Abstract
The WAVE-associated Rac GAP, WRP, is thought to regulate key aspects of synapse development and function and may be linked to mental retardation in humans. WRP contains a newly described inverse F-BAR (IF-BAR) domain of unknown function. Our studies show that this domain senses/facilitates outward protrusions analogous to filopodia and that the molecular basis for this is likely explained by a convex lipid-binding surface on the WRP IF-BAR domain. In dendrites the IF-BAR domain of WRP forms a bud on the shaft from which precursors to spines emerge. Loss of WRP in vivo and in vitro results in reduced density of spines. In vivo this is primarily a loss of mushroom-shaped spines. Developmentally, WRP function is critical at the onset of spinogenesis, when dendritic filopodia are prevalent. Finally, because WRP is implicated in mental retardation, behaviors of WRP heterozygous and null mice have been evaluated. Results from these studies confirm that loss of WRP is linked to impaired learning and memory.
Introduction
Dendritic spines are the postsynaptic structures responsible for the majority of excitatory synaptic transmission. As such they are being extensively studied for their roles in information storage and processing in response to experience. Spine formation and function is intimately coupled to remodeling of the spine actin cytoskeleton, which may translate information as changes in the membrane receptor composition and shape of the spine (Allison et al., 1998; Fischer et al., 2000; Fukazawa et al., 2003; Carlisle and Kennedy, 2005; Bourne and Harris, 2008; Wang et al., 2008). How this occurs during development and during plasticity is likely governed by unique signaling pathways to the spine actin cytoskeleton that is coupled to the membrane.
WAVE-1 functions as a scaffold for actin-based signaling in spines and proteomic analysis of the WAVE-1 signaling complex has identified WRP, a regulator of Rac and WAVE-1 signaling in neurons (Soderling et al., 2002). WRP binds WAVE-1 via its C-terminal SH3 domain, and genetic analysis in mice reveals that WAVE-1 regulates spine density (Soderling et al., 2007). Disruption of this signaling pathway results in multiple behavioral abnormalities, including deficits in sensorimotor gating and various learning and memory processes.
Little is known about the spatial targeting of WRP in neurons or how it may contribute to neuronal development. WRP and the srGAP family members have a conserved N-terminal Fes and CIP4 homology BAR-like (F-BAR) domain (Itoh et al., 2005; Aspenström, 2009). Several studies have shown this to be a lipid-binding domain that induces membrane invagination and regulates processes such as endocytosis (Takei et al., 1999; Kamioka et al., 2004; Itoh et al., 2005; Icking et al., 2006; Tsujita et al., 2006; Shimada et al., 2007). Surprisingly, a recent study of srGAP2 showed that its F-BAR domain enhances outward membrane protrusions (Guerrier et al., 2009). Because the membrane topology facilitated by the F-BAR domain of srGAP2 is opposite of other F-BAR domains, it has been termed an inverse F-BAR or inverse F-BAR (IF-BAR) domain (Carlson and Soderling, 2009). Yet, how common the IF-BAR domain is or what the physiological functions of these domains are remains very unclear.
A balanced chromosomal translocation within 3p25 from an individual with severe mental retardation has been shown to selectively disrupt the coding region of WRP within the IF-BAR domain (Endris et al., 2002). This observation implied WRP is linked to mental retardation in humans. That the IF-BAR domain may be physiologically important itself is suggested by natural splice variants of WRP, which include an isoform that encodes for just the IF-BAR domain (Endris et al., 2002). Thus, the current evidence raises three important questions. First, what is the cellular role of the WRP IF-BAR domain? Second, is the IF-BAR domain of WRP alone sufficient for any of the cellular functions of WRP? Third, is haploinsufficiency of WRP associated with cognitive deficits in a model organism?
Here, we analyze the role of WRP in spine development and test its role of cognitive functioning. The cumulative results of this study suggest a molecular mechanism whereby WRP regulates spine development in vivo during the induction of filopodia and support a possible link between WRP and human mental retardation.
Materials and Methods
Cell culture and transfections
3T3, Cos7, and HEK293T fibroblasts were cultured in DMEM supplemented with 10% FBS and 10 U/ml penicillin G (sodium salt) and 10 μg/ml streptomycin sulfate. Primary neuronal cultures from rat and mouse hippocampi were prepared as described previously (Banker and Goslin, 1988). Briefly, postnatal day 0 (P0) rodents were killed by rapid decapitation and hippocampal neurons were collected, pooled, and plated on poly-l-lysine-treated coverslips in 12-well plates at a density of 300,000 cells/well. Neurons were cultured in Neurobasal A medium supplemented with 2% (v/v) B-27 supplement and 1% (v/v) GlutaMAX (Invitrogen). Neuronal culture medium was refreshed every 5 d after plating with 1 ml of the above medium supplemented with 5 μm cytosine β-d-arabinofuranoside (Sigma-Aldrich) to restrict non-neuronal cell growth. Neuronal transfections were performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen) on either days in vitro (DIV) 5 or DIV9 after plating. Neuronal cultures were transfected with 2.0 μg of plasmid DNA consisting of 0.75 μg of a soluble tandem dimer Tomato (tdTomato) fluorescent protein-expressing plasmid and 1.25 μg of either a Cre-green fluorescent protein (GFP)-expressing plasmid or an empty sham vector. Rescue experimental cultures were transfected with 2.0 μg of plasmid DNA consisting of 0.50 μg of a soluble tdTomato-expressing plasmid and 0.75 μg of a Cre-GFP-expressing plasmid and 0.75 μg of a plasmid expressing either the GFP-tagged WRP IF-BAR domain, a V5-tagged full-length WRP construct, or a V5-tagged full-length WRP construct with the R211E/R212E mutation in the IF-BAR domain. HEK293 FreeStyle cells were grown in suspension in 293 Freestyle media (Invitrogen). Transfections in Cos7 and hippocampal neurons were performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Transfections in HEK293 FreeStyle cells were performed using 2 mg of 25 kDa linear polyethylemine (Polysciences) and 1 mg of DNA per liter of cells at a density of 1 × 106 cells/ml. Cells were harvested 3 d post-transfection.
Plasmid constructs
WRP IF-BAR mutant constructs were made using the Strategene QuikChange II XL Site-Directed Mutagenesis Kit according to the manufacturer's instructions using full-length WRP in pcDNA3.1, the WRP IF-BAR domain (residues 1–494 of WRP) in pBetaActin yellow fluorescent protein (YFP), or pGEX 4T-1 as the template. The yeast phosphatidylinositol 4,5-bisphosphate phosphatase Inp54 was a generous gift from Dr. John York (Duke University, Durham, NC). FBP17 was isolated by PCR from expressed sequence tag clone 6683892 (American Type Culture Collection, Manassas VA) and cloned into the HindIII site of the pBA-GFP vector. pCAG-Cre:GFP was supplied by Addgene (Cambridge, MA).
Antibodies and stains
For Western blot analyses, antibodies against GST (Millipore) and WRP (Soderling et al., 2007) were used. Flamingo Fluorescent Gel Stain (Bio-Rad) was used according to the manufacturer's instructions to fluorescently stain gels, which were imaged with the Typhoon 9400 Phosphorimager (GE Healthcare).
Microscopy and image analysis
Cells were prepared for microscopy by fixation in 4% paraforaldehyde/4% sucrose in PBS for 15 min at 37°C. Coverslips were mounted with FluorSave Reagent (Calbiochem). Images were taken on a Zeiss LSM 510 Meta, Zeiss LSM 710, or Leica SP2 microscope. All images were acquired using either a 63×/1.4 numerical aperture (NA) oil-immersion objective or a 100×/1.4 NA oil-immersion objective. Data collected were filtered in ImageJ (NIH) using Despeckle for the whole image. Maximum image projections and three-dimensional projections used in spine density calculations were also made in ImageJ from confocal z-series images. Additionally, dendrite lengths used to calculate spine density were measured in ImageJ using the NeuronJ plug-in (Meijering et al., 2004) from the maximum image projections. For statistical analysis, an unpaired t test with Welch correction was performed.
Spines were counted as dendritic protrusions measuring <3 μm in length and were classified into four qualitative categories based upon their visual appearance: mushroom, thin, stubby, and filopodial. Mushroom spines were defined as short spines with bulbous heads that had a width greatly exceeding the width and length of the neck. Thin spines were defined as longer spines with smaller bulbous heads having a width greater than the width of the neck but smaller than the length of the neck. Stubby spines were defined as a spine head attached to the dendritic shaft without any narrowing between the maximum head diameter and the contact point with the shaft. Filopodial spines were defined as long, slender projections from the dendritic shaft without any noticeable widening at the tip. A quantitative analysis of the spine head width and the spine neck length parameters for mushroom and thin spines was conducted to ensure there was no blurring of the two categories. Mushroom spines averaged 571 ± 7.2 nm in head width and 298 ± 4.4 nm in neck length, while thin spines averaged 382 ± 9.5 nm in head width and 671 ± 18.6 nm in neck length (pW = 0.0001, pN = 0.0001).
Line analyses were performed using MetaMorph (Molecular Devices). Briefly, a line analyzing fluorescence was drawn across the cell, and background readings were subtracted. The first 3 μm was considered to be the plasma membrane (PM). The average of the two highest fluorescent points from the plasma membrane was divided by the average fluorescence from the middle 3 μm of the cell. The PM index then is a ratio of the highest fluorescence in the plasma membrane to the average fluorescence in the cytoplasm of the cell.
Protein expression and purification
The IF-BAR domain of WRP, along with its respective point mutants, Rac, the F-BAR domains of srGAP1 and FNBP2, and FBP17, were all expressed as N-terminal GST fusions in bacteria (BL-21). Proteins were purified using glutathione-Sepharose (Sigma) at 4°C and eluted in 50 mm Tris, 150 mm NaCl, 3 mm DTT, and 25 mm reduced glutathione (Sigma) at pH 7.5. For lipid overlays, proteins were dialyzed in 1× PBS. For liposome cosedimentation studies, proteins were dialyzed in 0.1 m sucrose, 20 mm HEPES, 100 mm KCl, and 1 mm EDTA.
Full-length WRP V5/His and its respective point mutants were expressed in polyethylenimine-transfected 293 Freestyle cells and purified using nickel nitrilotriacetic acid agarose (Qiagen) at 4°C. Proteins were eluted in 10 mm sodium phosphate buffer, 1 m NaCl, 250 mm imidazole, and 5 mm β-mercaptoethanol at pH 7.3. Proteins were dialyzed in 0.1 m sucrose, 20 mm HEPES, 100 mm KCl, and 1 mm EDTA. The proteins were concentrated using a 100 kDa centrifugal filter unit (Millipore) and analyzed with Coomassie-stained SDS-PAGE gels.
Liposome preparation
Lipids were dried by evaporation and resuspended in 0.1 m sucrose, 20 mm HEPES, pH 7.4, 100 mm KCl, and 1 mm EDTA. After five cycles of freeze/thaw, large unilamellar vesicles were made by extrusion through a 100 nm pore diameter membrane. The lipids were then washed by the addition of extra buffer, pelleted by centrifugation at 100,000 × g for 1 h at 10°C, and resuspended in the same buffer. A phosphate assay was used to determine the final lipid concentration (Chen et al., 1956).
Lipid array overlays
Lipid overlays were performed according to the manufacturer's instructions (Echelon). Membranes prespotted with lipids were blocked in 3% fatty acid-free BSA (Sigma) in PBS with Tween 20 at room temperature for 1 h. Membranes were then incubated with 0.5 μg/ml the F-BAR WRP-GST or 1 μg/ml PLCδ-PH-GST overnight at 4°C and then Western blotted for GST.
Liposome sedimentation assays
Proteins were centrifuged at 100,000 × g for 1 h at 10°C. Five micrograms of protein was incubated with 100 μg of liposomes at room temperature in a final volume of 200 μl for 15 min. In negative controls, buffer (0.1 m sucrose, 20 mm HEPES, pH 7.4, 100 mm KCl, and 1 mm EDTA) was added instead of lipids. The reaction mixtures were then centrifuged at 100,000 × g for 1 h. The supernatant was separated from the pellet, and the pellet was resuspended with buffer in a volume equal to that of the supernatant. Affinity measurements were performed by cosedimentation assay using 297 nm WRP IF-BAR domain with varying concentrations of phosphatidylethanolamine/phosphatidylcholine/phosphatidylinositol 4,5-bisphosphate liposomes. Samples were then electrophoresed on a 7% SDS-polyacrylamide gel and analyzed by Western blot or Flamingo fluorescent stain.
Scanning electron microscopy
3T3 cells were transfected with GFP-tagged constructs (FBP17 and WRP IF-BAR) using Lipofectamine 2000 according to the manufacturer's directions (Invitrogen). After 24 h, the cells were sorted by fluorescent-activated cell sorting (FACS) for GFP expression using a BD FACS Aria II (A02) and replated onto 12 mm coverslips. The next day, cells were fixed in 2.5% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.3, for 1 h. The coverslips were then washed three times with water and then subjected to 1% osmium tetroxide in 0.1 m cacodylate buffer for 20 min. After washing three times with water, the samples were then subjected to dehydration in a graded ethanol series (50, 70, 80, 90, 95, and 100%). A BAL-TEC CPD030 Critical Point Dryer and Polaron E-5100 sputter coater were used to further dehydrate and coat the samples, after which the images were collected on a JEOL 6300 with the Orion Digital Micrography System.
Protein structure modeling
Template-based homology modeling was conducted using Phyre (Kelley and Sternberg, 2009). For this experiment, Phyre searched the Structural Classification of Proteins (SCOP) and Protein Data Bank databases for homologs of the WRP F-BAR domain using a profile–profile alignment algorithm. As expected, the top three high scoring alignments were the F-BAR domains of FBP17 (SCOP Code, c2eflA; E-value = 3.8−26), CIP4 (SCOP code, c2efkA; E-value = 9.8−26), and FCHo2 (SCOP code, c2v0oB; E-value = 1−20). Coordinates from the resulting model, constructed in Phyre by alignment-based threading of the WRP sequence into the solved structure of the FBP17 F-BAR domain, were downloaded and visualized using MacPyMOL (DeLano Scientific).
Animals
Conditional WRP knock-out animals were produced by homologous recombination at the Duke Transgenic Core Facility (Durham, NC). Briefly, a targeting construct was prepared by bacterial artificial chromosome recombineering, placing LoxP sites to flank exon 3 of WRP. Embryonic stem cells were selected for neomyocin resistance and screened for homologous recombination by PCR and Southern blot. All mice were housed in Duke University's Division of Laboratory Animal Resources facilities and all procedures were approved by the Duke University Institutional Animal Care and Use Committee and were in accordance with National Institutes of Health guidelines. For behavioral testing, Nestin-Cre positive (The Jackson Laboratory, stock no. 003771) male and female mice from heterozygous breeding pairs were transferred to the Duke University Medical School Mouse Behavioral and Neuroendocrine Analysis Core Facility at 3 months of age. Behavioral testing was conducted on eight or nine mice of each genotype composed of four or five males and females over the next 6 months in the order of the least to most stressful tests.
Behavioral testing
Novel object recognition test.
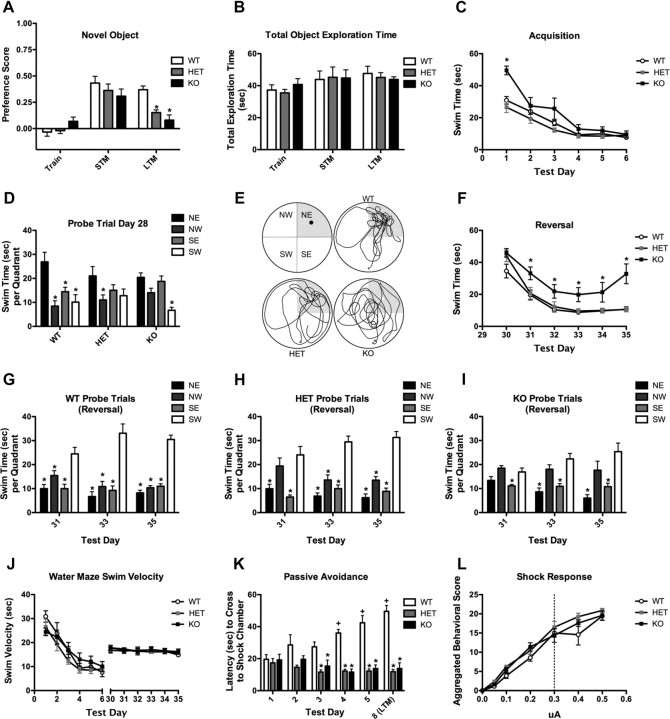
Testing was conducted on two consecutive days. On the first day, animals were individually placed into clear polycarbonate test boxes and presented with two identical objects for 10 min. Mice were then returned to their home cage for 20 min before being returned to the test arena for a 10 min short-term memory (STM) retention test, where one training object was replaced with a novel object. The animals were re-examined 24 h later for long-term memory (LTM) retention in a 10 min test with the familiar training object paired to a second novel object. Testing was conducted under indirect moderate illumination (80 lux) and filmed for later analyses using the Noldus Observer. Tapes were scored by a trained observer blind to the animal genotype or the test condition (STM or LTM). Preference for the novel object was expressed as a ratio of the total time spent with the familiar object subtracted from the total time spent with novel object, divided by the total time spent exploring both objects. Positive preference scores indicated a preference for the novel object, negative scores for the familiar, and scores approximating “0” indicated no preference. Repeated measures ANOVA (RMANOVA) was used to assess preference scores and total exploration time during STM and LTM tests; within-subject effects were designated as test time and between-subject effects as genotype. Significant test time by genotype interactions were decomposed with Bonferroni corrected pairwise comparisons. In all cases, data were shown as mean ± SEM and p < 0.05 was considered statistically significant; n = 8–12 mice per genotype.
Morris water maze.
Mice were handled and acclimated to water for 1 wk before the start of testing, and water maze learning was conducted using methods described previously (Rodriguiz et al., 2008; Roberts et al., 2009). The water maze was divided into four quadrants with a platform hidden 1 cm below the water surface in the northeast quadrant (see Fig. 7E). Water was made opaque with the addition of nontoxic white poster paint. Animals were trained daily to swim to the hidden platform over 6 d, with four 1 min test trials per day. Probe tests for the memory of the hidden platform were conducted 1 h following completion of the final test trial on days 2, 4, and 6. Remote memory was examined with a single probe test 21 d after the final acquisition test day (day 28). Following remote memory testing, animals were retrained to swim to the hidden platform for 1 d. On day 30, the platform location was changed to the southwest quadrant, and reversal learning was conducted over six additional days. Probe tests for the new platform location were conducted on days 31, 33, and 35. A separate set of mice was examined in the water maze using a platform made visible in the water by the placement of a flag marking the location. Performance of the mice on all test trials was assessed by swim time to the hidden platform from tracking profiles created by EthoVision (Noldus Information Technology). For probe tests, the total time mice spent swimming in each quadrant of the water maze was calculated for the 1 min trial. Learning curves for acquisition and reversal testing were assessed with RMANOVA, using test trial as the within-subject effect and genotype as the between-subject effect. Probe tests for the acquisition training and remote tests and for the reversal training were also analyzed with nested RMANOVA, using arena quadrant and test day as within-subject effects and genotype as the between-subject effects. In all cases, Bonferroni corrected post hoc comparisons were examined when significant interactions between the within-subject and between-subject effects were obtained. For all analyses, data were expressed as mean ± SEM and p < 0.05 was considered statistically significant; n = 8–9 mice/genotype.
Figure 7.
WRP heterozygous and knock-out mice are impaired in multiple learning and memory tests. A, Analysis of WRP WT, WRP heterozygous (HET), and WRP knock-out (KO) mice response in the novel object recognition test, n = 7–9 mice per genotype. No significant differences in object preferences were detected during training or STM. Both HET and KO mice were significantly impaired in the LTM preference test, indicating impaired memory in these animals. B, No differences among genotypes were found in the total object exploration time during the training, STM, or LTM, demonstrating that time spent in object exploration was not confounded with object preference. C, Graph depicting the swim time for each genotype to reach the platform during the acquisition phase (days 1–6) of water maze testing. While KO mice were impaired on day 1 compared to WT and HET mice, all three genotypes performed similarly on days 2–6. D, Remote memory was measured in a delayed probe trial (day 28). WT mice preferentially spent more time searching in the target (northeast) quadrant versus all other nontarget quadrants, whereas both HET and KO mice were impaired in the task. NE, Northeast; NW, northwest; SE, southeast; SW, southwest. E, Representative swim-trace patterns for WT, HT, and KO mice. Schematic of the four quadrants is shown in the top left panel. The target quadrant is shaded in each panel. F, Two days after the probe trial on day 28, mice were tested for their ability to relearn and remember a new position for the platform in the SW quadrant (SE). WRP KO mice were impaired on relearning compared to their WT and HET littermates, further supporting a learning and memory deficit in these mice. G–I, Graph of probe trials during the reversal test on days 31, 33, and 35. Wild-type mice (G) preferentially searched the target quadrant (i.e., SW) on each day. HET mice (H) showed a mild deficit on day 31, whereas KO mice (I) could not distinguish the target from the nontarget quadrants on each probe day. J, All genotypes had a similar swim velocity during the acquisition and reversal trials, demonstrating that swimming ability was not impaired in the mutants. K, Learning and memory processes were assessed in the five trial passive avoidance test. Mice were trained once each day for five consecutive days and then tested 72 h later on day 8 for LTM. Mean latency to enter the aversive chamber is shown for each genotype. While WRP WT mice learn to avoid the aversive chamber, as indicated by increased latency to enter the chamber, WRP HET and WRP KO mice are impaired in this response. L, Shock sensitivity for each genotype was determined by ethologically scoring to an increased range of foot shock. All three genotypes responded similarly to the aversive stimulus. The amperage used in K for passive-avoidance was 0.3 μA, as indicated by the dashed line. Data are expressed as mean ± SEM. n = 5–8 mice per genotype; *p < 0.05 compared to WT, + p < 0.05 compared to day 1.
Passive avoidance test.
The test was conducted in the two-chambered light–dark apparatus as described above (Med-Associates). Animals were given one conditioning trial per day, with 24 h interposed between each trial for five consecutive days. On each day, the animal was placed into the lighted chamber and after 5 s the door was opened to the darkened chamber. Once the mouse had crossed to the darkened chamber, the door was closed and a 5 s scrambled foot shock (0.3 mA) was given. The animal was removed immediately following the foot shock and returned to its home cage. To examine LTM processes, the mice were returned to the apparatus 72 h (day 8) after the final conditioning trial (day 5), and the latency to cross to the darkened chamber was recorded. Data were expressed as mean ± SEM and analyzed by RMANOVA with test trial [days 1, 2, 3, 4, 5 and 8 (72 h recall)] used as the within-subjects effect and genotype as the between-subjects effect. A posteriori tests were performed with Bonferroni pairwise corrected comparisons, and in all cases p < 0.05 was considered significant; n = 5–8 mice/genotype, *p < 0.05 compared to wild type (WT); +p < 0.05 compared to day 1.
Sensitivity to foot shock.
Mice were acclimated to the apparatus for 60 s. Animals were randomly presented six different intensities (0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 μA) of scrambled foot shock for 2 s. Behavioral responses were videotaped and subsequently scored using the Observer program (Noldus Information Technology) as described previously (Grove et al., 2004). RMANOVA was used to assess within-subject effects of shock intensity and between-subject effects of genotype. Bonferroni corrected pairwise comparisons were used for all post hoc analyses. In all cases, data are expressed as mean ± SEM, and p < 0.05 was considered statistically significant; n = 5–8 mice/genotype.
Results
Direct interaction between WRP and phosphoinositide-containing lipids
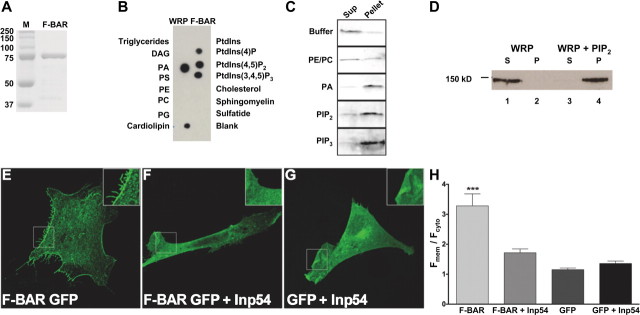
F-BAR domains are functionally diverse membrane-binding domains that recognize distinct membrane lipids. The lipid specificity of the F-BAR domain of WRP and its homologs is currently unknown. We initially assessed the specificity of the WRP F-BAR domain in vitro (Fig. 1A). Purified WRP was screened for its ability to directly bind lipids by overlay onto membranes containing an array of membrane lipid spots. After extensive washing, specific binding was detected using immunoblot analysis. Purified WRP F-BAR domain bound several lipids, including phosphatidic acid (PA), phosphatidylinositol (4,5)-bisphosphate (PIP2), and phosphatidylinositol 3,4,5-trisphosphate (PIP3), and cardiolipin (Fig. 1B). Importantly, binding was not observed with phosphatidylserine (PS), suggesting the WRP F-BAR domain does not bind all negatively charged lipids but instead displays specificity for binding. A positive control, the pleckstrin homology (PH) domain of protein kinase C delta (PKC-δ), only bound PIP2, suggesting that binding was specific under the conditions of this assay (supplemental Fig. S1A, available at www.jneurosci.org as supplemental material). The results from the overlay experiments were confirmed using a cosedimentation assay. This assay detects binding in solution using the more physiological presentation of lipids as liposomes. Liposomes containing 70% phosphatidylethanolamine (PE), 20% phosphatidylcholine (PC) and 10% of the indicated lipids were prepared by extrusion and incubated with the WRP F-BAR. Liposomes were pelleted by centrifugation, and equal volumes of the supernatant and pellet fractions were assayed for the levels of WRP F-BAR by Western blot analysis (Fig. 1C). WRP F-BAR was not enriched in the pellet fraction of either buffer alone or PE/PC-containing liposomes (Fig. 1C, top two panels). When PA, PIP2, or PIP3 were present in the liposomes, however, the WRP F-BAR domain was enriched in the liposome-containing pellet fraction (Fig. 1C, bottom three panels). These results confirm that the WRP F-BAR domain binds PA, PIP2, and PIP3 as demonstrated in Figure 1B.
Figure 1.
Analysis of WRP F-BAR membrane lipid interactions. A, Image of Coomassie-stained SDS-PAGE gel showing molecular weight markers (M) and purified GST-F-BAR domain. B, Representative immunoblot analysis of purified WRP F-BAR domain incubated on a membrane lipid array to detect WRP F-BAR and lipid interactions. Each lipid on the array is labeled. PtdIns, Phosphatidylinositol; DAG, diacylglycerol; Sulfatide, 3-sulfogalactosylceramide. C, Liposome cosedimentation analysis of WRP F-BAR with PE/PC liposomes containing 10% PA, PIP2, or PIP3). Negative controls of buffer alone and PE/PC only are also shown. Protein is visualized using an in-gel fluorescent stain. Sup, Supernatant. D, Cosedimentation analysis of purified full-length WRP. WRP is soluble in buffer alone (lanes 1,2), but cosedimentates with PIP2 in the pellet (lanes 3,4). S, Supernatant, P, pellet. E–G, Representative fluorescent images of Cos7 cells expressing F-BAR GFP (E), F-BAR GFP and the yeast PIP2 phosphatase Inp54 (F), or GFP alone and Inp54 (G). H, Graph depicting the ratio of fluorescence at the membrane (Fmem) to cytosolic fluorescence (Fcyt) localization of the F-BAR GFP in the presence or absence of coexpression with Inp54 or GFP with Inp54. Expression of Inp54 disrupts the membrane targeting of WRP F-BAR GFP. Data represent mean ± SEM, n = 10 for each point; ***p < 0.001, F-BAR GFP versus any other condition.
This result was also replicated for full-length WRP. Recombinant WRP was purified from suspension HEK293 cells and tested for its ability to bind liposomes containing PIP2. When incubated with buffer alone, WRP was completely soluble (Fig. 1D, lanes 1,2). By contrast, it was enriched in the pellet fractions when incubated with PIP2-containing liposomes (lanes 3,4). Thus, the ability of the WRP F-BAR domain to bind membrane lipids is recapitulated by full-length WRP.
To determine whether the affinity of the WRP F-BAR domain for lipids is within physiological ranges, binding constants were measured for PIP2 using the cosedimentation assay (supplemental Fig. S1B–D). Results of these binding assays determined that the apparent Kd for PIP2 binding is 0.76 ± 0.16 μm (supplemental Fig. S1B,C). At this concentration, PA bound the WRP F-BAR to a similar extent when compared to PIP2. PIP3 bound the WRP F-BAR slightly better than PA or PIP2, indicating that all three lipids have an apparent Kd for binding at or near 0.76 μm, although PIP3 may have a higher affinity (supplemental Fig. S1D). Thus, the WRP F-BAR domain is a high-affinity membrane lipid-binding domain comparable to the affinity of other phosphoinositide interaction domains (Itoh et al., 2001; Ford et al., 2002; Tsujita et al., 2006). These results imply the possibility that the membrane localization of WRP in cells is mediated by the specific interactions defined in vitro. To test this possibility, WRP F-BAR GFP was expressed in Cos-7 cells with and without the PIP2 phosphatase Inp54. Inp54 is a yeast phosphatase that is highly specific for PIP2. Expression of Inp54 in Cos-7 cells depletes cellular PIP2 by at least 50% (Raucher et al., 2000) and consequently also results in a loss of PIP3 over time. Analysis of the membrane and cytosolic levels of WRP F-BAR GFP in cells by quantitative fluorescence measurements showed a dramatic relocalization of WRP F-BAR GFP from the membrane to the cytosolic pool upon expression of Inp54 (Fig. 1E–H). These cell-based assays confirm the importance of the phosphoinositide and WRP F-BAR interaction for cellular membrane targeting. Furthermore, the WRP F-BAR domain promotes the formation of and is enriched in filopodial-like protrusions (Fig. 1E inset versus F,G insets).
The F-BAR domains of the srGAP family are all inverse F-BAR domains
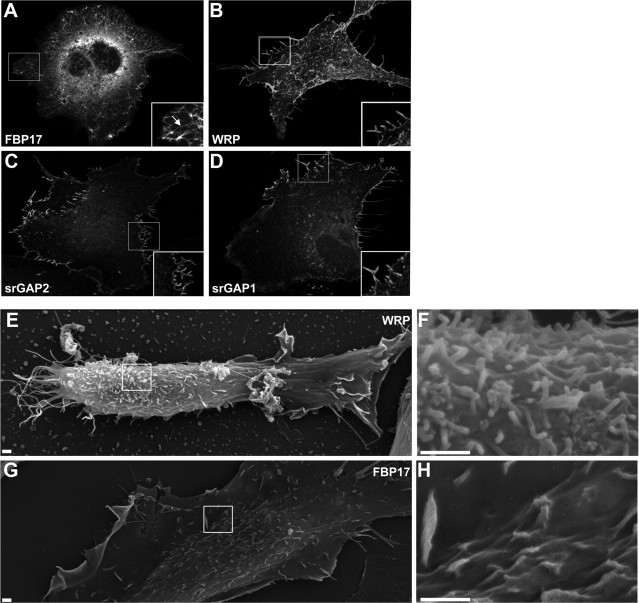
Analyses of the F-BAR domain in proteins such as FBP17 and srGAP2 have shown that it can either produce invagination or evagination of membranes (Kamioka et al., 2004; Tsujita et al., 2006; Carlson and Soderling, 2009; Guerrier et al., 2009). This possibility for WRP was tested in cell-based assays for WRP. Overexpression of FBP17 in Cos-7 cells induced membrane invagination as described previously (Fig. 2A, inset) (Kamioka et al., 2004; Tsujita et al., 2006). In contrast to FBP17, WRP F-BAR did not cause invagination but instead led to outward protrusions along the cell surface (Fig. 2B). These outward protrusions were very similar to those induced by the F-BAR domain of srGAP2, which also facilitated the appearance of outward “filopodial-like” structures when expressed in cells (Fig. 2C) (Guerrier et al., 2009). The srGAP1 F-BAR domain also led to similar membrane protrusions (Fig. 2D), suggesting that this family of Rho-GAPs collectively defines an “inverse F-BAR” or IF-BAR domain that is functionally distinct from other F-BAR domains such FBP17. We further analyzed the ability of the WRP IF-BAR domain to facilitate outward protrusions by scanning electron microscopy. These observations confirmed that unlike the FBP17 F-BAR domain, the WRP IF-BAR domain led to protrusions at the dorsal as well as lateral sides of cells (Fig. 2E–H).
Figure 2.
F-BAR domains of the srGAP/WRP family facilitate outward protrusions of the cell membrane. A–D, Cellular expression of several F-BAR domains in Cos7 cells. Fluorescent maximum projection images of Cos7 cell expressing FBP17-GFP (A), WRP F-BAR-GFP (B), srGAP2 F-BAR-GFP (C), or srGAP1 F-BAR-GFP (D). Boxed regions are depicted as inset images. Arrow in inset of A indicates tubulated membrane. E–H, Representative scanning electron micrographs of cell expressing WRP F-BAR GFP (E, F) or GFP FBP17 (G, H). Boxed regions in E and G represent regions shown in F and H. Scale bars, 1 μm.
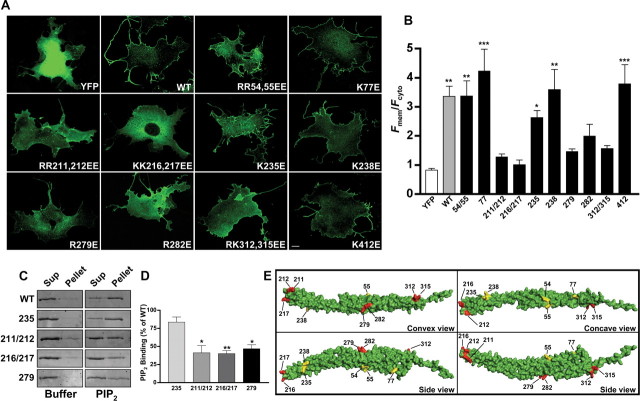
Divergence of the WRP IF-BAR from the canonical F-BAR domains of FBP17 and CIP4 was further suggested by multiple sequence comparisons (supplemental Fig. S2A, available at www.jneurosci.org as supplemental material). These alignments showed that most residues in FBP17 and CIP4 that were identified to interact with membrane lipids are not conserved in WRP. This lack of conservation was not attributable to the method of alignment, because residues involved in dimerization of FBP17 and CIP4 were well conserved within the WRP IF-BAR. Chemical cross-linking experiments suggest that WRP can dimerize/oligomerize, indicating this feature is indeed conserved between the WRP F-BAR and other F-BAR domains (supplemental Fig. S2B, left). Regarding residues involved in lipid binding, only three lipid binding residues in FBP17 (Shimada et al., 2007; Frost et al., 2008) were also found in WRP (Arg54, Arg55, and Lys77). Interestingly, increasing concentrations of KCl disrupt the ability of the WRP F-BAR domain to bind PIP2 in cosedimentation assays analogous to other F-BAR and I-BAR domains (supplemental Fig. S2C) (Saarikangas et al., 2009). This suggests that although there is little conservation of lipid-binding residues between other F-BAR domains and the WRP F-BAR domain, positively charged surface residues of the WRP F-BAR domain interact with phosphoinositides by electrostatic interactions. Thus, a mutagenesis screen of arginine and lysine residues within the WRP IF-BAR domain was performed to identify residues that are required for membrane binding as conducted previously for other F-BAR domains (Fig. 3) (Tsujita et al., 2006; Shimada et al., 2007; Frost et al., 2008). These screens identified eight amino acids that, when mutated to glutamic acid, abolished membrane targeting in cells (Fig. 3A,B). In vitro cosedimentation assays confirmed that these mutations did not completely inhibit F-BAR function but instead significantly reduced the ability of the WRP IF-BAR domain to directly bind purified liposomes (Fig. 3C,D). Additionally, chemical cross-linking also suggested that these mutations did not impair the ability of WRP to dimerize/oligomerize (supplemental Fig. S2B, right). Finally, the effect on GAP activity and the ability to associate with WAVE-1 for one of these mutations, R211E/R212E, was tested (supplemental Fig. S2D,E). These assays confirmed that the effect of these mutations was limited to disrupting the ability of WRP to bind membrane lipids.
Figure 3.
The WRP F-BAR membrane interaction interface maps to the presumed convex surface. A, Representative fluorescent images comparing the subcellular distribution of YFP, WT WRP F-BAR YFP, and several mutants of the WRP F-BAR domain. Identity of each mutant is indicated in the lower right quadrant of each panel. Scale bar, 10 μm. B, Graph comparing each construct against soluble YFP in cells using a ratio of fluorescence at the membrane (Fmem) to cytosolic fluorescence (Fcyt). Note that several mutants are not significantly different from soluble YFP. Data represents mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001, YFP versus any other construct. C, D, Analysis of purified mutant WRP F-BAR domain interactions with PIP2 by cosedimentation assay. C, Representative images of fluorescently stained SDS-polyacrylamide gels showing the amounts of wild-type WRP F-BAR domain (WT), or those of four mutants (K235E, R211E/R212E, K216E/K217E, R279E) in the supernatant (Sup) or pellet fractions. The results of cosedimentation assays with either buffer alone or PIP2-containing liposomes are shown. D, Graph showing the quantitative analysis of three independent experiments for the amount of each mutant protein cosedimentating with PIP2 as a percentage of the wild-type F-BAR domain. Data represent mean ± SEM; *p < 0.05, **p < 0.01, WT versus any other protein. E, Structural model of the WRP F-BAR domain monomer based on alignments to FBP17. Each view of the structure is labeled in the lower right quadrant. Locations of amino acid residues screened by mutagenesis are labeled in each view. Positions that affected membrane targeting are colored red, whereas positions that did not affect subcellular localization are colored yellow.
To approximate where in the WRP F-BAR domain these residues may reside, a structural model was constructed based upon the homology from the known structure of FBP17. Importantly, all eight mutations that disrupted membrane targeting were clustered on the convex surface and tip of the WRP F-BAR structural model (Fig. 3E, red). Residues that did not effect membrane localization in the WRP F-BAR domain (Arg54, Arg55, Lys77) but are required in FBP17 were located on the sides and concave surface (Fig. 3E yellow). These results, suggesting the predicted concave surface does not bind membrane, agree with our in vitro and in vivo data that show the WRP IF-BAR domain does not inwardly tubulate membranes but instead facilitates outward protrusions.
Regulation of dendritic spines by WRP
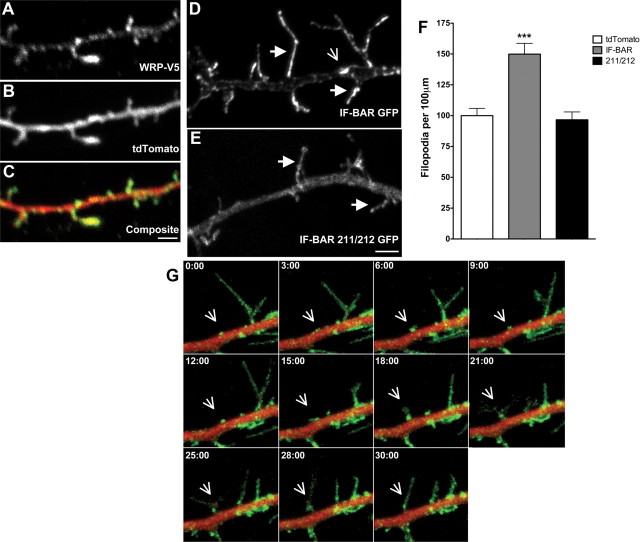
The physiological functions of WRP are likely to be neuronal, as it is almost exclusively expressed in neurons and it is implicated as a candidate gene for intellectual impairments associated with 3p deletions in humans (Endris et al., 2002; Soderling et al., 2002; Shuib et al., 2009). To examine this possibility, epitope-tagged WRP was expressed in neurons along with soluble tdTomato fluorescent protein and the localization of WRP was evaluated by immunofluorescence (Fig. 4A–C). WRP was enriched in dendritic filopodia (Fig. 4A) when compared to the soluble fill tdTomato (Fig. 4B). In a similar manner, the WRP IF-BAR domain was also enriched in filopodial protrusions but was largely absent from the dendritic shaft (Fig. 4D, solid arrows). In contrast, the IF-BAR 211/212 mutant that no longer binds lipids was soluble and not enriched in dendritic filopodia when compared to the dendritic shaft (Fig. 4E). Expression of the WRP IF-BAR domain also led to elevated numbers of dendritic filopodia when compared to neurons expressing either soluble tdTomato fluorescent protein or the IF-BAR 211/212 mutant (Fig. 4F). In addition to the enrichment of the IF-BAR domain in filopodia, there were also clusters of the IF-BAR domain along the surface of the dendritic shaft (Fig. 4D, open arrow). These clusters were further analyzed by live imaging in dendrites of the IF-BAR GFP along with soluble tdTomato fluorescent protein in dendrites (Fig. 4G, open arrow) (see also supplemental Movie 1, available at www.jneurosci.org as supplemental material). Remarkably, IF-BAR domain clusters formed buds along the shaft from which filopodia directly emerged. Together, these data suggest that the WRP IF-BAR domain may facilitate or sense initial membrane deformations on the dendritic shaft that are the sites from which filopodia emerge. Loss of WRP might therefore reduce the development of these structures.
Figure 4.
Localization of the WRP IF-BAR domain to filopodia in neurons requires membrane binding. A–C, Localization of WRP in dendritic filopodia in DIV9 cultured neurons. A, B, Fluorescent immunostaining of V5 epitope-tagged WRP (A) compared to the soluble fill tdTomato (B). C, Composite image showing the localization of WRP in filopodia and puncta along the shaft. D, Fluorescent image of WRP IF-BAR domain in DIV9 cultured neurons. Solid arrows indicate examples of dendritic filopodia showing an enrichment in fluorescent intensity. Open arrow is an example of a cluster of WRP along the shaft. E, Fluorescent image of WRP IF-BAR 211/212. Solid arrows indicate examples of dendritic filopodia, which do not show an enrichment in fluorescent intensity. F, Graph showing the quantitative analysis of dendritic protrusion density in neurons expressing tdTomato fluorescent protein alone or in combination with either WRP IF-BAR GFP or WRP IF-BAR 211/212 GFP. Data are expressed as the mean percentage of tdTomato protrusions ± SEM (%) from three independent experiments; ***p < 0.001, tdTomato versus IF-BAR GFP. G, Live imaging montage of WRP IF-BAR GFP versus soluble tdTomato. Open arrow indicates example cluster of WRP IF-BAR that form buds from which filopodia emerge. Time elapsed is indicated in the top left of each panel.
Because excitatory postsynaptic spines are thought to develop from dendritic filopodia, we next assessed the role of WRP in regulating spine density in vitro (Fig. 5). A conditional allele for WRP was produced by floxing the third exon (second coding exon) with homologous recombination (supplemental Fig. S3A-C, available at www.jneurosci.org as supplemental material). Crossing these mice into a cytomegalovirus-Cre line that expresses Cre in all tissues leads to a heritable deletion of the exon and a complete loss of WRP protein as assessed by immunoprecipitation and immunoblot analysis using a specific anti-WRP antibody (supplemental Fig. S3D, top). Other WRP-related family members are still expressed as determined by immunoblotting with an antibody that recognizes all members of the WRP and srGAP family (supplemental Fig. S3D, bottom). Histological examination of all three genotypes showed the WRP knock-out mice presented enlarged lateral ventricles, the etiology of which is unclear (supplemental Fig. S3E). Interestingly, enlarged lateral ventricles has been associated with terminal deletion of the short arm of chromosome 3 (p25-pter) in humans (Kariya et al., 2000). Although in some animals the cortex appeared thinner, layering of the cortex appeared normal, suggesting that WRP is not critical for cortical migration as previously shown for srGAP2 (supplemental Fig. S3F) (Guerrier et al., 2009).
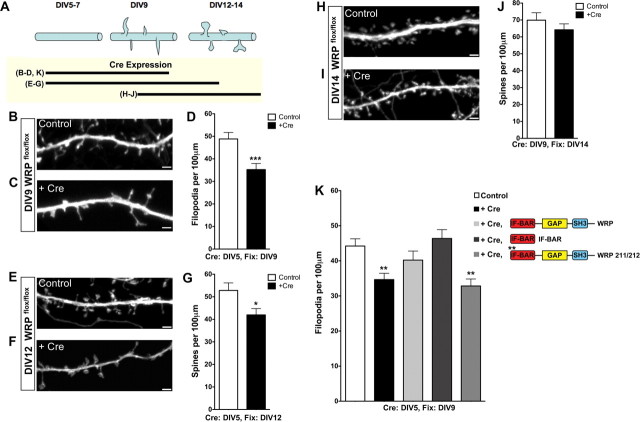
Figure 5.
WRP is required during the onset of spinogenesis. A, Top, Schematic diagram representing the time course of spine development in postnatal hippocampal neuronal cultures. Bottom, Lines represent the different time courses of Cre recombinase expression in B–J to analyze the role of WRP during various developmental stages. Representative images of soluble tomato fluorescent protein (tdTomato) expression to visualize dendritic filopodia in control (B, E, H) versus tdTomato and GFP-Cre recombinase-transfected (C, F, I) WRPflox/flox neurons. Time point (DIV) at which imaging was performed is indicated at the left of each image. In B, C, E, F, the transfection of tdTomato and GFP-Cre was at DIV5 before onset of spinogenesis, whereas in H and I tdTomato and GFP-Cre were transfected at DIV9 after initiation of spinogenesis. D, G, J, Graphs depicting quantitative analysis of dendritic filopodial densities (D) and spine densities (G, J) from control (tdTomato only) versus tdTomato and GFP-Cre-expressing neurons. K, Rescue of filopodia density upon loss of WRP. Control wild-type neurons have a higher density of filopodia compared to knock-out neurons after Cre expression. The reduced filopodial density in the knock-out neurons is rescued by the expression of full-length WRP or the isolated IF-BAR domain. In contrast, expression of WRP that contains mutations in the IF-BAR domain does not rescue knock-out neurons. Legend for K is in the upper right, along with a schematic of the various conditions. Data represent mean ± SEM; *p < 0.05, ***p < 0.001, control versus Cre-expressing neurons. D: n = 30 control, n = 28 +Cre; G, n = 21 control, n = 21 +Cre; J: n = 21 control, n = 22 +Cre; K: n = 30 control, 30 +Cre, 32 +Cre and WRP, 30 +Cre and IF-BAR, 27 +Cre and WRP 211/212. Analysis of significance in K was by ANOVA and Dunnett's multiple comparison post hoc test; **p < 0.01, control versus each condition.
Endogenous WRP protein is expressed throughout the developmental time period of synaptogenesis (supplemental Fig. S3G). Thus the role of WRP during spinogenesis was tested in vitro (Fig. 5) by transfecting cultured neurons from WRPflox/flox mice with GFP-Cre and the soluble fill tdTomato at different time points and then performing image analysis to assess the effect on spine formation and development. Cre-mediated recombination was examined in these primary cultures to ensure that excision of the floxed allele could occur under these experimental conditions (supplemental Fig. S3H). Neuronal cultures were first transfected at DIV5 and then analyzed at DIV9 (Fig. 5A–C), the time period over which the initiation of spinogenesis is maximal. Expression of GFP-Cre led to a significant reduction in dendritic filopodia at DIV9 when compared to neurons transfected with tdTomato alone (“control”) (Fig. 5D). Other measures of dendritic morphology were not significantly altered, including dendrite shaft width (average width: WT = 474 ± 32 nm; +Cre = 453 ± 19 nm). Because only a limited fraction of the neurons within the culture expressed Cre, these results suggest that WRP is required for efficient dendritic filopodial formation in a cell autonomous manner. The combined biochemical and cellular results of Figures 1–5D could suggest that the function of WRP in spine development is limited to the initiation but not maintenance of spines. To test this hypothesis we transfected homozygous floxed neurons with Cre either before or after the onset of dendritic filopodia and determined how the loss of WRP at different developmental stages in vitro would affect spine density. Transfection of Cre at DIV5 (before dendritic filopodial formation) resulted in a 20% reduction in spine density (Fig. 5E–G). In contrast, transfection of Cre at DIV9 (after dendritic filopodia appear) had no significant effect on spine density (Fig. 5H–J). Together, these results strongly support the hypothesis that WRP is required during the early (filopodial) stages of spine development. The role of the WRP IF-BAR domain was next directly tested in a rescue assay for filopodial density (Fig. 5K). As shown in Figure 5D, Cre expression in WRP floxed/floxed neurons lead to a significant reduction in filopodial density when compared to control neurons expressing only tdTomato. Re-expression of full-length WRP led to a return of filopodial density to a level that was not significant from control wild-type levels. Expression of the isolated IF-BAR domain also rescued filopodial protrusion density, whereas expression of full-length WRP that contained point mutations in the IF-BAR domain did not. Together, these data show the WRP IF-BAR domain is sufficient to rescue dendritic filopodial density in the absence of WRP. Thus, the loss of WRP in this assay is primarily due to the loss of the functional IF-BAR domain, which can support membrane protrusions during filopodial formation.
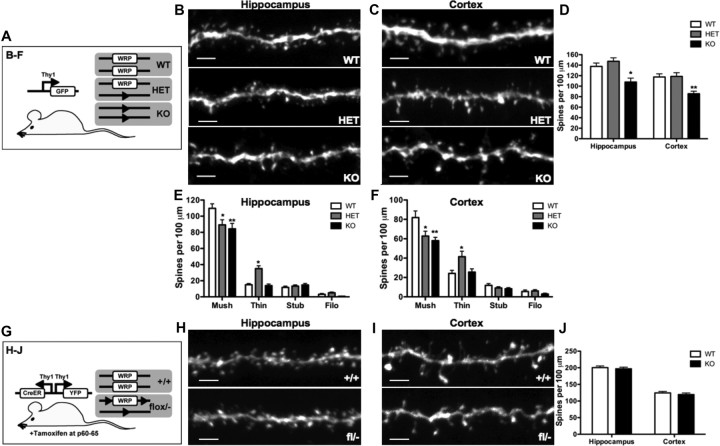
The observation that WRP regulates early spinogenesis in cultured neurons was next tested in vivo using WRP wild-type, heterozygous, and knock-out mice. WRP wild-type, heterozygous, and knock-out mice were crossed into Thy-GFP mice, which express GFP in a mosaic pattern within the hippocampus and cortex (Fig. 6A) (Feng et al., 2000). Confocal image analysis of dendritic sections from both the CA1 region of the hippocampus as well as layers IV/V of the cortex permitted the analysis of in vivo spine density and morphology between littermates of all three WRP genotypes (Fig. 6B–D). A complete loss of WRP resulted in a 24% reduction within CA1 (Fig. 6B) and a 30% reduction within the cortical layers IV/V of spine densities (Fig. 6C) when compared to wild-type and heterozygous mice (Fig. 6D). Spines in vivo display a range of morphologies, and this morphogenic variation is thought to reflect functional differences in spine strength and stability (Matus, 2000; Yuste and Bonhoeffer, 2001; Hayashi et al., 2004; Carlisle and Kennedy, 2005). Classes of spines based on morphology include mushroom-shaped, thin, stubby, and filopodial spines (Lendvai et al., 2000). Mushroom-type spines are believed to represent stable or mature spines that have increased synaptic strength compared to other types of spines. Therefore, the morphology of spines was also classified across all three genotypes (Fig. 6E,F). Surprisingly, although heterozygous mice had a similar total spine density compared to that of wild-type animals, the numbers of mushroom-shaped spines were reduced from wild-type levels and were closer to those of knock-out animals. This was due to an increase in the numbers of thin spines at the expense of the mushroom-type spines. Morphological differences were also observed in vitro between WRP wild-type and knock-out neurons. WRPflox/flox neurons transfected with Cre recombinase at DIV5 showed a significant reduction in the density of both mushroom shaped-spines (WT = 24.0 ± 2.4 spines/100 mm; +Cre = 13.2 ± 1.2 spines/100 mm, p = 0.0026) and thin spines (WT = 15.3 ± 1.2 spines/100 mm; +Cre = 7.8 ± 1.2 spines/100 mm, p = 0.0029) but no significant differences in the stubby or filopodial shaped spines (data not shown). These data, when combined with the in vitro spinogenesis results, suggest the spine abnormalities in the WRP mutant mice may have been due to an early developmental requirement for WRP function. To test this hypothesis, the WRP mice were bred with SLICKV mice, which coexpress CreERT2 along with soluble YFP in a mosaic pattern in neurons (Fig. 6G) (Young et al., 2008). Although our WRP antibodies were not useful for immunostaining to prove loss of WRP in Cre-positive neurons, we did bias the experimental design in two ways toward efficient loss of WRP. First, SLICKV-positive wild-type versus WRP mice that had one allele deleted and one allele floxed were treated with tamoxifen for 5 d by oral gavage starting a P60, an age after the majority of spines have developed. This treatment efficiently induces recombination of the single floxed allele in 95–96% of YFP-positive neurons in the hippocampus and cortex (Young et al., 2008) (Dr. Guoping Feng, McGovern Institute for Brain Research, Massachusetts Institute of Technology, Cambridge, Ma, personal communication). Secondly, treated mice were harvested after 2 weeks following the end of the tamoxifen treatment to allow for protein turnover of previously existing WRP. Comparison of the spine density between wild-type and the conditional WRP knock-outs showed no differences in spine density in the hippocampal CA1 region or in layers IV/V of the cortex (Fig. 6H–J). These data support the hypothesis that WRP is dispensable for maintenance of spine density but is required for spine development.
Figure 6.
WRP regulates mushroom spine development but not maintenance in vivo. A, Schematic depicting the Thy-1 GFP transgene crossed into the WRP WT, heterozygous (HET), and knock-out (KO mice. B, C, Maximum image projections of confocal images from secondary dendrites in the CA1 hippocampal region (B) and secondary dendrites of layer IV/V of the cortex (C). Genotype is indicated in bottom right corner. Scale bars, 3 μm. D, Graph depicting quantitative analysis from mice of spine densities from secondary hippocampal dendrites of WRP WT (n = 22), HET (n = 19), and KO (n = 20) and cortical dendrites of WRP WT (n = 19), HET (n = 19), and KO (n = 18). KO mice have a 24% reduction in spine density in the hippocampus and 30% reduction in the cortex compared to WT and HET littermates. E, F, Graph depicting quantitative analysis from mice of spine morphology from secondary hippocampal dendrites of WRP WT (n = 10), HET (n = 15), and KO (n = 11) (E) and cortical dendrites of WRP WT (n = 11), HET (n = 13), and KO (n = 10) (F). HET and KO mice have a 18.7 and 23.1% reduction in density mushroom type spines in the hippocampus and a 23.3 and 29.1% reduction in the cortex compared to WT. Data represents mean ± SEM; *p < 0.05, **p < 0.01, WT versus KO. G, Schematic depicting the SLICKV transgene crossed into the WRP WT, and KO mice. Time of tamoxifen injections (i.e., P60–65) is indicated at the bottom. H–J, Maximum image projections of confocal images from secondary dendrites in the CA1 hippocampal region (H) and secondary dendrites of layer IV/V of the cortex (I). WRP genotype is indicated in bottom right corner. Scale bars, 3μm. J, Graph depicting quantitative analysis of spine densities of secondary hippocampal dendrites of WRP WT (n = 20) and KO (n = 21) and cortical dendrites of WRP WT (n = 20) and KO (n = 20).
Role of WRP in long-term memory
Because the spine morphology analysis indicated that the circuitry of the WRP heterozygous and knock-out animals may be altered developmentally and because evidence suggests that WRP could be a determinant for mental retardation in patients with distal 3p microdeletions (Endris et al., 2002; Shuib et al., 2009), we also analyzed the behaviors of the WRP+/+, WRP−/+, and WRP−/− mice. All three genotypes performed equally well in tests of motor function and anxiety-like responses (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). For example, latencies to fall from the accelerating rotorod were similar across genotypes (supplemental Fig. S4A). The three genotypes were also indistinguishable in the open field, indicating that anxiety behaviors did not differ (supplemental Fig. S4B). In another test of anxiety-like behavior, responses were not differentiated by genotype in time spent in the lighted or darkened chamber in the light–dark emergence test (supplemental Fig. S4C), the latency to enter the lighted chamber (supplemental Fig. S4D), or the number of transitions between chambers in the light–dark emergence test (supplemental Fig. S4E).
In contrast, loss of WRP led to significant impairments in long-term memory as measured in multiple behavioral tests such as novel object recognition, water maze, and passive avoidance (Fig. 7). Novel object recognition examines hippocampal-dependent nonspatial learning and memory (Roberts et al., 2009). Mice were habituated to an enclosed chamber for 10 min each for 1 d. Following habituation to the empty test chamber, mice were allowed to explore two identical novel objects for 10 min during a single training session. No significant preference for exploring either object was observed (Fig. 7A, Train). Twenty minutes later, mice were allowed to explore one the familiar object versus a novel object in a test of short-term memory (Fig. 7A, STM). All three genotypes preferred the novel object, suggesting that short-term memory was intact. In contrast, when the test was repeated 24 h later, long-term memory was significantly impaired in both the WRP heterozygous and knock-out mice (Fig. 7A, LTM). These deficiencies were not due to differences in exploratory behavior, as total exploration time during the training (Train), short-term memory (STM), and long-term memory (LTM) tests were indistinguishable among genotypes (Fig. 7B).
A deficiency in learning and memory for both WRP heterozygous and knock-out mice was also observed in the Morris water maze test (Roberts et al., 2009) of spatial learning and memory. In this test, mice were trained to escape from a pool of water by swimming to a hidden platform using spatial cues, and performance was evaluated by their latency to reach the platform (swim time). On test day 1, WRP knock-out mice had a significantly longer swim time compared to wild-type and heterozygous littermates (Fig. 7C). This difference, however, was not observed on subsequent days, and by day 6 all three genotypes reached the platform within 10 s. Probe trials on days 2, 4, and 6 (supplemental Fig. S4F–H) further confirmed that all three genotypes performed similarly during the acquisition phase of the water maze, although WRP heterozygous mice were unable to distinguish between the target northeast quadrant versus the adjacent nontarget quadrants northwest and southeast on day 2 (supplemental Fig. S4G, day 2). Together, these data show that overall the three genotypes were able to learn and remember the location of the hidden platform over the 6 d testing period, but WRP heterozygous and knock-out mice showed some slight deficiencies in early training. To test long-term memory, a remote memory probe trial was conducted 21 d after the last acquisition trial on day 28. In this test the wild-type mice were able to remember and distinguish the target quadrant (northeast) from all other nontarget quadrants (Fig. 7D,E). In contrast, both the WRP heterozygous and knock-out mice did not remember or distinguish the target quadrant, showing that loss of WRP was associated with impaired remote memory. Reversal learning was also examined by training the mice to relearn the location of the platform in the quadrant opposite (southwest) to that of the initial target (northeast) (Fig. 7F–I). In reversal, the knock-out mice were significantly impaired relative to the wild-type and heterozygous mice (Fig. 7F). Probe trials were also performed on days 31, 33, and 35 of reversal (Fig. 7G–I). These probe trials confirmed that the wild-type mice could distinguish the new target from each of the nontarget quadrants (Fig. 7G), whereas the WRP knock-out mice could not (Fig. 7I). Interestingly, the WRP heterozygous mice also had some difficulty distinguishing the target from nontarget quadrants in the initial probe trial on day 31 (Fig. 7H). Impairments in the water maze were not due to altered swimming ability, as all three genotypes had similar swim velocities on each day of the acquisition and reversal tests (Fig. 7J). Together, these data demonstrate that both the WRP heterozygous and knock-out mice have impaired long-term memory in the Morris water maze; however, the knock-out animals are clearly more deficient.
WRP mice were tested in a third learning and memory test, passive avoidance (Fig. 7K,L). Mice were trained on this task for five consecutive days and were tested 72 h (day 8) after the final conditioning trial. On the first day of training, no genotype differences were discerned (Fig. 7K). However, on subsequent days the performance of WRP wild-type mice steadily improved, whereas latencies to cross to the darkened chamber for WRP heterozygous and knock-out animals remained low and did not change across training. On day 8 the latencies for the mutant mice were not significantly different from that of day 1 training. Hence, they were severely impaired. By contrast, WRP wild-type animals continued to avoid the chamber paired with foot shock, signifying intact LTM. One reason for the possible genotype effect could be attributed to differences in sensitivities to foot shock. When mice were evaluated for behavioral responses to an increasing intensity of foot shock, no genotype differences were observed up to 5 μAmps (Fig. 7L). Similarly, no genotype distinctions were detected in the latency to enter the darkened versus the lighted chamber in the absence of aversive stimuli (supplemental Fig. S4D). Thus, the impairments of the WRP heterozygous and knock-out animals cannot be attributed to genotype differences in motivation or sensitivities to foot shock. Spatial working memory was also evaluated in the WRP mutant animals using the Y-maze test. In this test, however, there was no genotype effect on performance, suggesting that working memory is intact in the WRP heterozygous and knock-out mice (supplemental Fig. S4I). Together, these behavioral data demonstrate that haploinsufficiency or complete loss of WRP can lead to deficiencies in multiple tests of learning and memory. These deficiencies appear to be strongest on long-term memory, as the mice are normal in short-term and working memory tests. These findings may bear some similarities to the cognitive impairments associated with microdeletions within 3p25-p26 in humans.
Discussion
Much progress has been made in understanding how the morphology and function of dendritic spines are altered by the actin cytoskeleton (Matus, 2000; Honkura et al., 2008; Wegner et al., 2008; Nadif Kasri et al., 2009). In comparison, relatively little is known about the signaling mechanisms that regulate the initial development of dendritic filopodia (Hotulainen et al., 2009). During synaptogenesis, the dendrites of neurons are known to develop numerous filopodia along the shaft. These filopodia are believed to probe the immediate environment for appropriate presynaptic contacts (Ziv and Smith, 1996; Fiala et al., 1998; McAllister, 2007; Toni et al., 2007). Upon contact, some filopodia are stabilized and may form the initial bridge between the dendritic and axonal shafts that is important for subsequent synapse development and maturation (Kayser et al., 2008; Lohmann and Bonhoeffer, 2008). The work presented here shows that WRP facilitates the development of dendritic filopodial protrusions and that loss of WRP leads to a reduction in these protrusions. Because dendritic filopodia may function in early synapse selection, this loss may be correlated to the reduction in mushroom-shaped spines in adult WRP mutant animals. Mechanistically, it appears that the WRP IF-BAR domain can facilitate remodeling of the membrane to initiate filopodial evagination, most likely by binding lipids via a convex surface. The finding that loss of WRP also leads to learning and memory deficits supports the putative linkage between haploinsufficiency of WRP and mental retardation in humans.
Molecular mechanisms linking WRP to early spine development
The anchoring of WRP to subcellular compartments is mediated by the IF-BAR domain via direct interactions with phosphoinositide-rich membranes. WRP binds to phosphoinositide-containing lipids by the N-terminal IF-BAR domain, and the affinity for this interaction (Kd, 0.76 μm) is comparable to other PH (0.1 μm for PLCδ1), and F-BAR domains (0.4 μm for FBP17) (Tsujita et al., 2006). Like other F-BAR domains, the WRP IF-BAR domain appears to interact with lipids via electrostatic contacts between positively charged surface residues and negatively charged lipid head groups. Yet, the interface by which the WRP IF-BAR domain binds lipids is likely to be distinct from the well characterized FBP17/CIP4 lipid interactive surface. Several observations support this notion. For example, most of the conserved arginine and lysine residues in FBP17 and CIP4 that bind to membrane are not conserved in WRP/srGAP IF-BAR domains. Only three residues in WRP are conserved with FBP17 and CIP4: Arg55, Arg56, and Lys77. Mutational analysis of these residues suggest they are not involved in lipid binding in WRP, as these mutants are still targeted to the membrane in cells. Further analysis identified a distinct and unique set of arginine and lysine residues that, when mutated to glutamic acid, reduced membrane and lipid binding as determined by in vivo and in vitro assays.
An additional line of distinction between the WRP IF-BAR and other F-BAR domains is suggested by mapping the location of mutations that disrupt lipid binding in WRP to a structural model of the WRP IF-BAR domain. This structural model was based on homology of the WRP IF-BAR domain to the solved structures of the F-BAR domains of FBP17, CIP4, and FCHo2. Final verification of the structure of the WRP IF-BAR domain, however, will have to be determined experimentally. Despite these limitations, the structural model presented here provides an estimation of the structure of the WRP IF-BAR domain. Using this model as an initial framework for interpreting the mutational data, it appears that all mutations that affect lipid binding are clustered at the predicted tip and convex surface of the WRP IF-BAR domain. This is in stark contrast to the lipid binding surface of all other characterized F-BAR domains, which all use the tip and concave surface. In FBP17 and CIP4, it is thought that membrane binding to the concave surface is part of the mechanism that enables these F-BAR domains to invaginate the membrane. We speculate that the proposed convex binding surface of WRP potentiates its subcellular targeting to outwardly projecting structures, such as dendritic filopodia. Thus, the combined mutational analysis and structural model suggests a mechanism of how the WRP IF-BAR domain may facilitate outward protrusions. Our studies of dendrites lacking WRP also show that dendritic protrusions still form, albeit at a significantly lower frequency, indicating that WRP-independent mechanisms must also exist. One attractive possibility is that other I-BAR (Mattila et al., 2003; Choi et al., 2005) or srGAP family members may function in the absence of WRP during spinogenesis. Recent analysis of the actin cytoskeleton by electron microscopy shows a highly branched network of actin filaments that fills dendritic buds and filopodial structures similar to those that the WRP IF-BAR domain prefers (Korobova and Svitkina, 2010). This highlights the likelihood that to produce outward protrusions, which are energetically unfavored, the cell probably coordinates membrane remodeling with the regulation of actin dynamics to maximize the efficiency of filopodial production. Thus, while our rescue data suggest the WRP IF-BAR domain is sufficient to rescue filopodia density in WRP-deficient neurons, it is quite possible WRP also coordinates actin dynamics through its Rac GAP domain and WAVE-1 binding SH3 domain (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Tashiro and Yuste, 2004). This regulation of actin could also contribute to the spine density and spine maturation and/or morphology differences we observe in vivo. Future work will be needed to examine the role of WAVE-1 in dendritic filopodia and spine morphology.
Role of WRP in long-term memory
The data presented above are consistent with the role of WRP early during synaptogenesis, but indicate that WRP is not required for maintenance of spine number after spines have developed. This fits with the reduced spine density in WRP-null mice, but normal density in mice with the conditional loss of WRP in adulthood. Our biochemical and cellular data also support the possibility that the IF-BAR domain alone has important neurophysiological functions. These results are consistent with the observation that a splice variant of WRP that only contains the IF-BAR domain exists. It also potentially explains studies of human mutations showing that truncation of WRP toward the end of the IF-BAR domain is not associated with mental retardation (Hamdan et al., 2009), whereas disruption of WRP within the IF-BAR domain is associated with retardation (Endris et al., 2002). Because different mutations in WRP lead to different phenotypes regarding mental retardation in humans, there is currently no clear consensus as to whether loss of WRP functions is causative for human cognitive impairments (Endris et al., 2002; Hamdan et al., 2009; Shuib et al., 2009). The behavioral results presented here clearly show that loss of WRP function by deletion of a critical exon within the IF-BAR domain leads to memory deficits in mice, both in heterozygous and knock-out animals. Thus, our data support the contention that haploinsufficiency of WRP in humans is associated with some forms of mental retardation. Interestingly, we did not observe impairments in tests of short-term memory and working memory. The WRP heterozygous and knock-out mice appeared to be most deficient in tests involving long-term memory. Numerous studies have observed altered density and morphology of spines in human patients of mental retardation, suggesting that altered circuitry underlying mental retardation is correlated with these abnormalities (Purpura, 1974; Ramakers, 2002; Nadif Kasri and Van Aelst, 2008). The reduced density of mushroom-shaped spines in the WRP heterozygous and WRP knock-out mice may be a contributing factor to the long-term memory dysfunction of these mice, analogous to density and behavioral alterations seen in dominant negative PAK transgenic animals (Hayashi et al., 2004). Furthermore, our previous analysis of mice expressing a mutant form of WAVE-1, which is impaired for WRP binding, showed the homozygous mice were also deficient in long-term memory (Soderling et al., 2007). Thus, the behavioral characterization of the WRP mice is consistent with our previous analysis of WAVE-1.
Why do the WRP knock-out mice have reduced densities of mushroom-shaped spines? One likely reason is due to the deficiencies in the initiation of spinogenesis during filopodial formation, which critically depends on the IF-BAR domain. Interestingly, the process of spinogenesis occurs early in the postnatal period, during which neuronal circuitry formation is fine-tuned (Konur and Yuste, 2004; Zuo et al., 2005). In the developing cortex, filopodial turnover is highly sensitive to perturbations in sensory experience, and this is thought to be important during synapse formation (Lendvai et al., 2000; Konur and Yuste, 2004). Because loss of WRP alters the normal course of spinogenesis, we speculate that this may permanently alter circuitry formation and function. Such an effect could underlie some of the behavioral deficiencies we observe. Interestingly, the WRP heterozygous mice also have reduced numbers of mushroom type spines, but this is due to an increase in the long/thin spines. Mechanistically, the basis for this difference is unclear, but it can imply that either a separate spine maturation defect is present or that the initiation of dendritic filopodia is sufficiently altered temporally or spatially to affect downstream maturation events. It is important to note, however, that behaviors such as learning and memory are complex, and it is quite possible that factors other than spine defects in the mutant mice also contribute to the phenotype. For example, the WRP knock-out animals have enlarged ventricles, and it is conceivable this morphology difference or other unknown differences also contribute to the phenotypes. It will also be of future interest to analyze the synaptic plasticity of the WRP mutant mice. Previous studies of the WAVE-1 mutant mice suggest late phase long-term potentiation could be especially affected (Soderling et al., 2007).
In summary, this study identifies a WRP-dependent molecular mechanism for potentiating membrane protrusions that likely facilitate the development of spines in vivo in mice. Furthermore, behavioral studies of the WRP heterozygous and knock-out mice reinforce the link between haploinsufficiency of WRP and cognitive impairments observed in 3p− syndrome in humans.
Footnotes
This work was supported by National Institutes of Health Grant R01-NS059957 (S.H.S.), March of Dimes Grant Basil O' Connor Starter Scholar Research Grant 5-FY07-671 (S.H.S.), Dana Foundation Grant (S.H.S.), and Intramural Research Program of the National Institute of Environmental Health Sciences Grant 1ZIAES100221-08 (S.M.D.). We thank Thomas McIntosh and Jihong Tong for their help with the liposome experiments, Tim Oliver for microscopy assistance, Hal Mekeel (Cell and Developmental Biology, University of North Carolina, Chapel Hill, NC) for assistance with SEM, the Duke Transgenic Mouse Facility of the Duke Comprehensive Cancer Center for embryonic stem cell selection and chimeric mouse production, and Guoping Feng for the Thy-1 GFP-O and SLICKV lines of mice. We also thank Jiechun Zhou, Matt Pease, and Caroline J. Kim for their assistance in the behavioral testing and Amar Doshi for assisting with histology.
References
- Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström P. Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol. 2009;272:1–31. doi: 10.1016/S1937-6448(08)01601-8. [DOI] [PubMed] [Google Scholar]
- Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Carlson B, Soderling SH. Mechanisms of cellular protrusions branch out. Dev Cell. 2009;17:307–309. doi: 10.1016/j.devcel.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, Toribara TY, Warner H. Microdetermination of phosphorus. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- Choi J, Ko J, Racz B, Burette A, Lee JR, Kim S, Na M, Lee HW, Kim K, Weinberg RJ, Kim E. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J Neurosci. 2005;25:869–879. doi: 10.1523/JNEUROSCI.3212-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, Maher ER, Tariverdian G, Kirsch S, Karch D, Rappold GA. The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci U S A. 2002;99:11754–11759. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Wagner U, Brinkhaus H, Matus A. Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat Neurosci. 2000;3:887–894. doi: 10.1038/78791. [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- Grove M, Demyanenko G, Echarri A, Zipfel PA, Quiroz ME, Rodriguiz RM, Playford M, Martensen SA, Robinson MR, Wetsel WC, Maness PF, Pendergast AM. ABI2-deficient mice exhibit defective cell migration, aberrant dendritic spine morphogenesis, and deficits in learning and memory. Mol Cell Biol. 2004;24:10905–10922. doi: 10.1128/MCB.24.24.10905-10922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Pellerin S, Dobrzeniecka S, Marineau C, Fombonne E, Mottron L, Lafrenière RG, Drapeau P, Lacaille JC, Rouleau GA, Michaud JL. No association between SRGAP3/MEGAP haploinsufficiency and mental retardation. Arch Neurol. 2009;66:675–676. doi: 10.1001/archneurol.2009.65. [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, Chattarji S, Kirkwood A, Tonegawa S. Altered cortical synaptic morphology and impaired memory consolidation in forebrain-specific dominant-negative PAK transgenic mice. Neuron. 2004;42:773–787. doi: 10.1016/j.neuron.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icking A, Schilling K, Wiesenthal A, Opitz N, Müller-Esterl W. FCH/Cdc15 domain determines distinct subcellular localization of NOSTRIN. FEBS Lett. 2006;580:223–228. doi: 10.1016/j.febslet.2005.11.078. [DOI] [PubMed] [Google Scholar]
- Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem. 2004;279:40091–40099. doi: 10.1074/jbc.M404899200. [DOI] [PubMed] [Google Scholar]
- Kariya S, Aoji K, Akagi H, Fukushima K, Chikumoto E, Ogawa T, Karaki M, Nishizaki K. A terminal deletion of the short arm of chromosome 3: karyotype 46, XY, del (3) (p25-pter); a case report and literature review. Int J Pediatr Otorhinolaryngol. 2000;56:71–78. doi: 10.1016/s0165-5876(00)00409-2. [DOI] [PubMed] [Google Scholar]
- Kayser MS, Nolt MJ, Dalva MB. EphB receptors couple dendritic filopodia motility to synapse formation. Neuron. 2008;59:56–69. doi: 10.1016/j.neuron.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Konur S, Yuste R. Developmental regulation of spine and filopodial motility in primary visual cortex: reduced effects of activity and sensory deprivation. J Neurobiol. 2004;59:236–246. doi: 10.1002/neu.10306. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvai B, Stern EA, Chen B, Svoboda K. Experience-dependent plasticity of dendritic spines in the developing rat barrel cortex in vivo. Nature. 2000;404:876–881. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- Lohmann C, Bonhoeffer T. A role for local calcium signaling in rapid synaptic partner selection by dendritic filopodia. Neuron. 2008;59:253–260. doi: 10.1016/j.neuron.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J Biol Chem. 2003;278:8452–8459. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Nadif Kasri N, Van Aelst L. Rho-linked genes and neurological disorders. Pflugers Arch. 2008;455:787–797. doi: 10.1007/s00424-007-0385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadif Kasri N, Nakano-Kobayashi A, Malinow R, Li B, Van Aelst L. The Rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev. 2009;23:1289–1302. doi: 10.1101/gad.1783809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purpura DP. Dendritic spine “dysgenesis” and mental retardation. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Roberts AC, Díez-García J, Rodriguiz RM, López IP, Luján R, Martínez-Turrillas R, Picó E, Henson MA, Bernardo DR, Jarrett TM, Clendeninn DJ, López-Mascaraque L, Feng G, Lo DC, Wesseling JF, Wetsel WC, Philpot BD, Pérez-Otaño I. Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation. Neuron. 2009;63:342–356. doi: 10.1016/j.neuron.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguiz RM, Gadnidze K, Ragnauth A, Dorr N, Yanagisawa M, Wetsel WC, Devi LA. Animals lacking endothelin converting enzyme-2 are deficient in learning and memory. Genes Brain Behav. 2008;7:418–426. doi: 10.1111/j.1601-183X.2007.00365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T, Yokoyama S. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Shuib S, McMullan D, Rattenberry E, Barber RM, Rahman F, Zatyka M, Chapman C, Macdonald F, Latif F, Davison V, Maher ER. Microarray based analysis of 3p25–p26 deletions (3p− syndrome) Am J Med Genet A. 2009;149A:2099–2105. doi: 10.1002/ajmg.a.32824. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Binns KL, Wayman GA, Davee SM, Ong SH, Pawson T, Scott JD. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat Cell Biol. 2002;4:970–975. doi: 10.1038/ncb886. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Yuste R. Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol Cell Neurosci. 2004;26:429–440. doi: 10.1016/j.mcn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Toni N, Teng EM, Bushong EA, Aimone JB, Zhao C, Consiglio A, van Praag H, Martone ME, Ellisman MH, Gage FH. Synapse formation on neurons born in the adult hippocampus. Nat Neurosci. 2007;10:727–734. doi: 10.1038/nn1908. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Edwards JG, Riley N, Provance DW, Jr, Karcher R, Li XD, Davison IG, Ikebe M, Mercer JA, Kauer JA, Ehlers MD. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Qiu L, Wang D, Zhao S, Gross J, Feng G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat Neurosci. 2008;11:721–728. doi: 10.1038/nn.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Lin A, Chang P, Gan WB. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005;46:181–189. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]