Abstract
Hepatitis E virus (HEV) is an important but extremely understudied human pathogen. Genotypes 1 and 2 are restricted to humans, whereas genotypes 3 and 4 are zoonotic, infecting both humans and pigs. This report describes, for the first time, the successful rescue of infectious HEV in vitro and in vivo from cloned cDNA of a genotype 4 human HEV (strain TW6196E). The complete genomic sequence of the TW6196E virus was determined and a full-length cDNA clone (pHEV-4TW) was assembled. Capped RNA transcripts from the pHEV-4TW clone were replication competent in Huh7 cells and infectious in HepG2/C3A cells. Pigs inoculated intrahepatically with capped RNA transcripts from pHEV-4TW developed an active infection, as evidenced by faecal virus shedding and seroconversion, indicating the successful rescue of infectious genotype 4 HEV and cross-species infection of pigs by a genotype 4 human HEV. To demonstrate the utility of the genotype 4 HEV infectious clone and to evaluate the potential viral determinant(s) for species tropism, four intergenotypic chimeric clones were constructed by swapping various genomic regions between genotypes 1 and 4, and genotypes 1 and 3. All four chimeric clones were replication competent in Huh7 cells, but only the two chimeras with sequences swapped between genotypes 1 and 4 human HEVs produced viruses capable of infecting HepG2/C3A cells. None of the four chimeras was able to establish a robust infection in pigs. The availability of a genotype 4 HEV infectious clone affords an opportunity to delineate the molecular mechanisms of HEV cross-species infection in the future.
Introduction
Hepatitis E virus (HEV) is endemic in many developing countries and is also responsible for sporadic cases of hepatitis E in some industrialized countries (Arankalle et al., 1994; Emerson & Purcell, 2003; Meng, 2010). HEV is classified in the genus Hepevirus of the family Hepeviridae (Meng et al., 2011a). The genome is a single-stranded, positive-sense RNA molecule of ~7.2 kb (Meng, 2010; Okamoto, 2007) and consists of a short 5′NCR, three ORFs and a short 3′NCR followed by a poly(A) tract (Chandra et al., 2008; Emerson et al., 2004a; Okamoto, 2007). A cap structure identified at the 5′ end of the HEV genome is essential for infectivity (Emerson et al., 2001; Zhang et al., 2001). ORF1 encodes a large non-structural protein with several putative functional domains (Ahmad et al., 2011). The activities of the methyl/guanylyltransferase, helicase and RNA-dependent RNA polymerase (RdRp) domains have been demonstrated experimentally (Karpe & Lole, 2010a, b; Magden et al., 2001; Rehman et al., 2008). A proline-rich hypervariable region (HVR) within ORF1 is dispensable for HEV replication in vitro but critical for infectivity in vivo (Pudupakam et al., 2009, 2011). The major intergenotypic sequence variations among HEV strains occur in the HVR, which differs by as much as 71 % in amino acid sequence among HEV isolates in different genotypes (Pudupakam et al., 2009, 2011). ORF2 encodes the capsid protein responsible for virion assembly, immunogenicity and host-cell receptor binding (He et al., 2008; Li et al., 1997, 2000; Meng et al., 2001; Riddell et al., 2000). A junction region (JR) between ORF1 and ORF2 contains a highly conserved stem–loop structure important for HEV replication (Cao et al., 2010; Huang et al., 2007). ORF3 encodes a small multifunctional protein (Chandra et al., 2008; Emerson et al., 2010b; Yamada et al., 2009). Whilst ORF3 is dispensable for virus replication in vitro (Emerson et al., 2006), it is required for infectivity in monkeys and pigs (Graff et al., 2005). The 3′NCR containing the poly(A) tract binds to the RdRp to direct the synthesis of complementary-strand viral RNA (Agrawal et al., 2001).
So far, at least four distinct genotypes of mammalian HEV have been recognized (Emerson et al., 2004a). Genotypes 1 and 2 are restricted to humans, whilst genotypes 3 and 4 are zoonotic (Meng, 2011; Pavio et al., 2010) and have been isolated from several animal species such as pig, deer, wild boar, rat, mongoose and rabbit (Cossaboom et al., 2011; Meng et al., 1997; Nakamura et al., 2006; Sato et al., 2011; Tei et al., 2003). Two putative new genotypes of mammalian HEV have been identified recently in rats from Germany and the USA (Johne et al., 2010; Purcell et al., 2011) and in wild boars from Japan (Sato et al., 2011). Non-human primates were infected experimentally with genotype 3 and 4 swine HEV (Arankalle et al., 2006; Meng et al., 1998b) and, conversely, pigs were experimentally infected with genotype 3 (Halbur et al., 2001) and 4 (Feagins et al., 2008) strains of human HEV. However, an attempt to infect pigs experimentally with genotype 1 and 2 strains of human HEV failed (Meng et al., 1998a). HEV infection in pigs is subclinical, although microscopic hepatitis lesions do develop in infected pigs (Halbur et al., 2001; Meng et al., 1997). HEV infection in humans is mostly asymptomatic or mild, with symptoms such as jaundice, abdominal pain, nausea and vomiting, although significant risk exists in pregnant woman (Navaneethan et al., 2008) and in patients with chronic liver diseases (Hamid et al., 2002; Ramachandran et al., 2004).
Molecular and biological studies of HEV have been limited due to the inability to propagate the virus efficiently in vitro, despite the recent successes in adapting HEV to grow in cell-culture systems (Shukla et al., 2011, 2012; Tanaka et al., 2009). So far, reverse genetics systems are available only for genotype 1 human HEV, genotype 3 human and swine HEV, and avian HEV. Unfortunately, an infectious cDNA clone for genotype 4 human or swine HEV has not been reported to date, even though their full-length sequences have been determined. Thus, the availability of an infectious clone of genotype 4 HEV will be critical for studying the mechanism of HEV cross-species infection in the future.
In this study, we report for the first time the successful construction and in vitro and in vivo characterization of an infectious cDNA clone of a genotype 4 human HEV (strain TW6196E). Additionally, we demonstrated the utility of this new genotype 4 HEV infectious clone for studying the potential viral determinants of cross-species infection by using intergenotypic chimeric viruses.
Results
Genomic organization of genotype 4 human HEV TW6196E
The TW6196E genome comprised 7256 nt, excluding the 3′ poly(A) tail, with 26 nt in the 5′NCR and 88 nt in the 3′NCR. Three major ORFs were identified: ORF1 was 5121 nt (nt 27–5147) and encoded a product of 1706 aa; ORF2 (nt 5186–7168) was 1983 nt and encoded 660 aa; and ORF3, starting 32 bases downstream of ORF1, comprised 339 nt (nt 5178–5516) and encoded a small protein of 112 aa. Sequence analysis revealed that genotype 4 HEV TW6196E shared ~74 % nucleotide sequence identity with genotype 1 HEV strains, 73 % with the genotype 2 Mexican strain, 75 % with genotype 3 strains and 94 % with genotype 4 strains.
Assembly of a full-length cDNA clone of genotype 4 human HEV TW6196E and demonstration of its replication competence in vitro
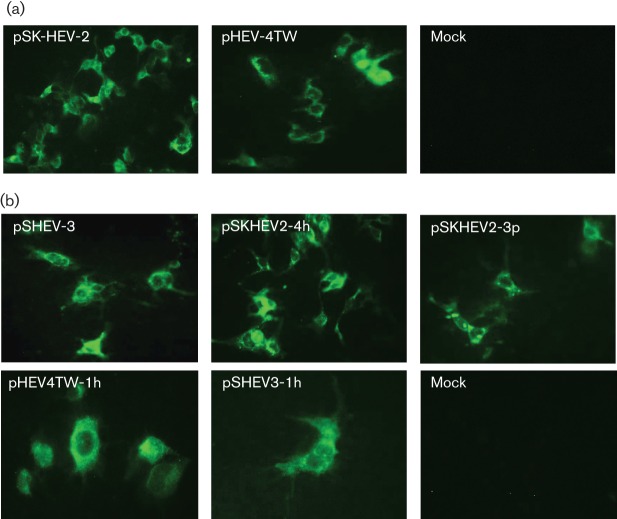
Three overlapping fragments covering the entire TW6196E viral genome were assembled by ligation at appropriate restriction enzyme sites (Fig. 1) and cloned into vector pGEM-7zf(−) to produce a full-length cDNA clone of HEV strain TW6196E, designated pHEV-4TW. The pHEV-4TW clone was positioned downstream of a T7 promoter and a unique SpeI restriction site, allowing linearization of the cDNA immediately downstream of the viral poly(A) sequence. Capped RNA transcripts from the pHEV-4TW clone were transfected into Huh7 cells to determine its replication competence in vitro. The Huh7 hepatocellular carcinoma cell line was used for these experiments, as it has been shown previously that transfected HEV genomic RNA replicates efficiently in Huh7 cells (Emerson et al., 2004b). RNA transcripts from the genotype 1 human HEV infectious clone pSK-HEV-2 were included as a positive control. As expected, ORF2 viral antigens were detected by immunofluorescent assay (IFA) in Huh7 cells transfected with the genotype 1 pSK-HEV-2 clone (Fig. 2a). Positive IFA signals were also detected in Huh7 cells transfected with capped RNAs from the pHEV-4TW clone, indicating the replication competence of this genotype 4 clone in cultured human liver cells (Fig. 2a). No detectable IFA signal was found in mock-transfected cells.
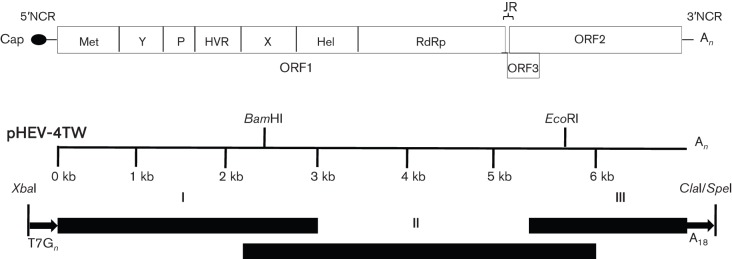
Fig. 1.
Construction of a full-length cDNA clone of genotype 4 human HEV strain TW6196E. The HEV genome consists of three ORFs (ORF1, ORF2 and ORF3), a JR, a short 5′NCR and 3′NCR, a 5′ cap structure and a 3′ poly(A) tract. The putative ORF1 functional domains are indicated: Met, methyltransferase; Y, Y domain; P, papain-like cysteine protease; HVR, hypervariable region; X, X domain; Hel, helicase; RdRp, RNA-dependent RNA polymerase. BamHI and EcoRI are unique restriction sites naturally present in the genomic sequence of strain TW6196E and were used to facilitate construction of the full-length cDNA clone. An XbaI site and a T7 promoter sequence were introduced at the 5′ end of fragment I. Eighteen As and the restriction sites SpeI and ClaI were introduced at the 3′ end of fragment III. Fragments I, II and III were ligated into the pGEM-7zf(−) vector to generate the full-length cDNA clone, designated pHEV-4TW.
Fig. 2.
Replication competence of the genotype 4 human HEV pHEV-4TW clone and intergenotypic chimeric viruses in Huh7 cells. (a) IFA staining for HEV ORF2 antigen in Huh7 cells transfected with capped RNA transcripts from the cDNA clones of genotype 1 human HEV (pSK-HEV-2) and genotype 4 human HEV (pHEV-4TW). (b) IFA staining for HEV ORF2 antigen in Huh7 cells transfected with capped RNA transcripts from the cDNA clones of genotype 3 swine HEV (pSHEV-3) and each of the four intergenotypic chimeric viruses (pSKHEV2-4h, pSKHEV2-3p, pHEV4TW-1h and pSHEV3-1h). Mock, Mock-infected cells.
Intergenotypic chimeric HEVs are replication competent in vitro
Capped RNA transcripts from the full-length cDNA clones of each of four HEV chimeras (pSKHEV2-4h, pSKHEV2-3p, pHEV4TW-1h and pSHEV3-1h; see Methods and Fig. 5) were transfected into Huh7 cells to determine their replication competence. The in vitro viability of the chimeras pSKHEV2-4h and pSKHEV2-3p has been demonstrated previously (Feagins et al., 2011) and was confirmed in this study (Fig. 2b; Table 1). ORF2-specific viral antigen was detected in the cytoplasm of Huh7 cells transfected with the genotype 3 HEV clone pSHEV-3, as well as in those transfected with the HEV chimeric clones pHEV4TW-1h and pSHEV3-1h (Fig. 2b; Table 1). No detectable IFA signal was found in mock-transfected cells. It is known that the ORF2 protein is synthesized from subgenomic RNA; thus, detection of ORF2 protein in Huh7 cells can be used as a surrogate marker for replication of the chimeric viruses. The results indicated that the ORF2 gene, along with the JR, ORF3 and 3′NCR, could be functionally exchanged between genotypes 1 and 4, and between genotypes 1 and 3, with respect to their ability to replicate in vitro.
Table 1. Replication competence in Huh7 cells and infectivity in HepG2/3A cells and in pigs of wild-type swine HEV genotype 3 and human HEV genotype 4, as well as intergenotypic chimeric HEV clones.
| Inoculum | Replication competence in Huh7 cells | Infectivity in HepG2/3A cells | Infectivity in pigs |
| pSK-HEV-2 (genotype 1 human HEV) | + | + | −* |
| pSHEV-3 (genotype 3 swine HEV) | + | + | + |
| pHEV-4TW (genotype 4 human HEV) | + | + | + |
| Chimera pSKHEV2-4h | + | + | − |
| Chimera pSKHEV2-3p | + | − | − |
| Chimera pHEV4TW-1h | + | + | − |
| Chimera pSHEV3-1h | + | − | −† |
The lack of infectivity in pigs of the genotype 1 human HEV was tested in a previous study (Meng et al., 1998a).
One pig inoculated with chimera pSHEV3-1h repeatedly tested positive for HEV RNA in bile collected during necropsy at 8 weeks p.i.
Recovery of infectious viruses in vitro from Huh7 cells transfected with genotype 4 pHEV-4TW and intergenotypic chimeric clones pSKHEV2-4h and pHEV4TW-1h
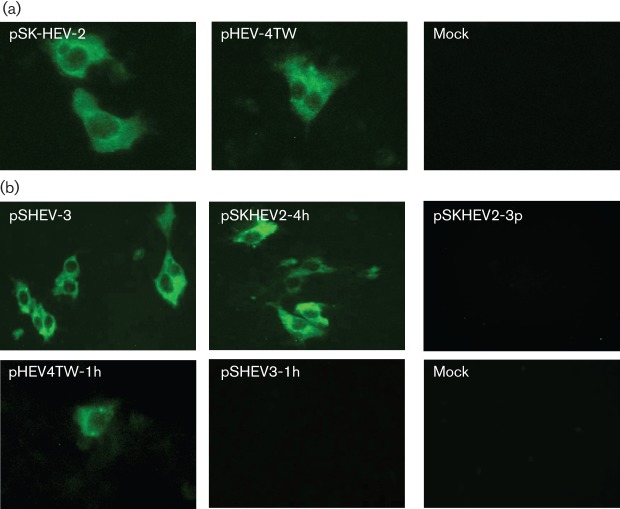
To determine whether the genotype 4 HEV pHEV-4TW clone was capable of producing infectious virus in vitro, HepG2/C3A cells were inoculated with lysates of Huh7 cells transfected with the capped RNA of clone pHEV-4TW. The HepG2/C3A hepatocellular carcinoma cell line was used for the in vitro infectivity assay, as previous studies have demonstrated that HepG2/C3A cells are infected more efficiently with HEV than Huh7 cells (Emerson et al., 2010b). Cytoplasmic fluorescence signals were detected by IFA in HepG2/C3A cells infected with lysates of Huh7 cells transfected with genotype 4 clone pHEV-4TW as well as with the wild-type genotype 1 clone pSK-HEV-2 (Fig. 3a; Table 1), indicating the presence of infectious virus particles derived from the genotype 4 HEV clone in Huh7 cell lysates. HepG2/C3A cells inoculated with lysates from mock-transfected cells did not show any evidence of infection.
Fig. 3.
Infectivity of the genotype 4 HEV pHEV-4TW clone and intergenotypic chimeric viruses in HepG2/C3A cells. (a) IFA staining for HEV ORF2 antigen in HepG2/C3A cells infected with lysates of Huh7 cells transfected with capped RNA transcripts from the cDNA clones of genotype 1 human HEV (pSK-HEV-2) and genotype 4 human HEV (pHEV-4TW). (b) IFA staining for HEV ORF2 antigen of HepG2/C3A cells infected with lysates of Huh7 cells transfected with capped RNA transcripts from the cDNA clone of the genotype 3 swine HEV (pSHEV-3) and each of the four intergenotypic chimeric viruses (pSKHEV2-4h, pSKHEV2-3p, pHEV4TW-1h and pSHEV3-1h).
To investigate further the effects of the JR, ORF3, ORF2 and 3′NCR exchange between HEV genotypes on viral particle production, we examined whether infectious chimeric virus particles could be recovered from the transfected Huh7 cells. Cells that were IFA positive for ORF2 protein were observed in HepG2/C3A cells inoculated with lysates of Huh7 cells transfected with chimeras pSKHEV2-4h and pHEV4TW-1h, as well as with the wild-type genotype 3 HEV pSHEV-3, indicating that assembly of chimeric infectious virus particles had occurred (Fig. 3b; Table 1). However, no detectable IFA signal was found in HepG2/C3A cells inoculated with lysates of Huh7 cells transfected with the RNA transcripts from chimeras pSKHEV2-3p and pSHEV3-1h (Fig. 3b; Table 1) or with lysates from mock-transfected cells.
Capped RNA transcripts from the genotype 4 HEV pHEV-4TW clone are infectious when injected intrahepatically into the livers of specific-pathogen-free (SPF) pigs
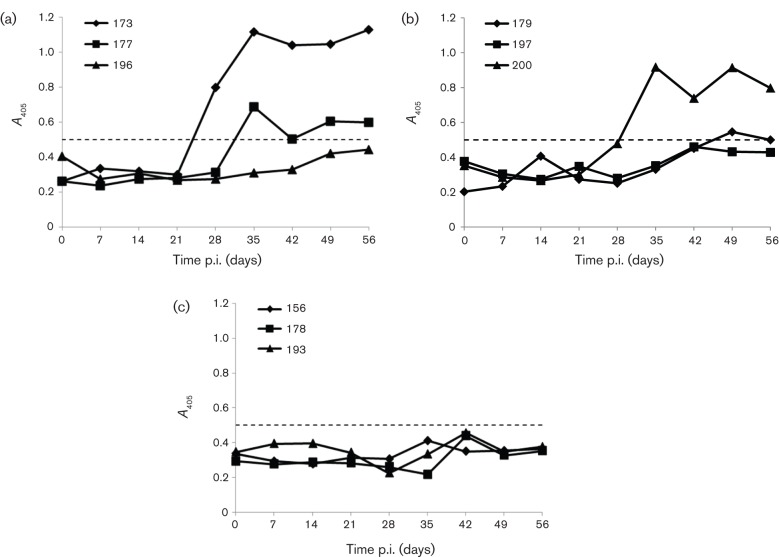
The in vivo infectivity of the genotype 4 HEV pHEV-4TW clone was tested by direct intrahepatic inoculation of capped RNA transcripts into the liver of SPF pigs. All three pigs in the wild-type genotype 3 HEV pSHEV-3 (positive control) group shed virus in their faeces starting at 1 week post-inoculation (p.i.). In addition, transient viraemia and seroconversion to HEV antibodies starting at 4 weeks p.i. were observed in pig #173 and #177 (Table 2; Fig. 4a). HEV RNA was also detected in a bile sample collected from a single pig (#196) during necropsy at 8 weeks p.i. (Table 2). Similarly to the genotype 3 swine HEV pSHEV-3 group, faecal virus shedding was detected in all three pigs inoculated with the genotype 4 human HEV pHEV-4TW clone starting as early as 1 week p.i. and lasting for 2–4 weeks (Table 2). Seroconversion to HEV antibodies was detected at 5–7 weeks p.i. in two of the three pigs (#179 and #200) inoculated with the genotype 4 HEV clone pHEV-4TW (Fig. 4b), indicating the successful rescue of infectious genotype 4 HEV in pigs from the cloned cDNA. Sequence analysis of the PCR-positive products confirmed that the viruses recovered from the infected pigs originated from their respective inocula (data not shown).
Table 2. Detection of HEV RNA in faecal/serum samples using ORF2-specific primers in pigs inoculated intrahepatically with capped RNA transcripts from genotype 3 swine HEV pSHEV-3, genotype 4 HEV pHEV-4TW and intergenotypic chimeras pHEV4TW-1h and pSHEV3-1h.
Pigs in the groups inoculated with the intergenotypic chimeric clones pSKHEV2-4h, pSKHEV2-3p and pHEV4TW-1h remained negative throughout the study and are not included in the table. Necropsy was at 56 days p.i. IC, Intestinal contents.
| Inoculum | Pig # | HEV RNA detection* | HEV RNA at necropsy | ||||||||||
| 0 | 7 | 14 | 21 | 28 | 35 | 42 | 49 | 56 | Liver | Bile | IC | ||
| pSHEV-3 | 173 | −/− | −/− | +/− | +/− | −/+ | −/− | −/− | −/− | −/− | − | − | − |
| 177 | −/− | −/− | −/− | −/− | +/+ | −/− | −/− | −/− | −/− | − | − | − | |
| 196 | −/− | +/− | −/− | −/− | −/− | −/− | −/− | +/− | −/− | − | + | − | |
| pHEV-4TW | 179 | −/− | −/− | −/− | −/− | −/− | +/− | +/− | −/− | +/− | − | − | − |
| 197 | −/− | +/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− | − | − | − | |
| 200 | −/− | +/− | +/− | +/− | +/− | −/− | −/− | −/− | −/− | − | − | − | |
| pSHEV3-1h | 157 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | − | + | − |
| 162 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | − | − | − | |
| 164 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | − | − | − | |
| PBS | 156 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | − | − | − |
| 178 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | − | − | − | |
| 193 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | − | − | − | |
Positive (+) or negative (–) HEV RNA detection in faecal/serum samples at the indicated days p.i.
Fig. 4.
Seroconversion to anti-HEV IgG in pigs inoculated with capped RNA transcripts from the genotype 3 swine HEV pSHEV-3 clone and genotype 4 human HEV pHEV-4TW clone. An ELISA (A405) was used to detect IgG anti-HEV antibodies in pigs #173, #177 and #196 inoculated intrahepatically with capped RNA transcripts from the wild-type pSHEV-3 clone (positive control) (a), pigs #179, #197 and #200 inoculated intrahepatically with capped RNA transcripts from the genotype 4 human HEV pHEV-4TW clone (b), and pigs #156, #178 and #193 inoculated intrahepatically with PBS (negative control) (c). The ELISA cut-off value is shown as a dotted line.
ORF2 and ORF3 genes from genotype 3 and 4 HEV fail to enable chimeric viruses based on the backbone of genotype 1 HEV to infect pigs
Capped RNA transcripts from the four intergenotypic chimeric viruses were inoculated intrahepatically into the livers of pigs to assess their infectivity in vivo. The cell lysates of Huh7 cells transfected with chimeric clones pSKHEV2-3p and pSKHEV2-4h failed to infect pigs in a previous study (Feagins et al., 2011). Due to the inefficient propagation of HEV in Huh7 cells, the amount of infectious virus that can be recovered from transfected Huh7 cells is very low and thus may not be sufficient to elicit an infection in pigs. Therefore, in this study, in addition to testing the infectivity of the two new chimeric viruses pHEV4TW-1h and pSHEV3-1h, we also re-evaluated the ability of chimeras pSKHEV2-4h and pSKHEV2-3p to infect pigs by direct intrahepatic inoculation of pigs with large quantities of capped RNA transcripts. All pigs inoculated with each of the four chimeras remained seronegative throughout the study (data not shown) and none shed virus in faeces or developed viraemia (data not shown), with the exception of a single pig inoculated with the chimera pSHEV3-1h, which repeatedly tested positive for HEV RNA in bile collected during necropsy at 8 weeks p.i. (Table 2). The positive PCR result from bile of the single pig was verified with a confirmatory nested RT-PCR using a different set of primers specific for the ORF1 gene (Table S1, available in JGV Online). The PCR product was also confirmed by sequencing. As expected, the three pigs in the negative-control group (#156, #178 and #193) remained negative for HEV antibodies and viral RNA throughout the study (Fig. 1c).
Discussion
The availability of infectious cDNA clones of genotypes 1 and 3 and avian HEV has provided ample opportunities to study the molecular mechanism of HEV replication and pathogenesis (Feagins et al., 2011; Pudupakam et al., 2011; Shukla et al., 2011). Unfortunately, an infectious cDNA clone of genotype 4 human or swine HEV has not been available. Therefore, the main objective of this study was to develop an infectious cDNA clone of a genotype 4 human HEV. We also aimed to utilize the newly developed genotype 4 HEV infectious clone to explore potential viral genetic determinants for species tropism using the intergenotypic chimeric virus strategy.
We demonstrated that capped RNA transcripts from the genotype 4 human HEV pHEV-4TW cDNA clone were replication competent in Huh7 cells and produced infectious virus particles capable of infecting HepG2/C3A cells. Importantly, we showed that SPF pigs inoculated intrahepatically with capped RNA transcripts from the genotype 4 HEV pHEV-4TW clone became infected, as evidenced by faecal virus shedding and seroconversion to HEV antibodies. Therefore, the results from this study indicated that the genotype 4 human HEV pHEV-4TW cDNA clone is infectious in vitro and in vivo, and is able to cross species barriers and infect pigs. The patterns of faecal virus shedding and seroconversion in pigs inoculated intrahepatically with RNA transcripts from the pHEV-4TW clone were similar to those inoculated with the genotype 3 swine HEV clone pSHEV-3 and with the live infectious virus stock of the genotype 4 human HEV (Feagins et al., 2008).
To demonstrate further the utility of the genotype 4 HEV infectious clone and to gain a better understanding of the molecular mechanism of HEV interspecies transmission, we subsequently constructed four intergenotypic chimeric viruses by swapping the ORF2 gene along with its JR, ORF3 and 3′NCR between genotype 1 and genotype 4, and between genotype 1 and genotype 3, HEV infectious clones. We showed that all four intergenotypic chimeric viruses (pSKHEV2-4h, pSKHEV2-3p, pHEV4TW-1h and pSHEV3-1h) were replication competent in Huh7 cells, indicating that the intergenotypic gene swaps did not disrupt virus replication competence. However, only chimeras pSKHEV2-4h and pHEV4TW-1h along with the wild-type genotype 3 and 4 clones produced infectious viruses capable of infecting HepG2/C3A cells. The ability of chimeras pSKHEV2-4h and pHEV4TW-1h to produce infectious viral particles in vitro suggests that the ORF2 gene, along with its adjacent JR, ORF3 and 3′NCR, is exchangeable between genotype 4 human HEV and genotype 1 human HEV. The lack of detectable infectivity in cells inoculated with chimeras pSKHEV2-3p and pSHEV3-1h could be explained by a low level of virus replication in the transfected Huh7 cells that led to reduced yields of infectious viruses. In addition, it is possible that the viral genes from genotype 1 human HEV may not be compatible with those from genotype 3 swine HEV, suggesting a need for homologous gene sequences for efficient assembly of infectious virus particles.
To determine further the infectivity of the four intergenotypic chimeric viruses, capped RNA transcripts from the four chimeras were inoculated intrahepatically into the livers of pigs. The RNA transcripts from chimeras pSKHEV2-4h and pSKHEV2-3p failed to infect pigs, as evidenced by a lack of faecal virus shedding, viraemia or seroconversion, which further confirmed the previous results in pigs inoculated with lysates of transfected cells (Feagins et al., 2011). The ORF2 capsid protein is thought to play an important role in HEV attachment and entry (He et al., 2008). However, the genotype 1 human HEV did not acquire the ability to infect pigs when its ORF2 capsid gene was replaced with that of the genotype 4 (chimera pSKHEV2-4h) or genotype 3 (chimera pSKHEV2-3p) HEV that naturally infects pigs. Binding to the cells followed by virus entry is just the first step required for virus replication. Once inside the cell, the virus must interact correctly with host-cell factors to reproduce its genome and package the progeny virions (de Chassey et al., 2008; Fernandez-Garcia et al., 2009; Panavas et al., 2005). Some of the host factors that are key determinants for virus replication are virus specific (Jiang et al., 2006; Quadt et al., 1993; Strauss & Strauss, 1994). The inability of the intergenotypic chimeras pSKHEV2-4h and pSKHEV2-3p to infect pigs could be explained by the lack of essential host factors in the swine cells required by the ORF1 non-structural proteins of the genotype 1 human HEV backbone in both chimeras. Non-structural viral genes dispensable for virus replication are also known to play a role in modulating the host immune response (Chen et al., 2010; Sun et al., 2010), and the viral mechanisms to counteract the host immune response can be species specific as well. The ORF1 non-structural proteins of the genotype 1 human HEV backbone in chimeras pSKHEV2-4h and pSKHEV2-3p may not contain species-specific amino acid residues important for evading the immune response in the pig, and therefore the virus cannot establish an infection in pigs. It has been demonstrated recently that insertion of a human rRNA sequence in the HVR of ORF1 from a genotype 3 HEV renders the virus capable of infecting cells of multiple species (Shukla et al., 2011), suggesting a potential role for ORF1, particularly the HVR, in interspecies infection.
Similarly, pigs inoculated with chimeric viruses pSHEV3-1h and pHEV4TW-1h containing the 5′NCR and ORF1 from genotype 3 and genotype 4 HEV, respectively, did not show any evidence of HEV infection, except for a single pig inoculated with chimera pSHEV3-1h, which had detectable HEV RNA in bile at necropsy. Capsid proteins determine the host range of many RNA viruses (Geissler et al., 1999; Moss & Racaniello, 1991). However, it has been demonstrated recently that strains Sar-55 and Akluj of human HEV can infect LLC-PK1 swine cells (Shukla et al., 2011), suggesting that human and swine HEV strains may share at least one cell receptor. The detection of HEV RNA in the bile of one pig (#157) inoculated with chimera pSHEV3-1h suggested that ORF2 protein from genotype 1 human HEV was able to bind to a swine cell receptor, although much less efficiently than the wild-type pSHEV-3 and pHEV-4TW virus. The lack of viraemia or faecal virus shedding in pig #157 may reflect an extremely low and undetectable level of HEV in the sera and faeces. ORF3 protein is responsible for virion egress from infected cells (Emerson et al., 2010a; Yamada et al., 2009), probably through interactions with cellular proteins (Korkaya et al., 2001; Surjit et al., 2006; Zafrullah et al., 1997). ORF3 from genotype 1 human HEV in chimeras pSHEV3-1h and pHEV4TW-1h may not interact efficiently with pig cellular proteins that are potentially involved in viral release, which may have a negative impact on the replication of the chimeric viruses. The stem–loop structures within the JR and 3′NCR of HEV are known to interact with the viral RdRp and possibly with host factors required for virus replication (Agrawal et al., 2001; Cao et al., 2010). The lack of viraemia and faecal virus shedding in the pigs inoculated with chimera pSHEV3-1h or pHEV4TW-1h may be a reflection of the functional importance of these species-specific protein–protein interactions during HEV replication.
In conclusion, we have constructed successfully, for the first time, an infectious cDNA clone of genotype 4 human HEV, and demonstrated that this full-length cDNA clone of the genotype 4 human HEV is infectious in cultured human liver cells and in pigs. Additionally, we demonstrated the utility of this new genotype 4 HEV infectious clone in studying the mechanism of HEV cross-species infection. Although the molecular mechanisms leading to interspecies transmission of HEV remain unknown, the results from this study suggest that species-specific interactions between viral and cellular proteins are probably important for successful replication of HEV in the host. Further in-depth structural and functional studies are required to define the sequence(s) that confer the ability of HEV to overcome the host range barriers.
Methods
Virus and cells.
The infectious stock of the genotype 4 human HEV strain TW6196E used in this study was prepared as a 10 % suspension of faecal material collected from an experimentally infected pig (Feagins et al., 2008). A subclone of a human hepatoma cell line (Huh7-S10-3) was maintained in Dulbecco’s modified Eagle’s medium containing 10 % FBS at 37 °C and 5 % CO2. Human HepG2/C3A (CRL-10741) hepatocellular carcinoma cells were purchased from the ATCC and propagated in Eagle’s minimum essential medium supplemented with 10 % FBS at 37 °C and 5 % CO2.
Determination of the complete genomic sequence of the genotype 4 human HEV TW6196E.
Total RNA was extracted from the TW6196E virus stock as described previously (Cooper et al., 2005; Huang et al., 2002) and used in 3′RACE, 5′RACE and primer walking to determine the complete genomic sequence of TW6196E. Briefly, a TW6196E ORF2-specific primer based on an available 346 bp sequence was used with 3′RACE to amplify the 3′ end of the viral genome. Degenerate primers were subsequently designed based on the sequences of the 3′RACE product and other known genotype 4 strains to amplify a 1–2 kb fragment upstream of the 3′-end RACE product using a SuperScript III One-step RT-PCR System with a Platinum Taq High Fidelity kit (Invitrogen). PCR products of the expected sizes were gel purified (Geneclean II kit; QBiogene), cloned into a TA vector (Invitrogen) and sequenced. The primer-walking method was used to amplify the majority of the genome (from the 5′ to the 3′ end), with the exception of the extreme 5′-end sequence, which was determined using 5′ RACE.
Construction of a full-length cDNA clone of genotype 4 human HEV TW6196E.
Three overlapping genomic fragments covering the entire viral genome were amplified by PCR using three different sets of PCR primers (Fig. 1). Fragment I was amplified with primers XbaIT7F and BamHI2450R (Table S1). The forward primer XbaIT7F contained an engineered XbaI site and a T7 promoter sequence followed by the extreme 5′ end of the viral genome. Fragment II, representing the middle portion of the viral genome, was amplified with primers BamHI2439F and EcoRI5865R, which partially overlapped fragments I and III. Fragment III was amplified with primers EcoRI5854F and ClaI7256R. The reverse primer ClaI7256R introduced 18 As and SpeI and ClaI sites at the 3′ end of the viral genome for linearization and cloning purposes. Fragments I and II were ligated simultaneously into pGEM-7zf(−) vector between the XbaI and EcoRI sites in a three-way ligation reaction. Fragment III was subsequently ligated into the pGEM-7zf(−) vector using the EcoRI site partially overlapping fragment II and the ClaI site in the pGEM-7zf(−) vector. A QuikChange Site-Directed Mutagenesis kit (Stratagene) was subsequently used to delete the built-in T7 promoter sequence in the pGEM-7zf(−) vector. Unwanted mutations in the clone were corrected by site-directed mutagenesis to produce a final full-length cDNA clone designated pHEV-4TW.
Construction of intergenotypic chimeric HEV cDNA clones.
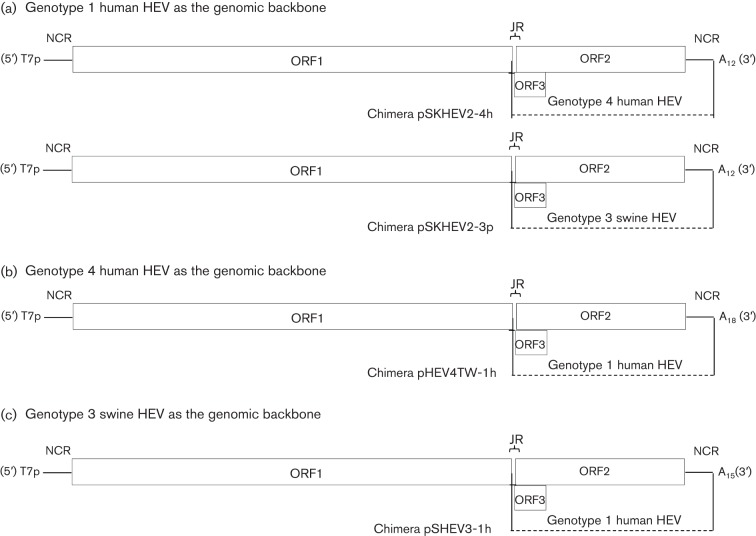
The pHEV-4TW clone from this study along with HEV genotype 1 human (pSK-HEV-2) and genotype 3 swine (pSHEV-3) HEV infectious clones (Emerson et al., 2001; Huang et al., 2005) were used as the backbones to construct four intergenotypic chimeric virus clones. The ORF2 gene along with its adjacent JR, ORF3 and 3′NCR were swapped between genotypes 1 and 4, and between genotypes 1 and 3, to produce four chimeric clones: pSKHEV2-4h, pSKHEV2-3p, pHEV4TW-1h and pSHEV3-1h (Fig. 5). By using the genotype 4 human HEV clone pHEV-4TW as the genomic backbone, chimera pHEV4TW-1h was generated by replacing the JR, ORF3, ORF2 and 3′NCR region of genotype 4 human HEV with that of genotype 1 human HEV (Fig. 5b). To construct the chimera pSHEV3-1h, the genotype 3 swine HEV infectious clone pSHEV-3 was used as the genomic backbone, in which the ORF2 gene along with the JR, ORF3 and 3′NCR region of the genotype 3 swine HEV was replaced with that of the genotype 1 human HEV (Fig. 5c). Standard and fusion PCRs with primer pairs SbfI-F/JR-R and JR-F/BglIIClaI-R (chimera pHEV4TW-1h), and ApaI-F/JR-R and JR-F/XbaI-R (chimera pSHEV3-1h) (Table S1) were used to produce the final fragments, which were cloned into the corresponding region of the genotype 4 or genotype 3 infectious clone HEV backbone. The construction of chimeras pSKHEV2-4h and pSKHEV2-3p has been described previously (Feagins et al., 2011) (Fig. 5a).
Fig. 5.
Schematic diagrams of the strategies used for construction of the four intergenotypic chimeric HEVs. (a) Two chimeric viruses were constructed using the genotype 1 human HEV infectious clone pSK-HEV-2 as the genomic backbone: chimera pSKHEV2-4h with the JR+ORF2+ORF3+3′NCR of genotype 4 human HEV replacing that of genotype 1 human HEV; and chimera pSKHEV2-3p with the JR+ORF2+ORF3+3′NCR of genotype 3 swine HEV replacing that of genotype 1 human HEV. (b) Chimera pHEV4TW-1h was constructed using the genotype 4 human HEV infectious clone pHEVTW-4 as the genomic backbone, with the JR+ORF2+ORF3+3′NCR of genotype 1 human HEV replacing that of genotype 4 human HEV. (c) Chimera pSHEV3-1h was constructed by using the genotype 3 swine HEV infectious clone pSHEV-3 as the genomic backbone, with the JR+ORF2+ORF3+3′NCR of genotype 1 human HEV replacing that of genotype 3 swine HEV.
In vitro RNA transcription.
The plasmid DNAs from each parental and chimeric clone were linearized with AclI (pSKHEV2-4h and pSKHEV2-3p), XbaI (pSHEV3-1h and pSHEV-3), BglII (pHEV4TW-1h) or SpeI (pHEV-4TW). The capped RNA transcripts for the in vivo infectivity assay were synthesized using an mMessage mMachine T7 kit (Ambion), as described previously (Córdoba et al., 2011), diluted with 3 vols cold RNase/DNase/proteinase-free PBS, frozen immediately on dry ice and used for direct intrahepatic inoculation of pigs the next day.
In vitro transfection.
Full-length capped RNA transcripts from genotype 3 pSHEV-3 and genotype 4 pHEV-4TW, and each of the four intergenotypic chimeric clones, were transfected into Huh7 cells, as described previously (Córdoba et al., 2011; Emerson et al., 2004b). At 3 days post-transfection at 34.5 °C, the cells were trypsinized and replated in eight-well LabTek chamber slides, and the incubation was continued at 34.5 °C for a further 3 days. The remaining cells were transferred to a T75 flask to produce virus stock.
Preparation of cell lysates and in vitro infectivity assays.
To generate virus stock for the in vitro infectivity assays, confluent monolayers of Huh7 cells in T75 flasks transfected with the genotype 3 swine HEV, genotype 4 human HEV and each of the four chimeric clones were harvested at 9 days post-transfection and centrifuged. The pellets were resuspended in 0.9 ml water. After three freeze–thaw cycles (−80 °C), 0.1 ml 10× concentrated PBS was added and debris was removed by centrifugation. Confluent monolayers of HepG2/C3A cells grown in eight-well glass chamber slides were inoculated with 100 µl each cell lysate. After 5 h incubation at 34.5 °C, 0.4 ml growth medium was added to each well and the cells were incubated for 6 days at 34.5 °C.
Immunofluorescence assay (IFA).
Transfected and infected cells were fixed and stained on day 6, as described previously (Córdoba et al., 2011; Emerson et al., 2004b). Briefly, cells were fixed with acetone, washed with PBS and incubated with chimpanzee 1313 anti-HEV convalescent-phase serum. Cells were rinsed with PBS and overlaid with Alexa Fluor 488-conjugated goat anti-human IgG (Molecular Probes). The slides were washed in PBS, and Vectashield fluorescent mounting medium (Vector Laboratories) was added for viewing by fluorescence microscopy.
Intrahepatic inoculation of SPF pigs with capped RNA transcripts derived from the genotype 4 human HEV pHEV-4TW and intergenotypic chimeric virus clones.
Twenty-one 4-week-old, HEV-seronegative SPF pigs were divided into seven groups (A–G) of three pigs each and each group was housed separately. Pigs were inoculated intrahepatically using an ultrasound-guided technique into four different sites of the liver, with ~200 µl capped RNA transcripts per injection site. The three pigs in group A were each injected with 0.8 ml of the capped RNA transcripts from the genotype 3 HEV pSHEV-3 clone (positive control). Group B pigs were each injected similarly with 0.8 ml capped RNA transcripts from the genotype 4 human HEV pHEV-4TW clone. Pigs in groups C–F were each injected with 0.8 ml capped RNA transcripts from the HEV chimeric clones pSKHEV2-4h, pSKHEV2-3p, pHEV4TW-1h and pSHEV3-1h, respectively. Pigs in group G were injected with PBS (negative control). The animals were monitored for evidence of HEV infection for a total of 8 weeks, at which time they were necropsied. Serum samples and faecal swab material were collected prior to inoculation and weekly thereafter. At necropsy, samples of serum, bile, liver and intestinal contents were collected from each pig.
Serology.
Serum samples were tested for anti-HEV IgG using an ELISA, essentially as described previously (Córdoba et al., 2011). Pre-immune and hyperimmune IgG anti-HEV swine sera were included as negative and positive controls, respectively. The ELISA cut-off value was calculated as the mean negative-control absorbance value plus 3 sd.
RNA extraction and RT-PCR.
Viraemia, faecal virus shedding and the presence of HEV RNA in bile, liver and intestinal content were tested by a nested RT-PCR using primers targeting the ORF2 gene (Table S1), as described previously (Córdoba et al., 2011). For detection of wild-type genotype 3 HEV pSHEV-3 and the chimeric pSKHEV2-3p, nested RT-PCR primer pairs 3pEX-F/3pEX-R (first round) and 3pIN-F/3pIN-R (second round) were used (Table S1). Primer pairs 4hEX-F/4hEX-R (first round) and 4hIN-F/4hIN-R (second round) were used for detection of wild-type genotype 4 HEV pHEV-4TW and chimeric pSKHEV2-4h. Primer pairs 1hEX-F/1hEX-R (first round) and 1hIN-F/1hIN-R (second round) were used to detect chimeras pHEV4TW-1h and pSHEV3-1h.
To confirm the positive PCR results, two additional sets of nested RT-PCR primers targeting a different genomic region were designed within the ORF1 gene of genotype 3 swine and genotype 4 human HEV (Table S1). The PCR products were sequenced to confirm that the viruses recovered from pigs originated from their respective inoculum.
Acknowledgements
We thank Drs Suzanne U. Emerson and Robert H. Purcell at the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA, for kindly providing us with the genotype 1 HEV clone pSK-HEV-2, Huh7 S-10 cells and the anti-HEV chimpanzee 1313 antiserum. We thank Dr E. Riedesel for assistance with the ultrasound-guided inoculation procedure, and Cody Branstad, Michelle Hemann and Kevin O’Neill for assistance with the animal work. This study was supported in part by grants from the National Institutes of Health (R01AI050611 and R01AI074667).
Footnotes
A supplementary table is available with the online version of this paper.
References
- Agrawal S., Gupta D., Panda S. K. (2001). The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282, 87–101 10.1006/viro.2000.0819 [DOI] [PubMed] [Google Scholar]
- Ahmad I., Holla R. P., Jameel S. (2011). Molecular virology of hepatitis E virus. Virus Res 161, 47–58 10.1016/j.virusres.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle V. A., Chadha M. S., Tsarev S. A., Emerson S. U., Risbud A. R., Banerjee K., Purcell R. H. (1994). Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc Natl Acad Sci U S A 91, 3428–3432 10.1073/pnas.91.8.3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle V. A., Chobe L. P., Chadha M. S. (2006). Type-IV Indian swine HEV infects rhesus monkeys. J Viral Hepat 13, 742–745 10.1111/j.1365-2893.2006.00759.x [DOI] [PubMed] [Google Scholar]
- Cao D., Huang Y.-W., Meng X.-J. (2010). The nucleotides on the stem–loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol 84, 13040–13044 10.1128/JVI.01475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Taneja S., Kalia M., Jameel S. (2008). Molecular biology and pathogenesis of hepatitis E virus. J Biosci 33, 451–464 10.1007/s12038-008-0064-1 [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhou X., Lunney J. K., Lawson S., Sun Z., Brown E., Christopher-Hennings J., Knudsen D., Nelson E., Fang Y. (2010). Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J Gen Virol 91, 1047–1057 10.1099/vir.0.016212-0 [DOI] [PubMed] [Google Scholar]
- Cooper K., Huang F. F., Batista L., Rayo C. D., Bezanilla J. C., Toth T. E., Meng X. J. (2005). Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol 43, 1684–1688 10.1128/JCM.43.4.1684-1688.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdoba L., Huang Y.-W., Opriessnig T., Harral K. K., Beach N. M., Finkielstein C. V., Emerson S. U., Meng X.-J. (2011). Three amino acid mutations (F51L, T59A, and S390L) in the capsid protein of the hepatitis E virus collectively contribute to virus attenuation. J Virol 85, 5338–5349 10.1128/JVI.02278-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom C. M., Córdoba L., Dryman B. A., Meng X.-J. (2011). Hepatitis E virus in rabbits, Virginia, USA. Emerg Infect Dis 17, 2047–2049 10.3201/eid1711.110428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugué S., Meiffren G., Pradezynski F., Faria B. F. & other authors (2008). Hepatitis C virus infection protein network. Mol Syst Biol 4, 230 10.1038/msb.2008.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Purcell R. H. (2003). Hepatitis E virus. Rev Med Virol 13, 145–154 10.1002/rmv.384 [DOI] [PubMed] [Google Scholar]
- Emerson S. U., Zhang M., Meng X.-J., Nguyen H., St Claire M., Govindarajan S., Huang Y. K., Purcell R. H. (2001). Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc Natl Acad Sci U S A 98, 15270–15275 10.1073/pnas.251555098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Anderson D., Arankalle A., Meng X.-J., Purdy M., Schlauder G. G., Tsarev S. A. (2004a). Hepevirus. In Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, pp. 851–855 Edited by Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. London: Elsevier/Academic Press [Google Scholar]
- Emerson S. U., Nguyen H., Graff J., Stephany D. A., Brockington A., Purcell R. H. (2004b). In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J Virol 78, 4838–4846 10.1128/JVI.78.9.4838-4846.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Nguyen H., Torian U., Purcell R. H. (2006). ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J Virol 80, 10457–10464 10.1128/JVI.00892-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Nguyen H. T., Torian U., Burke D., Engle R., Purcell R. H. (2010a). Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol 84, 9059–9069 10.1128/JVI.00593-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Nguyen H. T., Torian U., Burke D., Engle R., Purcell R. H. (2010b). Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol 84, 9059–9069 10.1128/JVI.00593-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins A. R., Opriessnig T., Huang Y. W., Halbur P. G., Meng X. J. (2008). Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J Med Virol 80, 1379–1386 10.1002/jmv.21223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins A. R., Córdoba L., Sanford B. J., Dryman B. A., Huang Y.-W., LeRoith T., Emerson S. U., Meng X.-J. (2011). Intergenotypic chimeric hepatitis E viruses (HEVs) with the genotype 4 human HEV capsid gene in the backbone of genotype 3 swine HEV are infectious in pigs. Virus Res 156, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia M. D., Mazzon M., Jacobs M., Amara A. (2009). Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5, 318–328 10.1016/j.chom.2009.04.001 [DOI] [PubMed] [Google Scholar]
- Geissler K., Schneider K., Fleuchaus A., Parrish C. R., Sutter G., Truyen U. (1999). Feline calicivirus capsid protein expression and capsid assembly in cultured feline cells. J Virol 73, 834–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Nguyen H., Yu C., Elkins W. R., St Claire M., Purcell R. H., Emerson S. U. (2005). The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J Virol 79, 6680–6689 10.1128/JVI.79.11.6680-6689.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbur P. G., Kasorndorkbua C., Gilbert C., Guenette D., Potters M. B., Purcell R. H., Emerson S. U., Toth T. E., Meng X. J. (2001). Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol 39, 918–923 10.1128/JCM.39.3.918-923.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid S. S., Atiq M., Shehzad F., Yasmeen A., Nissa T., Salam A., Siddiqui A., Jafri W. (2002). Hepatitis E virus superinfection in patients with chronic liver disease. Hepatology 36, 474–478 10.1053/jhep.2002.34856 [DOI] [PubMed] [Google Scholar]
- He S., Miao J., Zheng Z., Wu T., Xie M., Tang M., Zhang J., Ng M.-H., Xia N. (2008). Putative receptor-binding sites of hepatitis E virus. J Gen Virol 89, 245–249 10.1099/vir.0.83308-0 [DOI] [PubMed] [Google Scholar]
- Huang F. F., Haqshenas G., Guenette D. K., Halbur P. G., Schommer S. K., Pierson F. W., Toth T. E., Meng X. J. (2002). Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol 40, 1326–1332 10.1128/JCM.40.4.1326-1332.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Haqshenas G., Kasorndorkbua C., Halbur P. G., Emerson S. U., Meng X. J. (2005). Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J Virol 79, 1552–1558 10.1128/JVI.79.3.1552-1558.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Opriessnig T., Halbur P. G., Meng X. J. (2007). Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J Virol 81, 3018–3026 10.1128/JVI.02259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Serviene E., Gal J., Panavas T., Nagy P. D. (2006). Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol 80, 7394–7404 10.1128/JVI.02686-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Plenge-Bönig A., Hess M., Ulrich R. G., Reetz J., Schielke A. (2010). Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol 91, 750–758 10.1099/vir.0.016584-0 [DOI] [PubMed] [Google Scholar]
- Karpe Y. A., Lole K. S. (2010a). NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol 84, 3595–3602 10.1128/JVI.02130-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y. A., Lole K. S. (2010b). RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol 84, 9637–9641 10.1128/JVI.00492-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H., Jameel S., Gupta D., Tyagi S., Kumar R., Zafrullah M., Mazumdar M., Lal S. K., Xiaofang L. & other authors (2001). The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J Biol Chem 276, 42389–42400 10.1074/jbc.M101546200 [DOI] [PubMed] [Google Scholar]
- Li T. C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M. A., Uchida T., Takeda N., Miyamura T. (1997). Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol 71, 7207–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Riddell M. A., Seow H. F., Takeda N., Miyamura T., Anderson D. A. (2000). Recombinant subunit ORF2.1 antigen and induction of antibody against immunodominant epitopes in the hepatitis E virus capsid protein. J Med Virol 60, 379–386 [DOI] [PubMed] [Google Scholar]
- Magden J., Takeda N., Li T., Auvinen P., Ahola T., Miyamura T., Merits A., Kääriäinen L. (2001). Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol 75, 6249–6255 10.1128/JVI.75.14.6249-6255.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Dai X., Chang J. C., Lopareva E., Pillot J., Fields H. A., Khudyakov Y. E. (2001). Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288, 203–211 10.1006/viro.2001.1093 [DOI] [PubMed] [Google Scholar]
- Meng X. J. (2010). Recent advances in hepatitis E virus. J Viral Hepat 17, 153–161 10.1111/j.1365-2893.2009.01257.x [DOI] [PubMed] [Google Scholar]
- Meng X. J. (2011). From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res 161, 23–30 10.1016/j.virusres.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.-J., Purcell R. H., Halbur P. G., Lehman J. R., Webb D. M., Tsareva T. S., Haynes J. S., Thacker B. J., Emerson S. U. (1997). A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A 94, 9860–9865 10.1073/pnas.94.18.9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.-J., Halbur P. G., Haynes J. S., Tsareva T. S., Bruna J. D., Royer R. L., Purcell R. H., Emerson S. U. (1998a). Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol 143, 1405–1415 10.1007/s007050050384 [DOI] [PubMed] [Google Scholar]
- Meng X.-J., Halbur P. G., Shapiro M. S., Govindarajan S., Bruna J. D., Mushahwar I. K., Purcell R. H., Emerson S. U. (1998b). Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol 72, 9714–9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Anderson D. A., Arankalle V. A., Emerson S. U., Harrison T. J., Jameel S., Okamoto H. (2011a). Hepeviridae. London: Elsevier/Academic Press [Google Scholar]
- Moss E. G., Racaniello V. R. (1991). Host range determinants located on the interior of the poliovirus capsid. EMBO J 10, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Takahashi K., Taira K., Taira M., Ohno A., Sakugawa H., Arai M., Mishiro S. (2006). Hepatitis E virus infection in wild mongooses of Okinawa, Japan: demonstration of anti-HEV antibodies and a full-genome nucleotide sequence. Hepatol Res 34, 137–140 10.1016/j.hepres.2005.10.010 [DOI] [PubMed] [Google Scholar]
- Navaneethan U., Al Mohajer M., Shata M. T. (2008). Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int 28, 1190–1199 10.1111/j.1478-3231.2008.01840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. (2007). Genetic variability and evolution of hepatitis E virus. Virus Res 127, 216–228 10.1016/j.virusres.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Panavas T., Serviene E., Brasher J., Nagy P. D. (2005). Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A 102, 7326–7331 10.1073/pnas.0502604102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavio N., Meng X.-J., Renou C. (2010). Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet Res 41, 46 10.1051/vetres/2010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudupakam R. S., Huang Y. W., Opriessnig T., Halbur P. G., Pierson F. W., Meng X. J. (2009). Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J Virol 83, 384–395 10.1128/JVI.01854-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudupakam R. S., Kenney S. P., Córdoba L., Huang Y.-W., Dryman B. A., Leroith T., Pierson F. W., Meng X.-J. (2011). Mutational analysis of the hypervariable region of hepatitis E virus reveals its involvement in the efficiency of viral RNA replication. J Virol 85, 10031–10040 10.1128/JVI.00763-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell R. H., Engle R. E., Rood M. P., Kabrane-Lazizi Y., Nguyen H. T., Govindarajan S., St Claire M., Emerson S. U. (2011). Hepatitis E virus in rats, Los Angeles, California, USA. Emerg Infect Dis 17, 2216–2222 10.3201/eid1712.110482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadt R., Kao C. C., Browning K. S., Hershberger R. P., Ahlquist P. (1993). Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 90, 1498–1502 10.1073/pnas.90.4.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran J., Eapen C. E., Kang G., Abraham P., Hubert D. D., Kurian G., Hephzibah J., Mukhopadhya A., Chandy G. M. (2004). Hepatitis E superinfection produces severe decompensation in patients with chronic liver disease. J Gastroenterol Hepatol 19, 134–138 10.1111/j.1440-1746.2004.03188.x [DOI] [PubMed] [Google Scholar]
- Rehman S., Kapur N., Durgapal H., Panda S. K. (2008). Subcellular localization of hepatitis E virus (HEV) replicase. Virology 370, 77–92 10.1016/j.virol.2007.07.036 [DOI] [PubMed] [Google Scholar]
- Riddell M. A., Li F., Anderson D. A. (2000). Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J Virol 74, 8011–8017 10.1128/JVI.74.17.8011-8017.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Sato H., Naka K., Furuya S., Tsukiji H., Kitagawa K., Sonoda Y., Usui T., Sakamoto H. & other authors (2011). A nationwide survey of hepatitis E virus (HEV) infection in wild boars in Japan: identification of boar HEV strains of genotypes 3 and 4 and unrecognized genotypes. Arch Virol 156, 1345–1358 10.1007/s00705-011-0988-x [DOI] [PubMed] [Google Scholar]
- Shukla P., Nguyen H. T., Torian U., Engle R. E., Faulk K., Dalton H. R., Bendall R. P., Keane F. E., Purcell R. H., Emerson S. U. (2011). Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus–host recombinant. Proc Natl Acad Sci U S A 108, 2438–2443 10.1073/pnas.1018878108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P., Nguyen H. T., Faulk K., Mather K., Torian U., Engle R. E., Emerson S. U. (2012). Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86, 5697–5707 10.1128/JVI.00146-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. (1994). The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58, 491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Chen Z., Lawson S. R., Fang Y. (2010). The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol 84, 7832–7846 10.1128/JVI.00217-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Oberoi R., Kumar R., Lal S. K. (2006). Enhanced α1 microglobulin secretion from hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J Biol Chem 281, 8135–8142 10.1074/jbc.M509568200 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takahashi M., Takahashi H., Ichiyama K., Hoshino Y., Nagashima S., Mizuo H., Okamoto H. (2009). Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J Clin Microbiol 47, 1906–1910 10.1128/JCM.00629-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei S., Kitajima N., Takahashi K., Mishiro S. (2003). Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362, 371–373 10.1016/S0140-6736(03)14025-1 [DOI] [PubMed] [Google Scholar]
- Yamada K., Takahashi M., Hoshino Y., Takahashi H., Ichiyama K., Nagashima S., Tanaka T., Okamoto H. (2009). ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J Gen Virol 90, 1880–1891 10.1099/vir.0.010561-0 [DOI] [PubMed] [Google Scholar]
- Zafrullah M., Ozdener M. H., Panda S. K., Jameel S. (1997). The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol 71, 9045–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Purcell R. H., Emerson S. U. (2001). Identification of the 5′ terminal sequence of the SAR-55 and MEX-14 strains of hepatitis E virus and confirmation that the genome is capped. J Med Virol 65, 293–295 10.1002/jmv.2032 [DOI] [PubMed] [Google Scholar]