protein. The apparently co-ordinated regulation of putative virulence determinants that are not directly linked with virulence, at least for the lactating bovine mammary gland, may indicate that Vru additionally or historically coordinates gene expression in S. uberis for attributes other than virulence following intramammary infection of the lactating bovine mammary gland.
Abbreviations:
- CDS
coding sequence
- GAS
group A Streptococcus
- HRP
horseradish peroxidase
References
- Barnett T. C., Scott J. R. (2002). Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J Bacteriol 184, 2181–2191. 10.1128/JB.184.8.2181-2191.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57, 289–300. [Google Scholar]
- Botrel M. A., Haenni M., Morignat E., Sulpice P., Madec J. Y., Calavas D. (2010). Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog Dis 7, 479–487. 10.1089/fpd.2009.0425 [DOI] [PubMed] [Google Scholar]
- Bradley A. J., Leach K. A., Breen J. E., Green L. E., Green M. J. (2007). Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet Rec 160, 253–258. 10.1136/vr.160.8.253 [DOI] [PubMed] [Google Scholar]
- Bramley A. J. (1982). Sources of Streptococcus uberis in the dairy herd. I. Isolation from bovine faeces and from straw bedding of cattle. J Dairy Res 49, 369–373. 10.1017/S0022029900022500 [DOI] [PubMed] [Google Scholar]
- Caballero A. R., Lottenberg R., Johnston K. H. (1999). Cloning, expression, sequence analysis, and characterization of streptokinases secreted by porcine and equine isolates of Streptococcus equisimilis. Infect Immun 67, 6478–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon M. G., Scott J. R. (1987). Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci U S A 84, 8677–8681. 10.1073/pnas.84.23.8677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Bormann N., Cleary P. P. (1993). VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet 241, 685–693. 10.1007/BF00279912 [DOI] [PubMed] [Google Scholar]
- Cruz Colque J. I., Devriese L. A., Haesebrouck F. (1993). Streptococci and enterococci associated with tonsils of cattle. Lett Appl Microbiol 16, 72–74. 10.1111/j.1472-765X.1993.tb00346.x [DOI] [PubMed] [Google Scholar]
- Cunningham M. W. (2000). Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 13, 470–511. 10.1128/CMR.13.3.470-511.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. A., Kurian D., Ward P. N., Hunt L., Leigh J. A. (2010). Identification of sortase A (SrtA) substrates in Streptococcus uberis: evidence for an additional hexapeptide (LPXXXD) sorting motif. J Proteome Res 9, 1088–1095. 10.1021/pr901025w [DOI] [PubMed] [Google Scholar]
- Epperson W. B., Hoblet K. H., Smith K. L., Hogan J. S., Todhunter D. A. (1993). Association of abnormal uterine discharge with new intramammary infection in the early postpartum period in multiparous dairy cows. J Am Vet Med Assoc 202, 1461–1464. [PubMed] [Google Scholar]
- Field T. R., Ward P. N., Pedersen L. H., Leigh J. A. (2003). The hyaluronic acid capsule of Streptococcus uberis is not required for the development of infection and clinical mastitis. Infect Immun 71, 132–139. 10.1128/IAI.71.1.132-139.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer A., Schmidt K. H. (2000). Genetic organisation of the M protein region in human isolates of group C and G streptococci: two types of multigene regulator-like (mgrC) regions. Mol Gen Genet 262, 965–976. 10.1007/PL00008665 [DOI] [PubMed] [Google Scholar]
- Hill A. W., Leigh J. A. (1989). DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis. Epidemiol Infect 103, 165–171. 10.1017/S0950268800030466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillerton J. E., Shearn M. F., Teverson R. M., Langridge S., Booth J. M. (1993). Effect of pre-milking teat dipping on clinical mastitis on dairy farms in England. J Dairy Res 60, 31–41. 10.1017/S0022029900027321 [DOI] [PubMed] [Google Scholar]
- Hillerton J. E., Bramley A. J., Staker R. T., McKinnon C. H. (1995). Patterns of intramammary infection and clinical mastitis over a 5 year period in a closely monitored herd applying mastitis control measures. J Dairy Res 62, 39–50. 10.1017/S0022029900033653 [DOI] [PubMed] [Google Scholar]
- Hondorp E. R., McIver K. S. (2007). The Mga virulence regulon: infection where the grass is greener. Mol Microbiol 66, 1056–1065. 10.1111/j.1365-2958.2007.06006.x [DOI] [PubMed] [Google Scholar]
- Hughes T. R., Mao M., Jones A. R., Burchard J., Marton M. J., Shannon K. W., Lefkowitz S. M., Ziman M., Schelter J. M., et al. (2001). Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol 19, 342–347. 10.1038/86730 [DOI] [PubMed] [Google Scholar]
- Johansson L., Thulin P., Low D. E., Norrby-Teglund A. (2010). Getting under the skin: the immunopathogenesis of Streptococcus pyogenes deep tissue infections. Clin Infect Dis 51, 58–65. 10.1086/653116 [DOI] [PubMed] [Google Scholar]
- Kajfasz J. K., Martinez A. R., Rivera-Ramos I., Abranches J., Koo H., Quivey R. G., Jr, Lemos J. A. (2009). Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J Bacteriol 191, 2060–2068. 10.1128/JB.01609-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28, 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnby B., Booth N. A., Svensäter G. (2008). Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology 154, 924–931. 10.1099/mic.0.2007/013235-0 [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B., McIver K. S., Podbielski A. (2003). Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen–host interactions. Trends Microbiol 11, 224–232. 10.1016/S0966-842X(03)00098-2 [DOI] [PubMed] [Google Scholar]
- Kruze J., Bramley A. J. (1982). Sources of Streptococcus uberis in the dairy herd. II. Evidence of colonization of the bovine intestine by Str. uberis. J Dairy Res 49, 375–379. 10.1017/S0022029900022512 [DOI] [PubMed] [Google Scholar]
- Leigh J. A. (1994). Purification of a plasminogen activator from Streptococcus uberis. FEMS Microbiol Lett 118, 153–158. 10.1111/j.1574-6968.1994.tb06818.x [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Hodgkinson S. M., Lincoln R. A. (1998). The interaction of Streptococcus dysgalactiae with plasmin and plasminogen. Vet Microbiol 61, 121–135. 10.1016/S0378-1135(98)00179-5 [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Egan S. A., Ward P. N., Field T. R., Coffey T. J. (2010). Sortase anchored proteins of Streptococcus uberis play major roles in the pathogenesis of bovine mastitis in dairy cattle. Vet Res 41, 63. 10.1051/vetres/2010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J. B., Aziz R. K., Vicknair M. R., Nizet V., Buchanan J. T. (2008). Streptococcus iniae M-like protein contributes to virulence in fish and is a target for live attenuated vaccine development. PLoS ONE 3, e2824. 10.1371/journal.pone.0002824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Benavides M. G., Williamson J. H., Pullinger G. D., Lacy-Hulbert S. J., Cursons R. T., Leigh J. A. (2007). Field observations on the variation of Streptococcus uberis populations in a pasture-based dairy farm. J Dairy Sci 90, 5558–5566. 10.3168/jds.2007-0194 [DOI] [PubMed] [Google Scholar]
- McIver K. S., Myles R. L. (2002). Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol Microbiol 43, 1591–1601. 10.1046/j.1365-2958.2002.02849.x [DOI] [PubMed] [Google Scholar]
- Moshynskyy I., Jiang M., Fontaine M. C., Perez-Casal J., Babiuk L. A., Potter A. A. (2003). Characterization of a bovine lactoferrin binding protein of Streptococcus uberis. Microb Pathog 35, 203–215. 10.1016/S0882-4010(03)00150-5 [DOI] [PubMed] [Google Scholar]
- Olde Riekerink R. G., Barkema H. W., Kelton D. F., Scholl D. T. (2008). Incidence rate of clinical mastitis on Canadian dairy farms. J Dairy Sci 91, 1366–1377. 10.3168/jds.2007-0757 [DOI] [PubMed] [Google Scholar]
- Opdyke J. A., Scott J. R., Moran C. P., Jr (2001). A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol Microbiol 42, 495–502. 10.1046/j.1365-2958.2001.02657.x [DOI] [PubMed] [Google Scholar]
- Perez-Casal J., Caparon M. G., Scott J. R. (1991). Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol 173, 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A., Flosdorff A., Weber-Heynemann J. (1995). The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun 63, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A., Woischnik M., Pohl B., Schmidt K. H. (1996). What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol (Berl) 185, 171–181. 10.1007/s004300050028 [DOI] [PubMed] [Google Scholar]
- Rasmussen M., Edén A., Björck L. (2000). SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect Immun 68, 6370–6377. 10.1128/IAI.68.11.6370-6377.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosey E. L., Lincoln R. A., Ward P. N., Yancey R. J., Jr, Leigh J. A. (1999). PauA: a novel plasminogen activator from Streptococcus uberis. FEMS Microbiol Lett 178, 27–33. 10.1111/j.1574-6968.1999.tb13755.x [DOI] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., Barrell B. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. [DOI] [PubMed] [Google Scholar]
- Shum L. W., McConnel C. S., Gunn A. A., House J. K. (2009). Environmental mastitis in intensive high-producing dairy herds in New South Wales. Aust Vet J 87, 469–475. 10.1111/j.1751-0813.2009.00523.x [DOI] [PubMed] [Google Scholar]
- Smith A. J. (2000). The identification of genes required for growth of Streptococcus uberis in milk. PhD thesis, University of Reading, Reading, UK. [Google Scholar]
- Smith A. J., Ward P. N., Field T. R., Jones C. L., Lincoln R. A., Leigh J. A. (2003). MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect Immun 71, 4842–4849. 10.1128/IAI.71.9.4842-4849.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, 1–25. [DOI] [PubMed] [Google Scholar]
- Smyth G. K., Speed T. (2003). Normalization of cDNA microarray data. Methods 31, 265–273. 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- Terao Y., Kawabata S., Kunitomo E., Murakami J., Nakagawa I., Hamada S. (2001). Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol Microbiol 42, 75–86. 10.1046/j.1365-2958.2001.02579.x [DOI] [PubMed] [Google Scholar]
- Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., et al. (2001). Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506. 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
- Townsend J. P. (2003). Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics 4, 41. 10.1186/1471-2164-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasi J., Frykberg L., Carlsson L. E., Lindberg M., Guss B. (2000). M-like proteins of Streptococcus dysgalactiae. Infect Immun 68, 294–302. 10.1128/IAI.68.1.294-302.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. N., Leigh J. A. (2002). Characterization of PauB, a novel broad-spectrum plasminogen activator from Streptococcus uberis. J Bacteriol 184, 119–125. 10.1128/JB.184.1.119-125.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. N., Leigh J. A. (2004). Genetic analysis of Streptococcus uberis plasminogen activators. Indian J Med Res 119 (Suppl.), 136–140. [PubMed] [Google Scholar]
- Ward P. N., Field T. R., Ditcham W. G., Maguin E., Leigh J. A. (2001). Identification and disruption of two discrete loci encoding hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis. Infect Immun 69, 392–399. 10.1128/IAI.69.1.392-399.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. N., Field T. R., Rapier C. D., Leigh J. A. (2003). The activation of bovine plasminogen by PauA is not required for virulence of Streptococcus uberis. Infect Immun 71, 7193–7196. 10.1128/IAI.71.12.7193-7196.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. N., Holden M. T., Leigh J. A., Lennard N., Bignell A., Barron A., Clark L., Quail M. A., Woodward J., et al. (2009). Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genomics 10, 54. 10.1186/1471-2164-10-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. (2005). ProGenExpress: visualization of quantitative data on prokaryotic genomes. BMC Bioinformatics 6, 98. 10.1186/1471-2105-6-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Watson M. (2009). corna: testing gene lists for regulation by microRNAs. Bioinformatics 25, 832–833. 10.1093/bioinformatics/btp059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks R. N., Tikofsky L. L., Boor K. J. (2005). Ribotyping of Streptococcus uberis from a dairy’s environment, bovine feces and milk. Vet Microbiol 109, 257–265. 10.1016/j.vetmic.2005.05.008 [DOI] [PubMed] [Google Scholar]