Abstract
The PB1-F2 protein of the influenza A viruses (IAVs) can act as a virulence factor in mice. Its contribution to the virulence of IAV in swine, however, remains largely unexplored. In this study, we chose two genetically related H3N2 triple-reassortant IAVs to assess the impact of PB1-F2 in virus replication and virulence in pigs. Using reverse genetics, we disrupted the PB1-F2 ORF of A/swine/Wisconsin/14094/99 (H3N2) (Sw/99) and A/turkey/Ohio/313053/04 (H3N2) (Ty/04). Removing the PB1-F2 ORF led to increased expression of PB1-N40 in a strain-dependent manner. Ablation of the PB1-F2 ORF (or incorporation of the N66S mutation in the PB1-F2 ORF, Sw/99 N66S) affected the replication in porcine alveolar macrophages of only the Sw/99 KO (PB1-F2 knockout) and Sw/99 N66S variants. The Ty/04 KO strain showed decreased virus replication in swine respiratory explants, whereas no such effect was observed in Sw/99 KO, compared with the wild-type (WT) counterparts. In pigs, PB1-F2 did not affect virus shedding or viral load in the lungs for any of these strains. Upon necropsy, PB1-F2 had no effect on the lung pathology caused by Sw/99 variants. Interestingly, the Ty/04 KO-infected pigs showed significantly increased lung pathology at 3 days post-infection compared with pigs infected with the Ty/04 WT strain. In addition, the pulmonary levels of interleukin (IL)-6, IL-8 and gamma interferon were regulated differentially by the expression of PB1-F2. Taken together, these results indicate that PB1-F2 modulates virus replication, virulence and innate immune responses in pigs in a strain-dependent fashion.
Introduction
In pigs, influenza A virus (IAV) infections bear remarkable resemblance to influenza infections in humans with respect to the clinical presentations and pathological features of the disease. The A/swine/IA/15/30 (H1N1) virus, also known as classical swine H1N1 virus (cH1N1), was the first virus strain isolated and associated with the aetiology of swine influenza in this species (Shope, 1931). The cH1N1 viruses were the predominant IAV subtype circulating in North American swine populations for nearly 70 years (Vincent et al., 2008). Since the late 1990s, however, the epidemiology of swine influenza in North American pigs has changed significantly with the introduction of triple-reassortant (TR) influenza viruses. Initially introduced as H3N2 TR IAVs, these viruses contain gene segments from human-origin (HA, NA and PB1), avian-origin (PB2 and PA) and classical swine-origin (NS, NP and M) influenza viruses. Some of these viruses have expanded their host range and caused outbreaks in domestic turkeys in North Carolina, Minnesota and Ohio, USA, in 2003–2004 (Choi et al., 2004). A unique feature shared by TR viruses is the maintenance of the triple-reassortant internal gene (TRIG) cassette, which consists of the human-origin PB1, the avian-origin PA and PB2, and the swine-origin M, NP and NS gene segments. Currently, TR strains of the H3N2, H1N2 and H1N1 are enzootic in the swine population of the USA (Vincent et al., 2008). There have been at least three independent introductions of human H3N2 subtype viruses (as reassortants with the TRIG cassette) into the swine population, leading to phylogenetic clusters I, II, III and IV (the latter derived from cluster III) (Gramer et al., 2007; Kumar et al., 2011; Nfon et al., 2011; Richt et al., 2003; Webby et al., 2004).
The virulence of IAVs is considered a polygenic trait (Fukuyama & Kawaoka, 2011). One such virulence factor is PB1-F2, which is an 87–90 aa long protein encoded by an alternative (+1) ORF within the PB1 gene segment 2 (Chen et al., 2001). Full-length PB1-F2 is expressed by most avian IAVs, but it is truncated in cH1N1s and in human H1N1s isolated after 1950. Truncated versions of PB1-F2 have been also reported in some human H3N2 isolates and in TR swine viruses (Zell et al., 2007). Overall, the effects of PB1-F2 in relation to influenza–host interaction are not completely understood and appear to be cell-type-, virus-strain- and even host-specific. We have previously shown that restoring the PB1-F2 ORF in the context of a 2009 pandemic H1N1 strain has minimal effects in swine; however, the contribution of PB1-F2 to the virulence of IAVs in natural hosts (like birds and swine) has been limited (Marjuki et al., 2010; Pena et al., 2012; Schmolke et al., 2011).
In this study, we compared the effects of PB1-F2 expression, in the background of two TR H3N2 IAVs, with respect to virus replication and cell death using ex vivo infected porcine tissues and changes in pathogenicity and host responses in pigs.
Results
Generation of PB1-F2 recombinant influenza viruses
The sequences of the PB1-F2 proteins from viruses A/swine/Wisconsin/14094/99 (H3N2) (Sw/99) and A/turkey/Ohio/313053/04 (H3N2) (Ty/04) are 90 aa long and differ only at four amino acid positions from each other. The Sw/99 differs from Ty/04 at positions S23N, R44K, F57S and F83S (Fig. S1, available in JGV Online). The functional roles of these polymorphisms have not been established. The Ty/04 PB1-F2 is 54.4 % divergent from the 1918 H1N1 PB1-F2 and shares 74.4 % amino acid sequence identity with the 1968 A/Hong Kong/1/68 (H3N2) pandemic strain and 86.6 % with the A/Wuhan/359/95 (H3N2) seasonal strain (Fig. S1).
The Sw/99 or Ty/04 strains were cloned by reverse genetics (RG) and then the PB1-F2 ORF in each strain was abolished by mutating the ATG start codon to ACG and introducing two stop codons downstream of the gene. The strategy maintains the PB1 ORF wild-type (WT) sequence. Since the Sw/99 virus is of low virulence to pigs, we introduced the N66S mutation in the PB1-F2 as described by Conenello et al. (2007). The viruses used in this study are summarized in Table 1. Beyond PB1-F2, there are significant nucleotide and amino acid differences between the Ty/04 and Sw/99 viruses (Table S1); however, in this study we concentrated our efforts on determining the contribution of PB1-F2 for both of these strains and compared the effects on isogenic backgrounds.
Table 1. Influenza viruses used in this study.
| Virus | Acronym |
| A/turkey/Ohio/313053/04 (H3N2) | Ty/04 WT |
| A/turkey/Ohio/313053/04 (H3N2) PB1-F2 knockout | Ty/04 KO |
| A/swine/Wisconsin/14094/99 (H3N2) | Sw/99 WT |
| A/swine/Wisconsin/14094/99 (H3N2) PB1-F2 knockout | Sw/99 KO |
| A/swine/Wisconsin/14094/99 (H3N2) PB1-F2 N66S | Sw/99 N66S |
The effect of PB1-F2 ORF for polymerase activity in the background of the Sw/99 and Ty/04 polymerase complexes was assessed in vitro using a minireplicon assay as described previously (Pena et al., 2011). We found no differences in polymerase activity in the presence or absence of the PB1-F2 ORF in segment 2, suggesting that PB1-F2 does not affect polymerase activity of any of these H3N2 TR strains (data not shown).
More recently, the expression of a 12th gene product from influenza viruses has been described that, like PB1-F2, is expressed from an alternative AUG start codon in segment 2 (Wise et al., 2009). The resulting product is produced from the same PB1 ORF, but lacks the first 39 aa of PB1; it is termed N40. Removal of the PB1-F2 AUG increases expression of N40 in the context of the strain A/WSN/33 (H1N1) (Tauber et al., 2012). The Sw/99 and Ty/04 strains used in this study encode a potential N40 product and therefore we were interested in determining whether ablation of the PB1-F2 ORF would have any effects on its expression. Expression of the influenza virus polymerase complex was detected in cells infected with the isogenic Ty/04 and Sw/99 variants (Fig. S2). Additional analysis included isogenic viruses from the pandemic strain A/California/04/2009 (H1N1), which encode either a short 11 aa peptide (WT) or a full-length PB1-F2 ORF (90 aa, knock-in or KI) (Pena et al., 2012). Our results indicate that the Ty/04 KO (PB1-F2 knockout) and Sw/99 KO strains produce detectable levels of a protein product consistent with N40 (Fig. S2, upper panel, lanes 4 and 6, respectively). Densitometry analysis (data not shown), however, indicated that the proportion of N40 produced was higher in Sw/99 KO than in Ty/04 KO, although the biological significance of such an observation remains to be determined. In other recombinants (Sw/99 N66S, Ca/04 WT and Ca/04 KI), the presence or absence of PB1-F2 had negligible effects on N40 expression. It is interesting to note that Ty/04 KO consistently produced lower quantities of the PB1, PB2 and PA subunits, despite producing similar or higher levels of NP compared with other variants. Taken together, these results suggest that ablation of PB1-F2 affects N40 expression in a strain-dependent manner.
PB1-F2 affects infectivity and viability of porcine alveolar macrophages (PAMs)
As PB1-F2 has been shown to induce cell death in immune cells (Chen et al., 2001; McAuley et al., 2010a), we investigated whether the presence of PB1-F2 affects infectivity in and the viability of freshly isolated PAMs. The purity of PAM preparations was >98 %, as determined by flow cytometry using anti-porcine CD163 antibody (data not shown). Disruption of PB1-F2 expression in Sw/99 (Sw/99 KO) significantly reduced viral yields in PAMs from 6 to 48 h post-infection (p.i.) compared with the Sw/99 WT (P<0.05; Fig. 1a). Surprisingly, the presence of the N66S mutation (Sw/99 N66S) had detrimental effects on virus replication. Ty/04 WT replicated ~10-fold less in PAMs compared with Sw/99 WT and knocking out PB1-F2 (Ty/04 KO) had no effect on virus growth (Fig. 1b). The impact of Sw/99 PB1-F2 on PAM viability, assessed by the XTT colorimetric method, correlated with the replication of these viruses. Sw/99 WT caused more cytotoxicity to PAMs relative to Sw/99 KO and Sw/99 N66S viruses (Fig. 2a, c). PAM viability was also reduced upon infection with the Ty/04 isogenic viruses, but disruption of PB1-F2 ORF only impacted apoptotic cell death and not the overall viability as determined by the XTT method (Fig. 2b, c). It should be noted that cytotoxicity measured by the XTT assay was only detected when PAMs were infected with an m.o.i. of 10. Although the role of N40 overexpression by the Sw/99 KO strain for virus replication needs further investigation, the data suggest that PB1-F2 affects the replication and viability of PAMs in a strain-dependent manner.
Fig. 1.
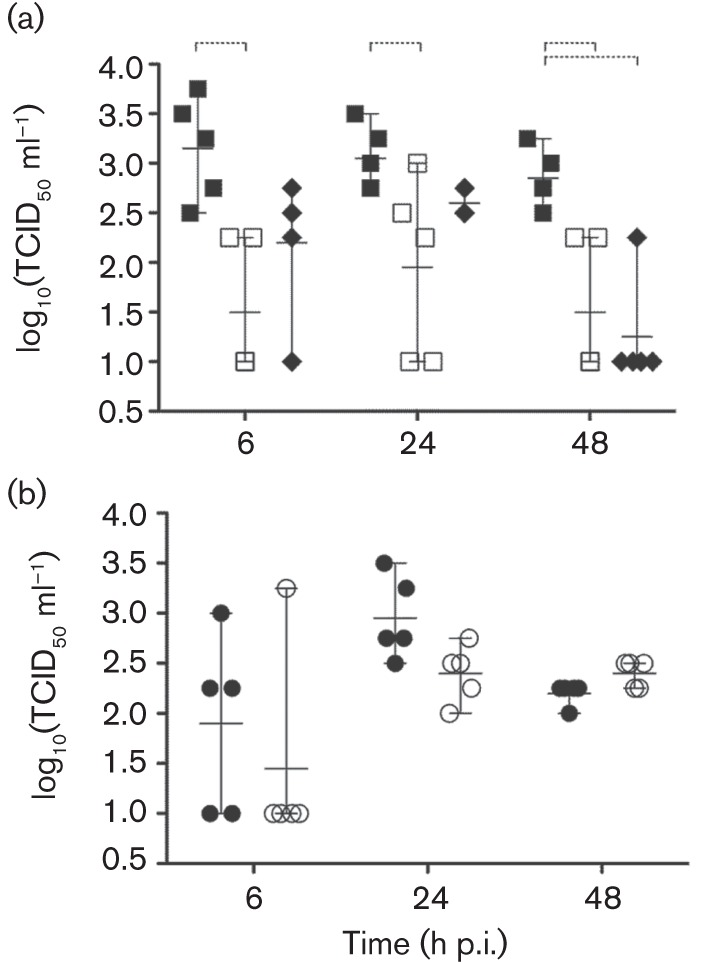
Infectivity of PAMs with PB1-F2 viruses. Virus titres at different times p.i. from PAMs previously infected with virus at an m.o.i. of 2. (a) Sw/99 strains: Sw/99 WT (▪), Sw/99 KO (□) Sw/99 N66S (♦). (b) Ty/04 strains: Ty/04 WT (•), Ty/04 KO (○). One-way ANOVA followed by Dunnett’s post-hoc test was used to determine significant differences between the WT and PB1-F2 modified viruses (dashed brackets, P<0.05). Means±sd of five independent experiments are shown.
Fig. 2.
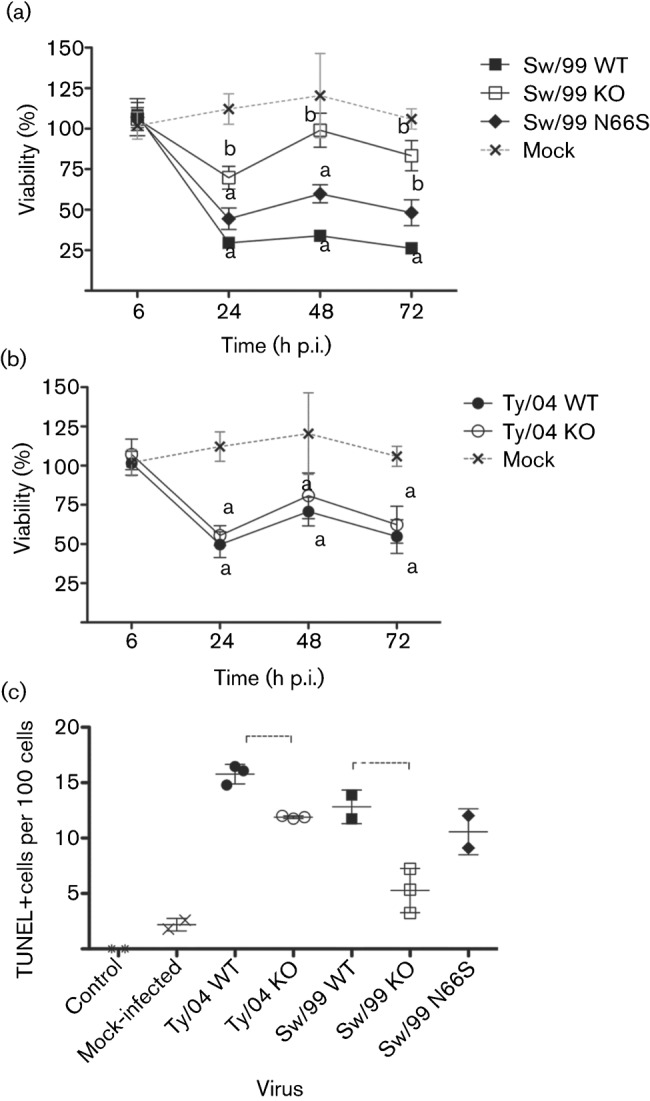
Viability of PAMs after infection with influenza viruses. (a) Sw/99 strains. (b) Ty/04 strains. PAMs were mock-infected or infected with PB1-F2 recombinant viruses at an m.o.i. of 10 and cell viability was measured by the XTT assay at 24, 48 and 72 h p.i. Each data point represents the mean±sd of three independent experiments. Different letters above data points indicate a statistically significant difference (P<0.05). (c) PAMs infected at an m.o.i. of 2 with the indicated viruses and assayed for apoptotic cell death using a TUNEL assay at 72 h p.i. Dashed brackets indicate a statistically significant difference (P<0.05).
PB1-F2 enhances replication of TR H3N2 IAV in porcine respiratory explants
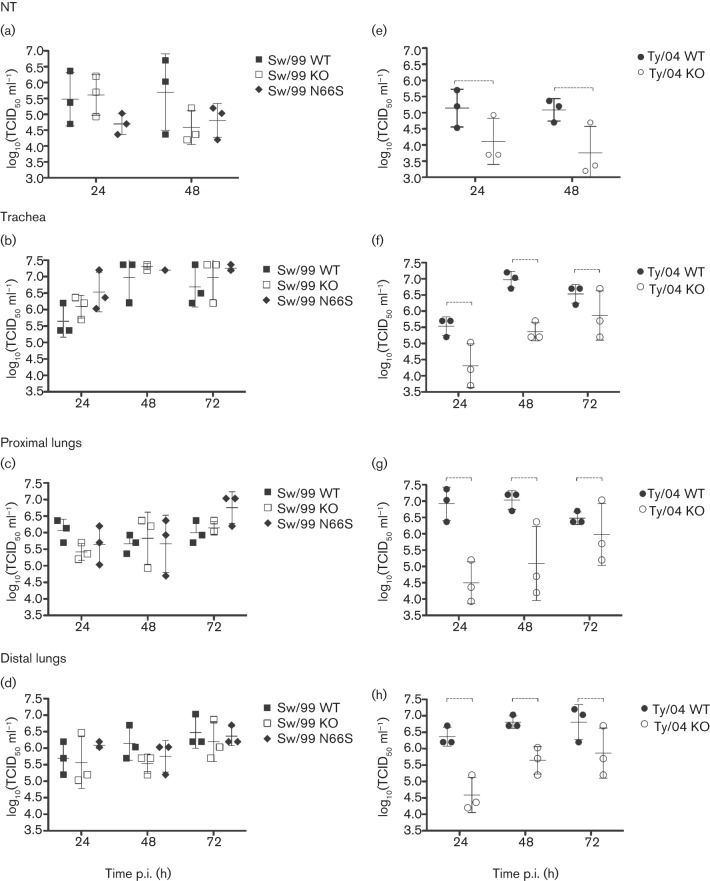
An ex vivo organ-culture model of the pig respiratory tract maintained in an air–liquid interface was used to study the replication of PB1-F2 recombinant viruses in a biologically relevant in vitro system. Tissue explants were prepared from nasal turbinates (NTs), trachea, proximal lung (close to the bronchi) and distal lung (close to the alveoli) from a healthy pig. In this system, PB1-F2 (absence or presence or the N66S mutation) had negligible effects on the replication of Sw/99 recombinants (Fig. 3a–d). However, Ty/04 KO had significantly decreased replication in respiratory explants compared with Ty/04 WT (P<0.05; Fig. 3e–h). Collectively, these data indicate strain-dependent effects of PB1-F2 in modulating virus production in swine respiratory tissues.
Fig. 3.
Replication of PB1-F2 viruses in porcine respiratory explants. NTs (a, e), tracheal (b, f), proximal lung (c, g) and distal lung (d, h) explants were infected with 106 TCID50 of each virus and titrated by TCID50. Values shown correspond to virus titre [log10(TCID50 ml−1)] from each replicate. Data are presented as means±sd of three independent experiments. Dashed brackets indicate a statistically significant difference (P<0.05).
Deletion of PB1-F2 increases virulence of Ty/04
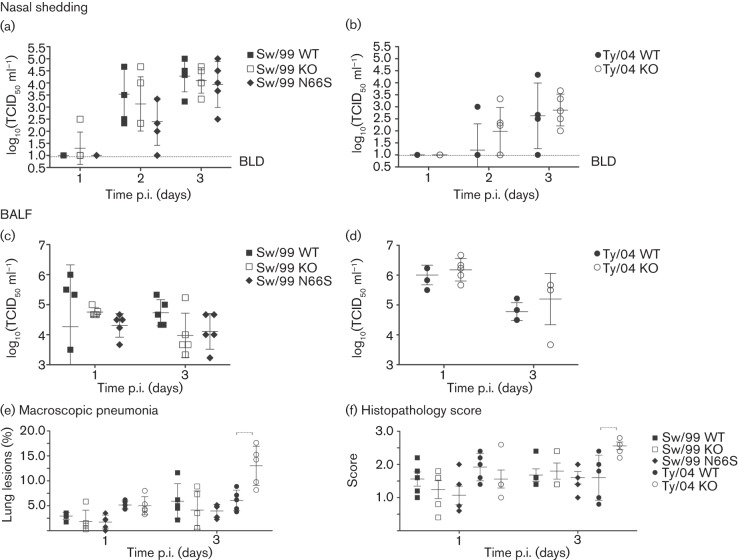
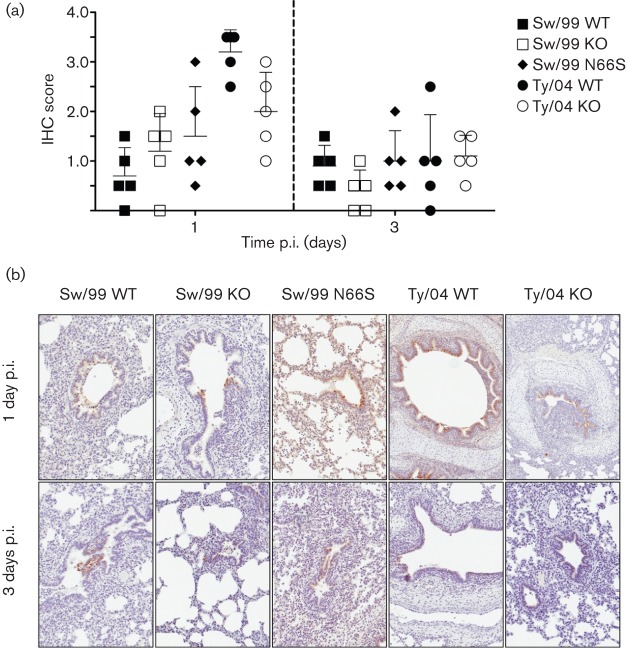
We next evaluated the effects of PB1-F2 in the virulence of TR H3N2 IAVs in vivo. Three-week-old cross-bred pigs were separated into five groups (10 pigs per group) and a control group (n = 5) and inoculated intratracheally with 105 TCID50 per animal (control animals received sterile medium). Five pigs from each group were euthanized at either 1 or 3 days p.i. (control pigs were sacrificed at 3 days p.i.). Sw/99 and Ty/04 recombinant viruses showed similar levels of nasal shedding and lung virus load irrespective of the presence of PB1-F2 ORF (or the N66S mutation) (Fig. 4a–d and data not shown). At necropsy, the absence of the PB1-F2 ORF or the N66S substitution had no effect on either macroscopic or microscopic lung pathology (Fig. 4e, f and data not shown) caused by the Sw/99 recombinants. Surprisingly, strain Ty/04 KO showed significantly increased macroscopic and microscopic pneumonia compared with Ty/04 WT (P<0.05; 3 days p.i.) (Fig. 4e, f). Contrary to what was expected, the presence of PB1-F2 may decrease the virulence of certain IAVs in pigs. Using immunohistochemistry (IHC), we observed that the levels of viral NP antigen correlated with the levels of virus replication observed for each virus (Fig. 5).
Fig. 4.
Replication and pathology induced by PB1-F2 recombinant viruses in swine. Pigs (n = 10 per group) were infected with 105 TCID50 virus and nasal swabs were collected from days 1 to 3 p.i. to measure virus shedding. Lungs were collected at 1 and 3 days p.i. (n = 5 day−1) for virus titration and pathological analysis. (a, b) Virus shedding in nasal secretions. (c, d) Pulmonary replication of PB1-F2 isogenic viruses in pigs (measured as virus titre in BALF). (e) Percentage of macroscopic lung lesions. (f) Lung histopathology scores. Data are presented as means±sd of five independent experiments. Dashed brackets indicate a statistically significant difference (P<0.05).
Fig. 5.
IHC of influenza viruses in swine lungs. (a) Anti-NP antigen IHC staining in lungs of infected pigs. Values are the mean±sd IHC scores based on the percentage of influenza-positive cells in the airway and lung interstitium, from five independent experiments. (b) Representative IHC slides depicting viral antigen primarily in airway epithelium at 1 and 3 days p.i.
PB1-F2 modulates the innate immune response in swine lungs
PB1-F2 strongly influences the early host responses during IAV infection (Le Goffic et al., 2011; McAuley et al., 2007) and therefore we measured the protein levels of ten porcine cytokines/chemokines [interleukin (IL)-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, tumour necrosis factor alpha (TNF-α), alpha interferon (IFN-α) and gamma interferon (IFN-γ)] in bronchoalveolar lavage fluid (BALF) collected from pigs sacrificed at 1 and 3 days p.i. (Fig. 6). Pigs infected with Ty/04 WT had higher levels of IFN-α and IL-1β at 1 day p.i. than those infected with Sw/99 WT. The profile of other cytokines/chemokines was not altered by the presence or absence of the PB1-F2 ORF in these strains (Fig. 6a, b and data not shown). Increased pulmonary levels of IL-6 were seen at 1 day p.i. during infection with Sw/99 WT compared with Sw/99 KO, suggesting differential regulation of this cytokine in the presence of PB1-F2 in pigs infected with this virus background (Fig. 6c). The N66S mutation resulted in no stimulation of IL-6 and higher stimulation of the IL-8 response compared with the Sw/99 WT (Fig. 6c, d). Ty/04 WT and Ty/04 KO strains stimulated IL-8 production at 1 day p.i., but only pigs infected with Ty/04 KO showed sustained responses of this cytokine by 3 days p.i. (Fig. 6d). Likewise, Ty/04 KO-infected pigs showed higher levels of IFN-γ at 3 days p.i. compared with pigs from other virus groups, which is consistent with the development of enhanced pneumonia in the former (Fig. 6e). Collectively, these results indicate that PB1-F2 differentially modulates the local levels of cytokines and chemokines during infection with TR H3N2 in pigs and that the heightened production of IL-8 and IFN-γ pro-inflammatory mediators correlated with the lung pathology developed in Ty/04 KO-infected animals.
Fig. 6.
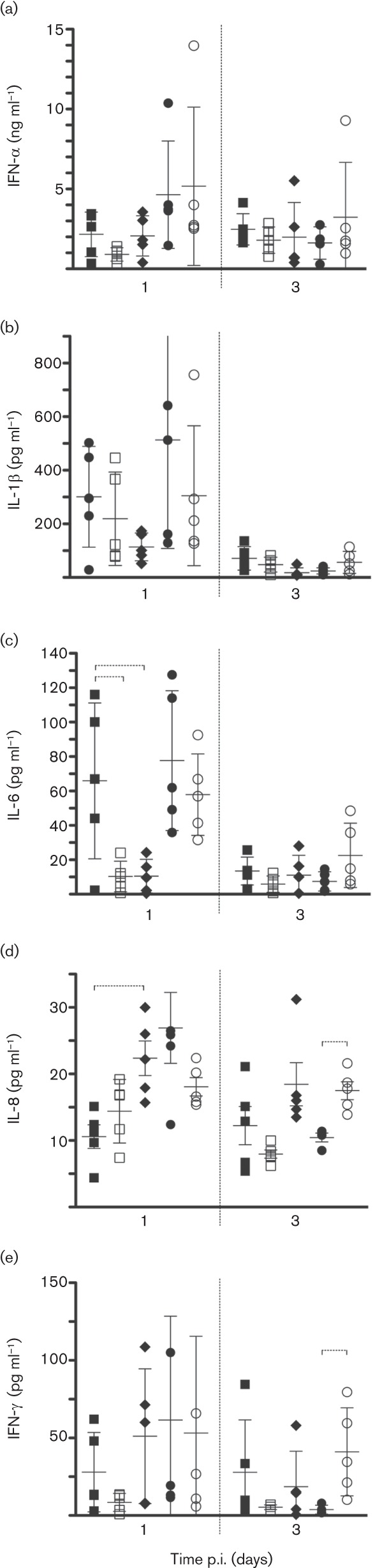
PB1-F2 modulates the innate immune response in swine lungs. Data represent cytokine/chemokine levels (by ELISA) in BALF from each pig in the respective groups (▪, Sw/99 WT; □, Sw/99 KO; ⧫, Sw/99 N66S; •, Ty/04 WT; ○, Ty/04 KO). (a) IFN-α; (b) IL-1β; (c) IL-6; (d) IL-8; (e) IFN-γ. Data are presented as means±sd of five independent experiments. Dashed brackets indicate a statistically significant difference (P<0.05).
Discussion
The phenotypes associated with the presence of PB1-F2 derived from TR H3N2 IAVs have not yet been elucidated. We created PB1-F2 null viruses in the background of two representative lineages of swine-origin North American TR H3N2 viruses: Sw/99 (cluster III) and Ty/04 (cluster IV). These viruses were chosen because they are genetically related and they both circulate in the swine population. Since Sw/99 is of low virulence for pigs (L. Pena & D. R. Perez, unpublished data), we studied the effects not only of removing PB1-F2, but also in the context of the virulence marker N66S. Previous studies have shown that removal of the PB1-F2 AUG increases expression of N40 (Tauber et al., 2012). A band consistent with the expression of N40 has been observed in influenza strains of human, avian and equine origin (Wise et al., 2009). A protein band consistent with N40 is readily detected when PB1-F2 is removed from the Sw/99 background, but only slightly when PB1-F2 is removed from the Ty/04 background. The pandemic Ca/04 strain, which naturally encodes only the first 11 aa of PB1-F2, does not show expression of N40 despite encoding such a polypeptide. In our previous report, we showed that restoring PB1-F2 in the Ca/04 background had minimal, although discernible, effects in pigs with respect to virulence (Pena et al., 2012). Our current Western blot assay indicates that these differences cannot be attributed to N40 expression because neither the Ca/04 WT nor the Ca/04 KI strains produce detectable N40. Whether the difference in N40 expression in the Sw/99 and Ty/04 strains could account for some of the differences observed in vitro and in vivo deserves further investigation beyond the scope of the current report.
IAVs infect alveolar macrophages in a variety of species, including mice, swine and humans (Reading et al., 2000; Rodgers & Mims, 1982; Seo et al., 2004; Upham et al., 2010; van Riel et al., 2011). In pigs, in vivo depletion of alveolar macrophages by chemical treatment unveiled that these cells are indispensable for controlling IAV infection (Kim et al., 2008). PB1-F2 has been suggested to influence virulence by destroying host immune cells such as macrophages (Chen et al., 2001; Lamb & Takeda, 2001; Zamarin et al., 2005). By infecting PAMs ex vivo with PB1-F2 recombinant viruses, we found that the presence of PB1-F2 was required for optimal replication of Sw/99, but not Ty/04 (Fig. 1). Likewise, PAM viability was affected by the presence of PB1-F2 in the Sw/99 strain (Fig. 2). Differences in apoptotic cell death were detected by TUNEL when PAMs were infected at an m.o.i. of 2, but this effect was only quantifiable by XTT assay when PAMs were infected at an m.o.i. of 10. It remains to be determined whether differences seen with the Sw/99 KO strain are due to lack of PB1-F2 expression or overexpression of N40 or a combination of both. Nevertheless, there was no correlation between the ability of PB1-F2 recombinant viruses to replicate in PAMs and the virulence to pigs. Disruption of PB1-F2 ORF had no effect in Sw/99 virulence as measured by virus shedding, viral lung load and lung pathology. These results are in agreement with those that showed no correlation between macrophage infectivity and/or cell death in vitro and the course of IAV disease in mice (McAuley et al., 2010a; Tate et al., 2010). In the background of Ty/04, PB1-F2 had no effect on virus replication, apoptosis or viability of PAMs, which would also suggest a limited, if any, role of N40 under these conditions.
Expression of full-length PB1-F2 as well as the N66S substitution has been shown to increase replication of certain IAV isolates in tissue culture (McAuley et al., 2010b; Schmolke et al., 2011; Smith et al., 2011). Our results support the hypothesis that PB1-F2 modulates virus replication in a strain-dependent manner. Although Ty/04 PB1-F2 did not affect polymerase activity, higher viral yields were observed in respiratory explants infected with Ty/04 WT relative to Ty/04 KO. This is consistent with expression levels of the polymerase subunits as revealed by Western blot analysis. In the backbone of Sw/99 viruses, neither deletion of PB1-F2 nor mutation of N66S affected viral growth kinetics in this system. These observations also emphasize a minor role of N40 in this system (Fig. 3). The replication of Sw/99 PB1-F2 recombinant viruses ex vivo paralleled the levels of virus shedding in nasal secretions and lung viral load in vivo. However, we found no differences between Ty/04 WT and Ty/04 KO regarding virus shedding in the upper respiratory tract and BALF titres, suggesting that results obtained from ex vivo and in vivo results do not always correlate with each other.
PB1-F2 proteins from human H3N2 IAV differ profoundly in their ability to induce inflammatory responses and pathology in lungs (McAuley et al., 2010a) with pro-inflammatory mutations in PB1-F2 mapped to P62L, H75R, Q79R and S82L (Alymova et al., 2011). The PB1-F2 sequences of both Sw/99 and Ty/04 present conserved residues in these positions, which include a mix of the pro-inflammatory motifs 62(L) and 82(L) as well as non-inflammatory mutations 75(H), 79(Q) (Fig. S1). The PB1-F2 deletion or N66S substitution in the background of Sw/99 did not alter virus-induced pneumonia in pigs. Unexpectedly, disruption of PB1-F2 ORF from the Ty/04 strain significantly aggravated both the microscopic and the macroscopic pneumonia caused by this virus at 3 days p.i. (P<0.05). The degree of pneumonia in Ty/04 KO cannot be explained by improved replication, as it replicated to similar levels as Ty/04 WT throughout the respiratory tract. Whether under these conditions N40 plays a role is not known; however, we would have also expected phenotypic differences using the Sw/99 KO virus, which expresses higher levels of N40 in vitro.
The pro-inflammatory cytokine response is critical for recruiting effector cells to the site of infection. Concomitantly with the development of severe pneumonia in Ty/04 KO-infected pigs, there was a significant increase in the pulmonary levels of IL-8 and IFN-γ at 3 days p.i. (P<0.05) compared with the Ty/04 WT group. IL-8 is a major chemokine involved in neutrophil recruitment to infection sites. Elevated levels of IL-8 have been implicated in the pathogenesis of virus-induced bronchiolitis in humans (Koh et al., 2007).
IFN-γ is an important immunoregulatory cytokine that is produced by T-cells, NK cells and macrophages. IFN-γ has pleiotropic effects on various cells of the immune systems and plays an important role in T-cell-mediated lung injury (Bruder et al., 2006). Influenza virus infection induces the local production of IFN-γ in the respiratory tract. Experiments in gene-targeted mice showed that IFN-γ plays no significant role in protection from lethal infection (Graham et al., 1993) and IAV has developed mechanisms to evade its antiviral activity by disrupting intracellular signalling pathways (Uetani et al., 2008). Recently, Le Goffic et al. (2011) reported that infection with the laboratory-adapted strain A/WSN/1933 (H1N1) in mice resulted in higher levels of IFN-γ relative to infection with an isogenic virus lacking PB1-F2. These results are in disagreement with the findings in pigs using a TR H3N2 IAV, in which deletion of PB1-F2 led to enhanced IFN-γ concentrations in BALF. The reason for this discrepancy is unclear, but it might be due to differences in virus strains and host systems used. There have been conflicting reports of the role of PB1-F2 in modulating type I IFNs, and thus the exact role of PB1-F2 in influencing the host innate immune response remains unresolved (Dudek et al., 2011; Le Goffic et al., 2010; Varga et al., 2011). It is possible that deregulation of the innate host defence through IL-8 and IFN-γ pathways might have played a role in the exacerbated immunopathology induced by Ty/04 KO virus. Should Ty/04 PB1-F2 have a direct effect in blocking or downregulating IL-8 and IFN-γ expression, this effect would explain the lessened pulmonary lesions developed in the Ty/04 WT group. These studies are beyond the scope of this work and warrant further investigation.
Finally, the potential role of PB1-F2 as a contributor of secondary bacterial infections must be noted. Studies in mice have shown that a virus encoding the 1918 PB1-F2 ORF led to increased virulence and more severe bacterial pneumonia (McAuley et al., 2007). In our studies, pigs are treated with antibiotic once prior to inoculation with influenza, in order to reduce bacterial contaminants that could obscure the specific effects provided by the virus. Thus, our studies were not designed to establish the role of PB1-F2 in secondary infections. Nevertheless, aerobic bacterial screening with BALF and colony counts were performed to rule out bacterial contribution to pneumonia lesions. We found no evidence of bacterial involvement in disease or pathology.
Further research efforts focusing on the modulation of the porcine immune system by PB1-F2, its interaction with host factors, and modulation of secondary bacterial pneumonia should shed light on the molecular mechanisms of PB1-F2 in a strain- and host-dependent manner.
Methods
Cell lines and virus strains.
Human embryonic kidney cells (293-T) were cultured in OptiMEM I (Gibco) containing 10 % FBS and antibiotics. Madin–Darby canine kidney (MDCK) cells were maintained in modified Eagle’s medium (MEM) (Sigma-Aldrich) supplemented with 5 % FBS (Sigma-Aldrich), 2 mM l-glutamine and antibiotics (100 U penicillin ml−1 and 100 µg streptomycin ml−1). A/swine/Wisconsin/14094/99 (H3N2) (Sw/99) (GenBank Taxonomy ID 136472) was a kind gift from Dr Sagar Goyal, University of Minnesota, Minneapolis, MN, USA. A/turkey/Ohio/313053/04 (H3N2) (GenBank Taxonomy ID 533026) was kindly provided by Dr Yehia Saif, Ohio State University, Wooster, OH, USA. Both viruses were successfully rescued from cloned cDNAs as described below and amplified in either MDCK cells (Sw/99) or embryonated eggs (Ty/04). Sw/99 and Ty/04 bearing mutations in PB1-F2 were generated by RG and are described below. The Sw/99 and Ty/04 WT PB1-F2 gene products were aligned with 1918 H1N1, 1957 H2N2 and 1968 H3N2 and previously characterized PB1-F2 proteins. Alignment was carried out using clustal w in megalign (Lasergene v. 8.1.5; dnastar). The A/California/04/2009 (H1N1) WT and knock-in variants (Ca/04 WT and Ca/04 KI, respectively) have been described previously (Pena et al., 2012).
RG and mutagenesis.
The eight-plasmid-based RG system was used to rescue WT A/swine/Wisconsin/14094/99 (H3N2) (Sw/99). The RG A/turkey/Ohio/313053/04 (H3N2) (Ty/04) has been described previously (Pena et al., 2011). A QuikChange II site-directed mutagenesis kit (Stratagene) was used according to manufacturer’s protocols to disrupt the PB1-F2 ORF of both Sw/99 and Ty/04, which display a low- and high-virulent phenotype for pigs, respectively (L. Pena & D. R. Perez, unpublished data). To ensure complete abolition of PB1-F2 translation, we mutated the ATG start codon to ACG and introduced two stop codons at amino acid positions 12 (C153G) and 58 (G291A) as described by Zamarin et al. (2006). The virulence marker N66S was introduced into the Sw/99 virus as described previously (Conenello et al., 2007). RG plasmids and recovered recombinant viruses were prepared as described by Hoffmann et al. (2000) and sequenced fully to confirm their identity.
Western blot analysis.
MDCK cells growing in 12-well plates were infected by the indicated virus strains at an m.o.i. of 1. After 1 h incubation at 35 °C, cells were washed two times with PBS to remove unattached viruses. After 24 h infection at 35 °C, cells were lysed with 2× Laemmli sample buffer (Bio-Rad) and incubated at 100 °C for 10 min. Western blot analysis was performed using standard techniques and incubating with different primary antibodies: anti-PB1 (goat) antibody targeting residues 50–100 of PB1 at a 1 : 100 dilution (Santa Cruz Biotechnology), anti-PB2 (goat) antibody targeting N terminus of PB2 at a 1 : 200 dilution (Santa Cruz Biotechnology), anti-PA (rabbit) antibody targeting residues 250–300 of PA at a 1 : 150 dilution (GenScript), anti-NP (mouse monoclonal) antibody at a 1 : 1000 dilution (2F4, made in house) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (mouse) antibody at a 1 : 3000 dilution (Santa Cruz Biotechnology). A corresponding HRP-conjugated secondary antibody at a 1 : 3000 dilution [donkey anti-goat IgG–HRP, donkey anti-rabbit IgG–HRP, donkey anti-mouse IgG–HRP (Santa Cruz Biotechnology)] was used. Immunoreactive proteins were visualized using West Pico (Pierce) enhanced chemiluminescence substrate and autoradiography.
Isolation, infection and viability of porcine alveolar macrophages (PAMs).
PAMs were isolated from five healthy pigs as described previously (Loving et al., 2006). PAMs were seeded at a density of 1×105 cells per well in a 96-well flat-bottom plate with a final volume of 100 µl PAM complete medium. At 24 h after plating, the medium was removed and PAMs were infected with 100 µl of the recombinant viruses diluted in PAM medium at an m.o.i. of 2 or 10. PAM viability was measured by the XTT colorimetric method using a Cell Proliferation kit II (Roche Molecular Biochemicals) according to the manufacturer’s protocol. At the indicated time points, 20 µl XTT working solution (1 mg ml−1), prepared immediately before use, was added to each well and the microplate was incubated for an additional 2 h at 37 °C and read using an ELISA plate reader (SpectraMax M5; Molecular Devices) with a 450 nm optical filter. PAM viability was determined as a relative percentage of the mock-treated non-infected control. The infectivity of PB1-F2 recombinant viruses was tested in freshly isolated PAMs seeded on six-well plates in 3 ml MEMα (Invitrogen) supplemented with 10 % FBS and antibiotics (100 U penicillin ml−1 and 100 µg streptomycin ml−1). PAMs were incubated overnight and infected at an m.o.i. of 2 with MEM without trypsin. Following 1 h adsorption at 37 °C, PAMs were washed three times with PBS and the culture was replenished with 3 ml MEMα supplemented with 10 % FBS and antibiotics. At the indicated time points, 300 µl supernatant was collected to assess virus replication. Virus yield in PAMs was determined by standard TCID50 in MDCK cells.
TUNEL assay in ex vivo infected PAMs.
Terminal deoxynucleotidyltransferase-mediated dUTP nick end labelling (TUNEL) was used to quantify the levels of apoptotic cell death induced by recombinant viruses in ex vivo infected PAMs. Apoptotic cells were detected using an In situ Cell Death Detection kit, Fluorescein (Roche Applied Science) according to manufacturer’s instructions. Briefly, PAMs were infected at an m.o.i. of 2 and, at 72 h p.i., cells were fixed with 4 % paraformaldehyde and permeabilized in PBS containing 0.1 % Triton X-100 and 0.1 % sodium citrate. Cells were then incubated in the TUNEL reaction mixture in the dark at 37 °C for 60 min followed by detection of DNA strand breaks in apoptotic cells by fluorescence microscopy. A minimum of 600 cells was counted in a blinded fashion from more than 10 random microscopic fields and the percentage of cell apoptosis was determined.
Virus titration.
Viral stocks and virus present in biological samples were titrated on MDCK cells and the TCID50 ml−1 was determined by the method of Reed & Muench (1938). The end-point viral titre was determined by a haemagglutination assay using 0.5 % turkey red blood cells.
Animal studies.
Swine pathogenicity studies were conducted at the National Animal Disease Center in Ames, IA, USA, under protocols approved by the US Department of Agriculture’s Agricultural Research Service (USDA-ARS) Animal Care and Use Committee. Porcine explants were prepared under protocols approved by the Institutional Animal Care and Use Committee, University of Maryland. Animal studies adhered strictly to the US Animal Welfare Act laws and regulations. Where indicated, animals were humanely euthanized with Beuthanasia-D (Intervet/Schering-Plough) at a dosage of 1 ml per 4.5 kg body weight.
Three-week-old cross-bred pigs were obtained from a high-health herd free of swine influenza virus and porcine reproductive and respiratory syndrome virus. All pigs were treated once, at 11 days prior to inoculation with influenza virus, with Ceftiofur crystalline free acid (5 mg kg−1; Pfizer Animal Health) to reduce bacterial contaminants prior to the start of the experiment. Fifty pigs were divided randomly in five groups (n = 10) and housed in separate isolation rooms. Pigs were infected intratracheally with 1×105 TCID50 of each of the recombinant viruses diluted in 2 ml MEM using previously described protocols and monitored for signs of disease and virus shedding (Pena et al., 2012; Vincent et al., 2010).
Pathological examination of swine lungs.
At necropsy, lungs were removed in toto and evaluated to determine the percentage of the lung affected by purple-red, consolidated lesions that are typical of influenza virus infection in pigs. The percentage of the surface affected with pneumonia was calculated based on weighted proportions of each lobe to the total lung volume as previously described (Halbur et al., 1995). Each lung was then lavaged with 50 ml MEM to obtain BALF. A single veterinary pathologist scored all lungs and was blinded to the treatment groups.
Isolation, culture and infection of porcine respiratory explants.
Porcine nasal turbinates (NTs), tracheal and lung explants were prepared as described in detail previously (Pena et al., 2012; Van Poucke et al., 2010). At 24 h of culture, explants were washed with PBS and 106 TCID50 of recombinant viruses diluted in 500 µl explant medium were deposited in the upper compartment for 1 h at 37 °C. Subsequently, explants were washed three times with PBS and the culture was replenished with 500 µl explant medium. At 24, 48 and 72 h p.i., 100 µl supernatant was removed to assess virus yields.
Histopathology and immunohistochemistry.
Tissue samples were collected, fixed and stained as described previously (Pena et al., 2012). Microscopic lesions were evaluated by a board-certified veterinary pathologist blinded to treatment groups. Scoring of lesions was based on scales adapted from Gauger et al. (2011). Influenza virus type A-specific antigen was detected in lung tissues using a previously described immunohistochemical (IHC) method with minor modifications (Pena et al., 2012; Vincent et al., 1997).
Quantification of cytokine/chemokine protein levels in bronchoalveolar lavage fluid.
Levels of nine porcine cytokines/chemokines (IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, TNF-α and IFN-γ) in BALF were determined by multiplex ELISA following the manufacturer’s recommendations (SearchLight, Aushon Biosystems). Levels of IFN-α protein were measured by ELISA using F17 mAb, K9 mAb and recombinant porcine IFN-α (R&D Systems) as described previously (Brockmeier et al., 2009).
Statistical analysis.
All statistical analyses were performed using GraphPad Prism software v. 5.00 (GraphPad Software). Comparison between two treatment means was achieved using a two-tailed Student’s t-test, whereas multiple comparisons were carried out by ANOVA. The differences were considered statistically significant at P<0.05.
Acknowledgements
We are indebted to Annabelle Pascua Crusan for her help with explant studies. We thank Michelle Harland, Hillary Horst, Gwen Nordholm, Brian Pottebaum, Jason Crabtree and Jason Huegel for technical assistance with the swine study. We would like to thank Susan Brockmeier for doing the bacteriological screening. This research was made possible through funding by a CDC-HHS grant (1U01CI000355), an NIAID-NIH grant (R01AI052155), a CSREES-USDA grant (2005-05523), an NIAID-NIH contract (HHSN266200700010C) and USDA-ARS.
Footnotes
Two supplementary figures and a supplementary table are available with the online version of this paper.
References
- Alymova I. V., Green A. M., van de Velde N., McAuley J. L., Boyd K. L., Ghoneim H., McCullers J. A. (2011). Immunopathogenic and anti-bacterial effects of the H3N2 influenza A virus PB1-F2 map to amino acid residues 62, 75, 79, and 82. J Virol 85, 12324–12333 10.1128/JVI.05872-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmeier S. L., Lager K. M., Grubman M. J., Brough D. E., Ettyreddy D., Sacco R. E., Gauger P. C., Loving C. L., Vorwald A. C. & other authors (2009). Adenovirus-mediated expression of interferon-alpha delays viral replication and reduces disease signs in swine challenged with porcine reproductive and respiratory syndrome virus. Viral Immunol 22, 173–180 10.1089/vim.2008.0075 [DOI] [PubMed] [Google Scholar]
- Bruder D., Srikiatkhachorn A., Enelow R. I. (2006). Cellular immunity and lung injury in respiratory virus infection. Viral Immunol 19, 147–155 10.1089/vim.2006.19.147 [DOI] [PubMed] [Google Scholar]
- Chen W., Calvo P. A., Malide D., Gibbs J., Schubert U., Bacik I., Basta S., O’Neill R., Schickli J. & other authors (2001). A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7, 1306–1312 10.1038/nm1201-1306 [DOI] [PubMed] [Google Scholar]
- Choi Y. K., Lee J. H., Erickson G., Goyal S. M., Joo H. S., Webster R. G., Webby R. J. (2004). H3N2 influenza virus transmission from swine to turkeys, United States. Emerg Infect Dis 10, 2156–2160 10.3201/eid1012.040581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conenello G. M., Zamarin D., Perrone L. A., Tumpey T., Palese P. (2007). A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3, e1414–e1421 10.1371/journal.ppat.0030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek S. E., Wixler L., Nordhoff C., Nordmann A., Anhlan D., Wixler V., Ludwig S. (2011). The influenza virus PB1-F2 protein has interferon-antagonistic activity. Biol Chem 392, 1135-1144 [DOI] [PubMed] [Google Scholar]
- Fukuyama S., Kawaoka Y. (2011). The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol 23, 481–486 10.1016/j.coi.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger P. C., Vincent A. L., Loving C. L., Lager K. M., Janke B. H., Kehrli M. E., Jr, Roth J. A. (2011). Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (δ-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 29, 2712–2719 10.1016/j.vaccine.2011.01.082 [DOI] [PubMed] [Google Scholar]
- Graham M. B., Dalton D. K., Giltinan D., Braciale V. L., Stewart T. A., Braciale T. J. (1993). Response to influenza infection in mice with a targeted disruption in the interferon gamma gene. J Exp Med 178, 1725–1732 10.1084/jem.178.5.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramer M. R., Lee J. H., Choi Y. K., Goyal S. M., Joo H. S. (2007). Serologic and genetic characterization of North American H3N2 swine influenza A viruses. Can J Vet Res 71, 201–206 [PMC free article] [PubMed] [Google Scholar]
- Halbur P. G., Paul P. S., Frey M. L., Landgraf J., Eernisse K., Meng X. J., Lum M. A., Andrews J. J., Rathje J. A. (1995). Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 32, 648–660 10.1177/030098589503200606 [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Kawaoka Y., Hobom G., Webster R. G. (2000). A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97, 6108–6113 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Lee Y. W., Lee K. J., Kim H. S., Cho S. W., van Rooijen N., Guan Y., Seo S. H. (2008). Alveolar macrophages are indispensable for controlling influenza viruses in lungs of pigs. J Virol 82, 4265–4274 10.1128/JVI.02602-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh Y. Y., Jung E., Koh J. Y., Kim J. Y., Yoo Y., Kim C. K. (2007). Bronchoalveolar cellularity and interleukin-8 levels in measles bronchiolitis obliterans. Chest 131, 1454–1460 10.1378/chest.06-0188 [DOI] [PubMed] [Google Scholar]
- Kumar S. R., Deflube L., Biswas M., Shobana R., Elankumaran S. (2011). Genetic characterization of swine influenza viruses (H3N2) isolated from Minnesota in 2006–2007. Virus Genes 43, 161–176 10.1007/s11262-011-0618-4 [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Takeda M. (2001). Death by influenza virus protein. Nat Med 7, 1286–1288 10.1038/nm1201-1286 [DOI] [PubMed] [Google Scholar]
- Le Goffic R., Bouguyon E., Chevalier C., Vidic J., Da Costa B., Leymarie O., Bourdieu C., Decamps L., Dhorne-Pollet S., Delmas B. (2010). Influenza A virus protein PB1-F2 exacerbates IFN-β expression of human respiratory epithelial cells. J Immunol 185, 4812–4823 10.4049/jimmunol.0903952 [DOI] [PubMed] [Google Scholar]
- Le Goffic R., Leymarie O., Chevalier C., Rebours E., Da Costa B., Vidic J., Descamps D., Sallenave J. M., Rauch M. & other authors (2011). Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. PLoS Pathog 7, e1002202 10.1371/journal.ppat.1002202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving C. L., Brockmeier S. L., Ma W., Richt J. A., Sacco R. E. (2006). Innate cytokine responses in porcine macrophage populations: evidence for differential recognition of double-stranded RNA. J Immunol 177, 8432–8439 [DOI] [PubMed] [Google Scholar]
- Marjuki H., Scholtissek C., Franks J., Negovetich N. J., Aldridge J. R., Salomon R., Finkelstein D., Webster R. G. (2010). Three amino acid changes in PB1-F2 of highly pathogenic H5N1 avian influenza virus affect pathogenicity in mallard ducks. Arch Virol 155, 925–934 10.1007/s00705-010-0666-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley J. L., Hornung F., Boyd K. L., Smith A. M., McKeon R., Bennink J., Yewdell J. W., McCullers J. A. (2007). Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2, 240–249 10.1016/j.chom.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley J. L., Chipuk J. E., Boyd K. L., Van De Velde N., Green D. R., McCullers J. A. (2010a). PB1-F2 proteins from H5N1 and 20th century pandemic influenza viruses cause immunopathology. PLoS Pathog 6, e1001014 10.1371/journal.ppat.1001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley J. L., Zhang K., McCullers J. A. (2010b). The effects of influenza A virus PB1-F2 protein on polymerase activity are strain specific and do not impact pathogenesis. J Virol 84, 558–564 10.1128/JVI.01785-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nfon C., Berhane Y., Zhang S., Handel K., Labrecque O., Pasick J. (2011). Molecular and antigenic characterization of triple-reassortant H3N2 swine influenza viruses isolated from pigs, turkey and quail in Canada. Transbound Emerg Dis 58, 394–401 10.1111/j.1865-1682.2011.01219.x [DOI] [PubMed] [Google Scholar]
- Pena L., Vincent A. L., Ye J., Ciacci-Zanella J. R., Angel M., Lorusso A., Gauger P. C., Janke B. H., Loving C. L., Perez D. R. (2011). Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol 85, 456–469 10.1128/JVI.01503-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena L., Vincent A. L., Loving C. L., Henningson J. N., Lager K. M., Lorusso A., Perez D. R. (2012). Restored PB1-F2 in the 2009 pandemic H1N1 influenza virus has minimal effects in swine. J Virol 86, 5523–5532 10.1128/JVI.00134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading P. C., Miller J. L., Anders E. M. (2000). Involvement of the mannose receptor in infection of macrophages by influenza virus. J Virol 74, 5190–5197 10.1128/JVI.74.11.5190-5197.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty per cent endpoints. Am J Hyg 27, 493–497 [Google Scholar]
- Richt J. A., Lager K. M., Janke B. H., Woods R. D., Webster R. G., Webby R. J. (2003). Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol 41, 3198–3205 10.1128/JCM.41.7.3198-3205.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers B. C., Mims C. A. (1982). Influenza virus replication in human alveolar macrophages. J Med Virol 9, 177–184 10.1002/jmv.1890090304 [DOI] [PubMed] [Google Scholar]
- Schmolke M., Manicassamy B., Pena L., Sutton T., Hai R., Varga Z. T., Hale B. G., Steel J., Pérez D. R., García-Sastre A. (2011). Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog 7, e1002186 10.1371/journal.ppat.1002186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S. H., Webby R., Webster R. G. (2004). No apoptotic deaths and different levels of inductions of inflammatory cytokines in alveolar macrophages infected with influenza viruses. Virology 329, 270–279 10.1016/j.virol.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Shope R. E. (1931). The etiology of swine influenza. Science 73, 214–215 10.1126/science.73.1886.214 [DOI] [PubMed] [Google Scholar]
- Smith A. M., Adler F. R., McAuley J. L., Gutenkunst R. N., Ribeiro R. M., McCullers J. A., Perelson A. S. (2011). Effect of 1918 PB1-F2 expression on influenza A virus infection kinetics. PLOS Comput Biol 7, e1001081 10.1371/journal.pcbi.1001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M. D., Pickett D. L., van Rooijen N., Brooks A. G., Reading P. C. (2010). Critical role of airway macrophages in modulating disease severity during influenza virus infection of mice. J Virol 84, 7569–7580 10.1128/JVI.00291-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber S., Ligertwood Y., Quigg-Nicol M., Dutia B. M., Elliott R. M. (2012). Behaviour of influenza A viruses differentially expressing segment 2 gene products in vitro and in vivo. J Gen Virol 93, 840–849 10.1099/vir.0.039966-0 [DOI] [PubMed] [Google Scholar]
- Uetani K., Hiroi M., Meguro T., Ogawa H., Kamisako T., Ohmori Y., Erzurum S. C. (2008). Influenza A virus abrogates IFN-γ response in respiratory epithelial cells by disruption of the Jak/Stat pathway. Eur J Immunol 38, 1559–1573 10.1002/eji.200737045 [DOI] [PubMed] [Google Scholar]
- Upham J. P., Pickett D., Irimura T., Anders E. M., Reading P. C. (2010). Macrophage receptors for influenza A virus: role of the macrophage galactose-type lectin and mannose receptor in viral entry. J Virol 84, 3730–3737 10.1128/JVI.02148-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poucke S. G., Nicholls J. M., Nauwynck H. J., Van Reeth K. (2010). Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol J 7, 38 10.1186/1743-422X-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Leijten L. M., van der Eerden M., Hoogsteden H. C., Boven L. A., Lambrecht B. N., Osterhaus A. D., Kuiken T. (2011). Highly pathogenic avian influenza virus H5N1 infects alveolar macrophages without virus production or excessive TNF-alpha induction. PLoS Pathog 7, e1002099 10.1371/journal.ppat.1002099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z. T., Ramos I., Hai R., Schmolke M., García-Sastre A., Fernandez-Sesma A., Palese P. (2011). The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog 7, e1002067 10.1371/journal.ppat.1002067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent L. L., Janke B. H., Paul P. S., Halbur P. G. (1997). A monoclonal-antibody-based immunohistochemical method for the detection of swine influenza virus in formalin-fixed, paraffin-embedded tissues. J Vet Diagn Invest 9, 191–195 10.1177/104063879700900214 [DOI] [PubMed] [Google Scholar]
- Vincent A. L., Ma W., Lager K. M., Janke B. H., Richt J. A. (2008). Swine influenza viruses: a North American perspective. Adv Virus Res 72, 127–154 10.1016/S0065-3527(08)00403-X [DOI] [PubMed] [Google Scholar]
- Vincent A. L., Lager K. M., Faaberg K. S., Harland M., Zanella E. L., Ciacci-Zanella J. R., Kehrli M. E., Jr, Janke B. H., Klimov A. (2010). Experimental inoculation of pigs with pandemic H1N1 2009 virus and HI cross-reactivity with contemporary swine influenza virus antisera. Influenza Other Respir Viruses 4, 53–60 10.1111/j.1750-2659.2009.00121.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby R. J., Rossow K., Erickson G., Sims Y., Webster R. (2004). Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res 103, 67–73 10.1016/j.virusres.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Wise H. M., Foeglein A., Sun J., Dalton R. M., Patel S., Howard W., Anderson E. C., Barclay W. S., Digard P. (2009). A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83, 8021–8031 10.1128/JVI.00826-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D., García-Sastre A., Xiao X., Wang R., Palese P. (2005). Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1, e4 10.1371/journal.ppat.0010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D., Ortigoza M. B., Palese P. (2006). Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol 80, 7976–7983 10.1128/JVI.00415-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R., Krumbholz A., Eitner A., Krieg R., Halbhuber K. J., Wutzler P. (2007). Prevalence of PB1-F2 of influenza A viruses. J Gen Virol 88, 536–546 10.1099/vir.0.82378-0 [DOI] [PubMed] [Google Scholar]