Abstract
Members of the family Picornaviridae are important pathogens of humans and animals, although compared with the thousands of known bird species (>10 000), only a few (n = 11) picornaviruses have been identified from avian sources. This study reports the metagenomic detection and complete genome characterization of a novel turkey picornavirus from faecal samples collected from eight turkey farms in Hungary. Using RT-PCR, both healthy (two of three) and affected (seven of eight) commercial turkeys with enteric and/or stunting syndrome were shown to be shedding viruses in seven (88 %) of the eight farms. The viral genome sequence (turkey/M176/2011/HUN; GenBank accession no. JQ691613) shows a high degree of amino acid sequence identity (96 %) to the partial P3 genome region of a picornavirus reported recently in turkey and chickens from the USA and probably belongs to the same species. In the P1 and P2 regions, turkey/M176/2011/HUN is related most closely to, but distinct from, the kobuviruses and turdivirus 1. Complete genome analysis revealed the presence of characteristic picornaviral amino acid motifs, a potential type II-like 5′ UTR internal ribosome entry site (first identified among avian-origin picornaviruses) and a conserved, 48 nt long ‘barbell-like’ structure found at the 3′ UTR of turkey/M176/2011/HUN and members of the picornavirus genera Avihepatovirus and Kobuvirus. The general presence of turkey picornavirus – a novel picornavirus species – in faecal samples from healthy and affected turkeys in Hungary and in the USA suggests the worldwide occurrence and endemic circulation of this virus in turkey farms. Further studies are needed to investigate the aetiological role and pathogenic potential of this picornavirus in food animals.
Introduction
The family Picornaviridae is currently divided into 12 genera: Aphthovirus, Avihepatovirus, Cardiovirus, Enterovirus, Erbovirus, Hepatovirus, Kobuvirus, Parechovirus, Sapelovirus, Senecavirus, Teschovirus and Tremovirus (Knowles et al., 2012). Picornaviruses are small, non-enveloped viruses with single-stranded, positive-sense genomic RNA. In general, the genomes (7.2–9.1 kb long) have a common organizational pattern. The single ORF that encodes the polyprotein is flanked by 5′ and 3′ UTRs. The viral polyprotein has been divided into three regions: P1, P2 and P3. The P1 region encodes the viral capsid proteins (VP4-VP2-VP3-VP1), whilst the P2 and P3 regions encode proteins involved in protein processing (2Apro, 3Cpro and 3CDpro) and genome replication (2B, 2C, 3AB, 3BVPg, 3CDpro, 3Dpol) (Racaniello, 2007). In addition, aphthoviruses, cardioviruses, erboviruses, kobuviruses, sapeloviruses, senecaviruses, teschoviruses and ‘turdiviruses’ (proposed) encode a leader (L) protein before the P1 region.
Members of the family Picornaviridae are important pathogens associated with several diseases. Picornavirus infections sometimes cause severe disorders of the gastrointestinal tract and the respiratory, neural, hepatocellular and circulatory systems in humans and animals (Racaniello, 2007; Alexandersen et al., 2012). In addition, approximately three-quarters of all emerging infectious disease agents (including viruses) in humans are thought to be zoonotic in origin (Woo et al., 2006). Among animal species, birds are a well-known reservoir of emerging infectious diseases such as avian influenza virus (H5N1), West Nile virus and Japanese encephalitis virus in humans (Woo et al., 2006, 2010). Beside the zoonotic agents, duck hepatitis virus (DHAV-1) in the genus Avihepatovirus (Kim et al.; 2006) and the recently identified turkey hepatitis virus (THV) (Honkavuori et al., 2011) circulate among domesticated poultry and may cause significant economical losses. Investigation of novel avian viruses, especially from domestic birds, and comparative analysis of their genomes is required to identify unrecognized pathogens and emerging viruses.
Compared with the thousands of known bird species (>10 000), only a few picornaviruses (n = 11) have been described from avian sources. A minority of these picornaviruses have been known for a relatively long time, namely avian encephalomyelitis virus (genus Tremovirus) (Marvil et al., 1999), avian sapelovirus (formerly named duck picornavirus TW90A) in the genus Sapelovirus (Tseng & Tsai, 2007), THV (genus ‘Megrivirus’) (Honkavuori et al., 2011) and DHAV-1 (genus Avihepatovirus). Most of the recently identified avian picornaviruses – two novel species (turdivirus 1 and 2) from wild birds of the family Turdidae (Woo et al., 2010), and pigeon picornavirus A and B from pigeons (Kofstad & Jonassen 2011) – fall in an as-yet-unassigned, but distinct, picornavirus genus. Very recently, a quail picornavirus in common quail (Pankovics et al., 2012) and two novel picornaviruses, tentatively named chicken and turkey galliviruses (ChGV and TuGV, respectively), were discovered in chickens and turkeys (Farkas et al., 2012). The complete genome sequences of pigeon picornavirus A, ChGV and TuGV are not known; for TuGV, only a partial P3 sequence is available.
This study reports the detection and complete genome characterization of a turkey picornavirus, related to the recently identified picornavirus TuGV (Farkas et al., 2012), from faecal samples of healthy and affected commercial turkey (Meleagris gallopavo) collected from different turkey flocks in Hungary. This picornavirus is related distantly to members of the genus Kobuvirus and turdivirus 1 (in the proposed picornavirus genus ‘Orthoturdivirus’) and represents a novel picornavirus species.
Results
Complete genome acquisition and characterization of turkey picornavirus
In faecal sample M176, following viral metagenomic analysis (Kapoor et al., 2008) we found 15 sequence reads that were assembled into eight contigs (Fig. 1) covering 42 % of a picornaviral genome related to turdivirus 1 (GenBank accession no. GU182406) as the closest match using a blast search. The complete RNA genome of this turkey picornavirus (turkey/M176/2011/HUN) – which follows the common picornavirus genome organization: 5′ UTR-L-P1(VP0-3-1)-P2(2A-B-C)-P3(3A-B-C-D)-3′ UTR – consists of 8496 nt, excluding the poly(A) tail (Fig. 1). A large ORF of 7425 nt, which encodes a potential polyprotein precursor of 2474 aa, preceded at the 5′ UTR by 761 nt and followed at the 3′ UTR by 310 nt and a poly(A) tail, was found. The L protein is 450 nt (150 aa) long. The complete P1 (2562 nt; 854 aa), P2 (2025 nt; 675 aa) and P3 (2391 nt; 796 aa) regions show highest amino acid sequence identity to turdivirus 1 (GenBank accession no. GU182406) and porcine kobuvirus (EU787450) (Table 1). In the P3 region, the study sequence shows 77 and 96 % amino acid identity to ChGV (GenBank accession no. JF424824; available sequence length was 3219 nt) and TuGV (GenBank accession numbers JF424828–JF424830; available sequence lengths varying between 1018 and 1020 nt). Possible cleavage sites of the polyprotein were Q/G (Gln-Gly) in all cases, except for Q1711/N and Q1997/S between 2C/3A and 3C/3D, respectively (Fig. 1), supported by NetPicoRNA prediction (Blom et al., 1996). The base composition of coding region was found to be 22.7 mol% A, 20.1 mol% G, 28.2 mol% C and 29.0 mol% U.
Fig. 1.
Predicted genome organization of turkey picornavirus turkey/M176/2011/HUN (GenBank accession no. JQ691613). P1 represents viral structural proteins and P2 and P3 represent non-structural proteins. Nucleotide (upper numbers) and amino acid (lower numbers) lengths are indicated in each gene box. Conserved picornaviral amino acid motifs and predicted potential N-terminal cleavage sites along the coding region are indicated below and above the bar. Positions of the amino acid motifs are indicated by the position of the first amino acid of the motif. Above the gene box, sequence contigs acquired from pyrosequencing are indicated by grey bars.
Table 1. Genomic features of turkey picornavirus (turkey/M176/2011/HUN; GenBank accession no. JQ691613) and representatives of existing or proposed picornavirus genera.
Amino acid sequence identity is shown as a percentage, based on the P1, P2, P3, 3Cpro and 3Dpol regions. Bold indicates the two highest levels of amino acid identity.
| Picornavirus genus | Virus | Genome features | Pairwise amino acid identity (%) | ||||||
| GenBank accession no. | Size (nt) | G+C content | P1 | P2 | P3 | 3Cpro | 3Dpol | ||
| Aphthovirus | Foot-and-mouth disease virus C | NC_002554 | 8115 | 0.54 | 11 | 14 | 21 | 12 | 33 |
| Avihepatovirus | Duck hepatitis A virus 1 | NC_008250 | 7687 | 0.43 | 9 | 10 | 17 | 16 | 19 |
| Cardiovirus | Encephalomyocarditis virus 1 | NC_001479 | 7835 | 0.49 | 10 | 13 | 21 | 9 | 31 |
| Enterovirus | Human enterovirus C PV-1 | NC_002058 | 7440 | 0.46 | 11 | 12 | 22 | 10 | 29 |
| Erbovirus | Equine rhinitis B virus 2 | NC_003077 | 8821 | 0.50 | 11 | 10 | 18 | 8 | 28 |
| Hepatovirus | Hepatitis A virus 1 | NC_001489 | 7478 | 0.38 | 8 | 12 | 18 | 7 | 24 |
| Kobuvirus | Porcine kobuvirus 1 | EU787450 | 8210 | 0.52 | 18 | 27 | 40 | 21 | 53 |
| Parechovirus | Human parechovirus 2 | NC_001897 | 7348 | 0.39 | 9 | 10 | 16 | 7 | 22 |
| Sapelovirus | Porcine sapelovirus 1 | NC_003987 | 7491 | 0.41 | 12 | 10 | 21 | 12 | 28 |
| Senecavirus | Seneca valley virus 1 | NC_011349 | 7310 | 0.51 | 9 | 12 | 21 | 12 | 31 |
| Teschovirus | Porcine teschovirus 1 | NC_003985 | 7117 | 0.45 | 11 | 11 | 21 | 10 | 30 |
| Tremovirus | Avian encephalomyelitis virus 1 | NC_003990 | 7055 | 0.45 | 9 | 13 | 20 | 10 | 25 |
| ‘Salivirus’ | Salivirus 1 | GQ179640 | 7982 | 0.56 | 16 | 24 | 35 | 20 | 44 |
| ‘Megrivirus’ | Turkey hepatitis virus 1 | HM751199 | 9075 | 0.46 | 11 | 18 | 26 | 16 | 36 |
| ‘Orthoturdivirus’ | Turdivirus 1 | GU182406 | 8035 | 0.58 | 18 | 32 | 45 | 25 | 62 |
| ‘Paraturdivirus’ | Turdivirus 2 | GU182408 | 7641 | 0.47 | 13 | 19 | 33 | 23 | 41 |
Analysis of 5′ and 3′ UTRs
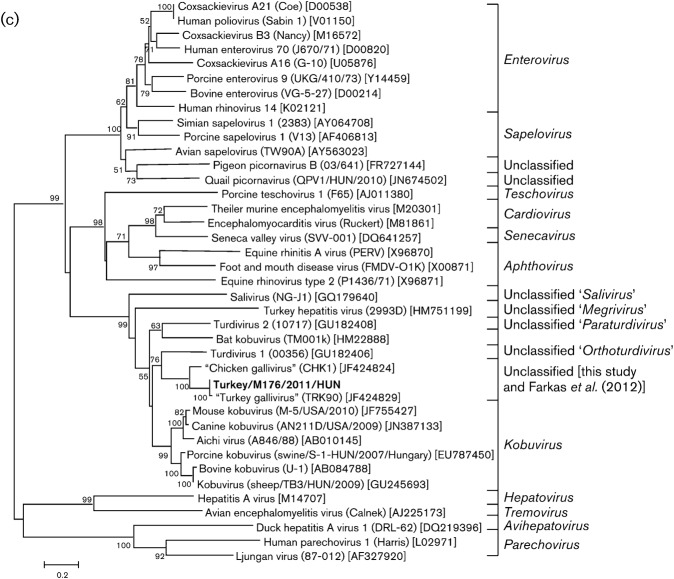
The predicted length of the 5′ UTR is 761 nt (Fig. 2). Seventeen AUG motifs were identified in the 5′ UTR. The last in-frame AUG is the predicted initiation codon, at position 762–764. This AUG codon is preceded by a significant polypyrimidine tract [p(Y) tract]. The longest p(Y) tract (UCUUUUCCUCUUCUUUUUCUUUU) is located upstream in the RNA between nt 640 and 662. Using a blastn search, high (89 %) sequence similarity was found between nt 543–579 of turkey/M176/2011/HUN and the apical region of the J domain of the internal ribosomal entry site (IRES) of encephalomyocarditis virus (EMCV; GenBank accession no. HM641897) in the genus Cardiovirus. Based upon these data and the predicted secondary structure of the 5′ UTR IRES, turkey/M176/2011/HUN has a potential type II-like IRES. This type of IRES comprises five major core domains from H to L and conserved motifs that are also recognizable in the study sequence (Fig. 2). As for EMCV, specific binding between turkey/M176/2011/HUN domain J and the p(Y) tract-binding protein (PTB), and between domains J/K and initiating factor eIF4G and eIF4A, are possible (Fig. 2). A difference was observed between the two virus genomes in the position of the pyrimidine-rich region preceding domain L in the study sequence (Fig. 2). In addition, six or more contiguous cytosines (C) could not be identified in the determined length of the 5′ UTR of turkey/M176/2011/HUN.
Fig. 2.
Predicted RNA secondary structure of the IRES of the turkey picornavirus (turkey/M176/2011/HUN; GenBank accession no. JQ691613). The complete structure of the 5′ UTR, including domains A–L and the type II IRES, has been annotated as proposed previously (inset). The central 5′ IRES domains of turkey picornavirus are labelled from H to L to maintain the continuity of the current nomenclature. The positions of conserved type II IRES motifs and the predicted polyprotein AUG start codon are indicated by shaded boxes. Nucleotide sequence similarity in the apical region of domain J between EMCV (genus Cardiovirus) and turkey picornavirus is boxed and indicated by a black arrow). Analogous to EMCV, possible specific binding between domain J and PTB is indicated by a white arrow.
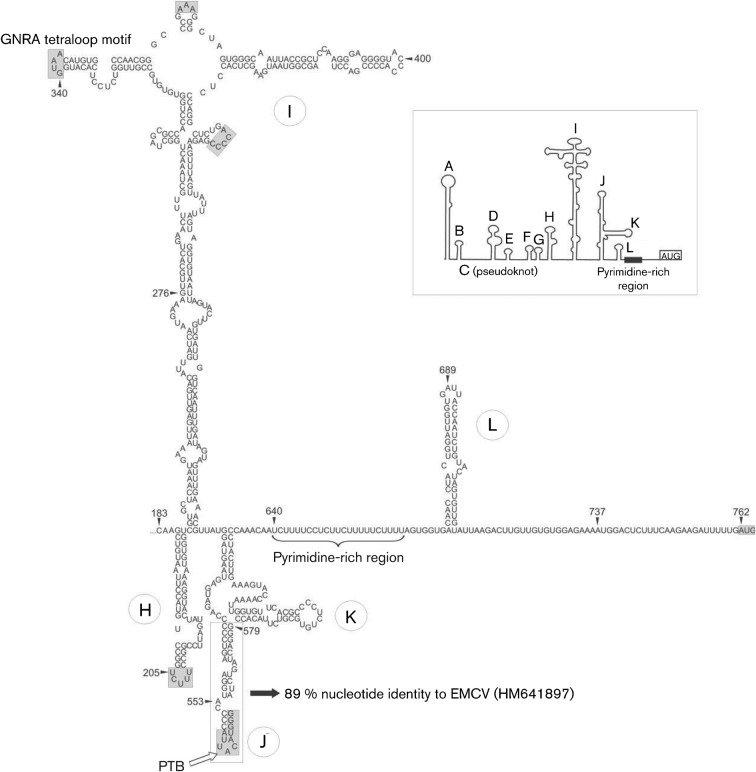
The 310 nt long 3′ UTR sequence of turkey/M176/2011/HUN was similar in length (309 and 311 nt) and nucleotide sequence (73 and 96 % nucleotide identity) to ChGV strain CHK1/USA/2010 (GenBank accession no. JF424824) and TuGV strain TRK90/USA/2010 (JF424829). The predicted secondary RNA structure of the 3′ UTR shows the presence of multiple stem–loops (Fig. 3a) and a 48 nt long ‘barbell-like’ structure (Fig. 3a, b) of the 3′ UTRs with a relatively short p(Y) tract at the upper loop region of the turkey/M176/2011/HUN, which was highly similar in position and nucleotide sequence (UUUCUUU, identical nucleotides) to the core binding site of PTB located at the 3′ UTR of murine norovirus 1 isolate Mu/NoV/GV/MNV1/2002/USA (GenBank accession no. AY228235.2) (Bailey et al., 2010). As well as being found in turkey/M176/2011/HUN, the ‘barbell-like’ structure was also recognizable in members of the genera Avihepatovirus and Kobuvirus, with a 9+6 nt long identical nucleotide motif in the lower loop regions (Fig. 3b–d). The position of the conserved ‘barbell-like’ structure at the 3′ UTRs of different picornaviruses shows significant variability. The first nucleotide of this structure was 227 nt from the stop codon in DHAV-1 (GenBank accession no. DQ249299; 3′ UTR length 317 nt), 56, 70, 72 nt in Aichi virus (FJ890523; 3′ UTR length 218 nt), mouse kobuvirus (JF755427; 3′ UTR length 241 nt) and canine kobuvirus (JN088541; 3′ UTR length 243 nt), respectively, and 167, 171, 171 nt in ChGV (JF424824; 3′ UTR length 310 nt), TuGV (JF424829; 3′ UTR length 314nt) and the study sequence, respectively (Fig. 3d).
Fig. 3.
Conserved motif analysis of the 3′ UTR genome region. (a, c) The predicted secondary 3′ UTR structures of turkey/M176/2011/HUN (a) and of representative members of the genera Avihepatovirus and Kobuvirus (c). Dotted boxes indicate the conserved ‘barbell-like’ structure. (b) Detailed structure of the ‘barbell-like’ formation of the study sequence 3′ UTR. Grey boxes indicate identical nucleotides of different picornavirus species. (d) Sequence alignment of partial 3′ UTRs including the ‘barbell-like’ structure (dotted box) of duck hepatitis A virus 1 (DHAV-1; GenBank accession no. DQ249299), Aichi virus (AiV; FJ890523), mouse kobuvirus (MoKV; JF755427), canine kobuvirus (CaKV; JN088541), turkey gallivirus (TuGV; JF424829), chicken gallivirus (ChGV; JF424824) and the study sequence M176 (turkey/M176/2011/HUN; JQ691613). Note that the numbering of the 3′ UTR alignment begins from the STOP codon.
Analysis of coding regions (L, P1, P2 and P3)
Turkey/M176/2011/HUN encodes a 150 aa L protein upstream of the capsid-encoding genome region; it exhibits no significant sequence similarity to picornaviruses, including aphtho-, cardio-, erbo-, sapelo-, tescho- and turdiviruses. Homologous sequences were also not found using a conserved domains search in cdd (Marchler-Bauer et al., 2011). Neither the catalytic dyad (Cys and His) of papain-like thiol proteases found in the foot-and-mouth disease virus L protein (Gorbalenya et al., 1991) nor a presumed zinc-binding motif, Cys-His-Cys-Cys, found in Theiler’s murine encephalomyelitis virus and quail picornavirus (Chen et al., 1995; Pankovics et al., 2012) was present.
The P1 genome region is located downstream of the L protein-encoding site. As in kobu-, avihepato-, parecho- and turdiviruses, the potential cleavage site between the VP4 and VP2 is not recognizable and turkey/M176/2011/HUN virions are probably built from only three capsid monomers (VP0-VP3-VP1). Multiple potential myristylation motifs (GxxxS, where x is a non-conserved amino acid) (Chow et al., 1987) were recognized in the N-terminal end of VP4 (Fig. 1).
The P2 genome region encodes the non-structural proteins 2A-2B-2C. The 2A protein of turkey/M176/2011/HUN contains the highly conserved picornaviral H-box/NC regions (Fig. 1) (Hughes & Stanway, 2000). Hydrophobic domains were observed in the 2B peptide, with the unique hydrophobic polyproline (P7×) stretch in the C-terminal end (Fig. 1). Similarly to other picornaviruses, the 2C protein of turkey/M176/2011/HUN possesses the highly conserved GxxGXGKS (X, uncharged; x, variable) motif for NTP-binding sites and the DDLxQ motif for putative helicase activity (Gorbalenya et al., 1989; Gorbalenya et al., 1990) (Fig. 1).
The non-structural proteins of the P3 genome region comprise 3A-3BVPg-3Cpro-3Dpol. The short 3B protein (small genome-linked protein or VPg) of turkey/M176/2011/HUN contains no sequence repeats. The conserved tyrosine residue at the third position of VPg proteins of all known picornaviruses was present (e.g. Aichi virus; Yamashita et al., 1998). The conserved catalytic triad (H, E/D, C) of the 3C viral cysteine-active-centre protease was seen in turkey/M176/2011/HUN with a modification (I instead of E/D) (Bazan & Fletterick, 1988); the active-site cysteine in motif GxCG (x, variable) was also present (Gorbalenya et al., 1989) (Fig. 1). The RNA-dependent RNA polymerase (3Dpol) contained highly conserved motifs (KDELR, GGxPSG, YGDD and FLKR) (Fig. 1).
Phylogenetic analysis of turkey picornavirus
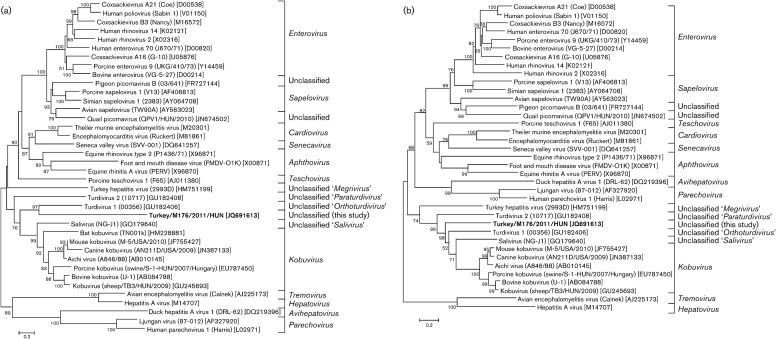
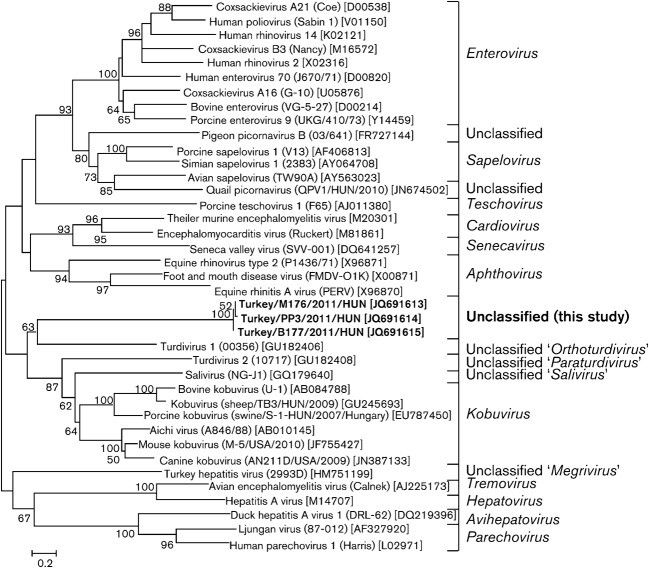
The relatively high nucleotide and amino acid sequence identity at the P3 genome region of turkey/M176/2011/HUN to TuGV and ChGV is also observable in the P3 phylogenetic tree, where the galliviruses and the study sequence cluster together and form a distinct lineage among the other representative members of the family Picornaviridae. The position of turkey/M176/2011/HUN in the phylogenetic trees based on deduced amino acid sequences constructed from the other genome regions (P1, P2) shows a consistent relationship to turdivirus 1 (GenBank accession no. GU182406) of the proposed genus ‘Orthoturdivirus’ and a distant relationship to viruses of the genus Kobuvirus and to klasseviruses/saliviruses (Fig. 4).
Fig. 4.
Phylogenetic relationship between turkey picornavirus (turkey/M176/2011/HUN; GenBank accession no. JQ691613; indicated in bold), representative members of the 12 picornavirus genera and unassigned picornaviruses, based on amino acid sequences of the different picornavirus coding regions: P1 (a), P2 (b) and P3 (c).
Identification and phylogenetic analysis of additional turkey picornavirus sequences
A specific primer pair was used to amplify the complete VP1 of turkey picornavirus to screen faecal samples from healthy and affected commercial meat turkeys collected from an additional seven turkey farms (Table 2). All tested faecal samples were RT-PCR-positive for turkey picornavirus except for samples B195 and PP1, which were collected from 7-day-old turkeys (Table 2). Two VP1-positive samples (from one affected and one healthy turkey) were selected randomly for sequence and phylogenetic analysis. Pairwise comparisons of the three acquired VP1 sequences show 95–96 % nucleotide and 98 % amino acid sequence identity. This high degree of nucleotide and amino acid sequence similarity was also noticeable in the phylogenetic tree (Fig. 5), where all three turkey picornavirus sequences from Hungary grouped together and formed a distinct cluster among picornaviruses. The distinction of the study sequences from the closest available relative, turdivirus 1, was based on the high sequence difference, where the VP1 study sequences (265 aa) show only 18 % amino acid sequence identity to turdivirus 1 (GenBank accession no. GU182406).
Table 2. Epidemiological and clinical background of tested turkeys from eight turkey farms in Hungary, and diagnostic results of the collected faecal samples for turkey picornavirus.
| Farm location | Sample ID | Age (days) | Symptoms | Routine diagnostic tests* | VP1 RT-PCR† | GenBank accession no. |
| Bogyiszló | B-177 | 8 | Catarrhal enteritis, foamy/watery caecum content | par, −bact, − | Positive | JQ691615 |
| B-415 | 14 | Rickets | par, −bact, − | Positive | ||
| Szalánta | B-195 | 7 | Catarrhal enteritis, foamy/watery caecum content | par, −bact, − | Negative | |
| Karakószörcsök | B-232 | 21 | Growth depression | par, −bact, − | Positive | |
| Nagyalásony | B-308 | 47 | Growth depression, skeletal disorders | par, −bact, − | Positive | |
| Bakonypölöske | B-356 | 14 | Uneven growth, catarrhal enteritis, foamy/watery caecum content | par, −bact, + (E. coli bacteraemia) | Positive | |
| Bikal | B-407 | 42 | Skeletal disorders | par, −bact, − | Positive | |
| Nemesbőd | PP-1 | 7 | Healthy | None | Negative | |
| PP-2 | 14 | Healthy | None | Positive | ||
| PP-3 | 21 | Healthy | None | Positive | JQ691614 | |
| Bögöte | M176 | 20 | Growth depression | par, −bact, − | Positive | JQ691613§ |
par, Pathogen parasite diagnostics based on microscopic observation using native faecal samples; bact, routine bacterial culture of heart blood, and liver or bone marrow.
For the primer sequences, see Table S1 (available in JGV Online).
Complete genome.
Fig. 5.
Unrooted phylogenetic tree based on the deduced VP1 capsid protein sequences of the study sequences (in bold) and representative members of the family Picornaviridae.
Discussion
Using a metagenomics and RT-PCR approach, we report the first complete genome characterization and an initial prevalence study of a turkey picornavirus in Hungary that showed a high degree of sequence similarity at the partial P3 genome region to picornaviruses reported by Farkas et al. (2012), tentatively named galliviruses. Galliviruses are present in chicken and turkey flocks in the USA (Farkas et al., 2012), although the complete genome sequence of both ChGV (GenBank accession no. JF424824; longest available sequence length 3219 nt) and TuGV (JF424828–JF424830; longest available sequence length 1020 nt) remain unknown. Because of the high nucleotide and amino acid sequence identity of turkey/M176/2011/HUN to TuGV at the available partial P3 genome region, we refer to this group of novel picornaviruses as galliviruses.
Taxonomic classification of turkey picornavirus
Complete genome analysis of the turkey picornavirus, turkey/M176/2011/HUN, showed that the closest relatives were turdivirus 1 (proposed genus ‘Orthoturdivirus’) and porcine kobuvirus (genus Kobuvirus). According to the current criteria of new genus definition from the Picornaviridae Study Group of the International Committee on Taxonomy of Viruses (http://www.picornastudygroup.com/definitions/genus_definition.htm), novel picornavirus genera are defined by amino acid identity in the P1, P2 and P3 regions of <40, <40 and <50 %, respectively. Based upon this theoretical agreement – and the supporting result of the phylogenetic analysis – turkey picornavirus, together with galliviruses (Farkas et al., 2012), probably represents a novel picornavirus genus.
Analysis of the 5′ UTR structure of turkey picornavirus
Analysis of the 5′ UTR revealed that turkey picornavirus has a type II-like IRES – first identified among avian origin picornaviruses – based upon sequence and secondary structural similarities to cardioviruses, and particularly to EMCV. Initiation of translation on these IRESs begins with specific binding of the eIF4G to J–K domains, which is stimulated by eIF4A (Yu et al., 2011). In addition to canonical translation factors, type II IRESs also require IRES trans-acting factors (ITAFs), such as PTB, that are hypothesized to promote local conformational changes of domain J and to bring it into closer proximity to the base of domain I (Yu et al., 2011). Structural analogies to the EMCV IRES predict similar translation initiation in turkey picornavirus. On the other hand, the cardiovirus 5′ UTR contains a poly(C) tract between domains C and D of the 5′ UTR that varies in length (from 80 to 250 nt) and is associated with virulence (Racaniello, 2007). Whether turkey picornavirus contains a poly(C) tract – which we could not determine by 5′ RACE – currently remains unknown.
Conserved motif analysis in the viral polyprotein of turkey picornavirus
During the genome analysis, characteristic picornaviral motifs (e.g. Rhv-like capsid domains, 2C, 3C and 3D functional domains) were recognized in the viral polyprotein, with minor variance observed in some motifs (e.g. isoleucine instead of the common glutamic acid or aspartic acid in the 3C catalytic triad or more than one potential myristylation motif at the N-terminal end of VP0). The role and functionality of these variations remain to be elucidated.
Analysis of the 3′ UTR ‘barbell-like’ structure
A series of stem–loops, similar to that described by Farkas et al. (2012), was identified at the 3′ UTR of turkey/M176/2011/HUN. The highly structured heteropolymeric regions of picornaviral 3′ UTRs are very diverse and their functions are not yet completely understood, although some evidence indicates that the 3′ UTRs could contain specific binding sites for viral and/or cellular proteins (Rohll et al., 1995). The presence of a nearly identical p(Y) tract located at the same position (upper loop region of the 3′ UTR) in murine noroviruses (MNVs) and in turkey/M176/2011/HUN indicates that it could serve as a core binding site for the cellular poly(C)-binding proteins (PCBP1 and 2) and/or PTB (Makeyev & Liebhaber, 2002; Sawicka et al., 2008), as demonstrated previously for MNVs (Bailey et al., 2010). Further studies with functional analyses will be required to identify cellular or viral factors that might interact with this region.
A 48 nt long ‘barbell-like’ structure with two identical motifs at the lower loop was identified in galliviruses, in turkey/M176/2011/HUN and in members of two other picornavirus genera. Studies of the picornaviral 3′ UTR revealed the presence of a ‘kissing’ interaction between different 3′ UTR stem–loops (Pilipenko et al., 1996), and of 3′ UTR–5′ UTR IRES interactions (Liu et al., 2009) with complementary base pairing. A genome-wide complement sequence search to find sections of sequence corresponding to the two identical motifs of the ‘barbell-like’ structure was unsuccessful. The possible function of this conserved structure is unknown, although the highly conserved nature of this ‘barbell-like’ motif suggests a pivotal role in the viral reproduction process.
Turkey picornavirus in healthy and affected turkeys
The general presence of turkey picornavirus in faecal samples of healthy and affected turkeys in Hungary in the USA (Farkas et al., 2012) strongly suggests the worldwide occurrence and endemic circulation of this virus. The presence of this virus in healthy turkeys does not rule out its pathogenic potential related to stunting syndrome and/or poult enteritis complex, similar to other enteric viral pathogens (e.g. turkey astro-, rota- and reoviruses), as these viruses are also detectable in apparently healthy turkey flocks (Pantin-Jackwood et al., 2007, 2008; Day et al., 2010). Furthermore, it is also possible that this virus could contribute to the development of symptoms as part of co-infections with other pathogens (Day et al., 2010). A clear connection between turkey picornavirus infection and clinical outcomes will require more detailed investigation, including experimental animal infection studies and periodic surveys.
The family Picornaviridae is currently divided into 12 genera; two (Avihepatovirus and Tremovirus) include only avian picornaviruses. Due to the increasing number of novel picornaviruses of different (e.g. avian) origins, the taxonomy of the family Picornaviridae is evolving rapidly. Among the four novel picornavirus genera proposed in 2011, one includes avian picornavirus(es) (THV in the proposed genus ‘Megrivirus’) (Knowles et al., 2012). Furthermore, two of the three recently identified avian picornaviruses (turdiviruses 1 and 2) are probably the type species of two novel genera, ‘Orthoturdivirus’ and ‘Paraturdivirus’ (Woo et al., 2010) and the third, quail picornavirus (Pankovics et al., 2012), stands at the borderline of a potential novel picornavirus genus. Turkey picornavirus, together with galliviruses (Farkas et al., 2012), is a member of another potential genus of avian picornaviruses. The accelerating pace of novel avian picornavirus characterization indicates that there are still many unidentified and highly divergent avian picornaviruses circulating among domestic and wild birds.
Methods
High-throughput pyrosequencing and analysis.
A faecal sample (M176) collected in April 2011 from a commercial meat turkey (M. gallopavo) showing stunting syndrome in a turkey farm located in north-west Hungary was selected for viral metagenomic analysis. PBS-diluted specimens were passed through a 0.45 µm sterile filter and centrifuged at 6000 g for 5 min. The pellet was mixed with a mixture of nucleases to enrich for particle-protected nucleic acids (Victoria et al., 2009). Nucleic acids were extracted using a QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer’s instructions. Viral nucleic acid libraries containing both RNA and DNA molecules were constructed by sequence-independent random RT-PCR amplification, as described by Victoria et al. (2009). 454 pyrosequencing using 454 GS FLX technology was then performed as described previously (Kapoor et al., 2008; Victoria et al., 2009). The pyrosequencing reads and assembled sequence contigs were compared with the GenBank nucleotide and protein databases using blastn and blastx, respectively.
Complete genome acquisition of turkey picornavirus.
Specific primer pairs (Table S1, available in JGV Online) were designed based on the sequence contigs from the pyrosequencing reads to determine the complete nucleotide sequence of the turkey picornavirus using RT-PCR and long-range RT-PCR. RNA was extracted from 150 µl faecal suspension [35–40 % (v/v) in 0.1 M PBS] using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RT-PCR and long-range RT-PCR were performed as described previously (Reuter et al., 2002; Boros et al., 2012). The 5′ and 3′ ends of the genome were determined by using a 5′/3′ RACE PCR kit (Roche) as described previously (Boros et al.; 2011). PCR products were sequenced directly with a BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems) using the primer-walking method and run on an automated sequencer (ABI PRISM 310 Genetic Analyzer; Applied Biosystems).
Detection of turkey picornavirus in different turkey flocks.
Additional faecal samples from seven different turkey flocks located in western Hungary were collected in 2011 from turkeys of different ages and showing different symptoms, and three samples (PP1–3) were collected from a healthy turkey for three consecutive weeks after hatching (Table 2). Native faecal samples were initially tested by microscopic observation for pathogen parasites and by routine bacterial culture of heart blood, and liver or bone marrow. RNA samples were tested for turkey picornavirus by RT-PCR as described above and using specific primer pairs targeting the VP1 region (Table S1).
Sequence and phylogenetic analysis.
Sequences from representative members of different picornavirus genera were obtained from GenBank and the study sequences were aligned using clustal_x software (version 2.0.3) (Thompson et al., 1997); similarity calculations were performed by using GeneDoc software (version 2.7) (Nicholas & Nicholas, 1997). Phylogenetic trees of the amino acid alignments were created using the neighbour-joining method based on the Jones–Taylor–Thornton matrix-based model of mega software (version 5) (Tamura et al., 2011). Bootstrap values (based on 1000 replicates) for each node are given when >50 %. The secondary structures of 5′ and 3′ UTRs were predicted using the Mfold program (Zuker, 2003) and the two-dimensional model was drawn by using the Corel Draw Graphics Suite (version 12). The complete genome sequence of novel turkey picornavirus was submitted to GenBank with accession numbers JQ691613–JQ691615.
Acknowledgements
This work was supported by a grant from the Hungarian Scientific Research Fund (OTKA, K83013). G. R. was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and E. D. by NHLBI grant R01HL083254.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Alexandersen S., Knowles N. J., Dekker A., Belsham G. J., Zhang Z., Koenen F. (2012). Picornaviruses. In Diseases of Swine, 10th edn, pp. 587–620 Edited by Zimmerman J. J., Karriker L. A., Ramirez A., Schwartz K. J., Stevenson G. W. Chichester, UK: Wiley [Google Scholar]
- Bailey D., Karakasiliotis I., Vashist S., Chung L. M., Rees J., McFadden N., Benson A., Yarovinsky F., Simmonds P., Goodfellow I. (2010). Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J Virol 84, 2859–2870 10.1128/JVI.02053-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F., Fletterick R. J. (1988). Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A 85, 7872–7876 10.1073/pnas.85.21.7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Hansen J., Brunak S., Blaas D. (1996). Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci 5, 2203–2216 10.1002/pro.5560051107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros Á., Pankovics P., Simmonds P., Reuter G. (2011). Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS ONE 6, e29145 10.1371/journal.pone.0029145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros Á., Pankovics P., Knowles N. J., Reuter G. (2012). Natural interspecies recombinant bovine/porcine enterovirus in sheep. J Gen Virol 93, 1941–1951 10.1099/vir.0.041335-0 [DOI] [PubMed] [Google Scholar]
- Chen H. H., Kong W. P., Roos R. P. (1995). The leader peptide of Theiler’s murine encephalomyelitis virus is a zinc-binding protein. J Virol 69, 8076–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow M., Newman J. F., Filman D., Hogle J. M., Rowlands D. J., Brown F. (1987). Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327, 482–486 10.1038/327482a0 [DOI] [PubMed] [Google Scholar]
- Day J. M., Ballard L. L., Duke M. V., Scheffler B. E., Zsak L. (2010). Metagenomic analysis of the turkey gut RNA virus community. Virol J 7, 313 10.1186/1743-422X-7-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T., Fey B., Hargitt E., III, Parcells M., Ladman B., Murgia M., Saif Y. (2012). Molecular detection of novel picornaviruses in chickens and turkeys. Virus Genes 44, 262–272 10.1007/s11262-011-0695-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Blinov V. M., Koonin E. V. (1989). Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett 243, 103–114 10.1016/0014-5793(89)80109-7 [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Wolf Y. I. (1990). A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett 262, 145–148 10.1016/0014-5793(90)80175-I [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Lai M. M. (1991). Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett 288, 201–205 10.1016/0014-5793(91)81034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkavuori K. S., Shivaprasad H. L., Briese T., Street C., Hirschberg D. L., Hutchison S. K., Lipkin W. I. (2011). Novel picornavirus in Turkey poults with hepatitis, California, USA. Emerg Infect Dis 17, 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P. J., Stanway G. (2000). The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J Gen Virol 81, 201–207 [DOI] [PubMed] [Google Scholar]
- Kapoor A., Victoria J., Simmonds P., Slikas E., Chieochansin T., Naeem A., Shaukat S., Sharif S., Alam M. M. & other authors (2008). A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A 105, 20482–20487 10.1073/pnas.0807979105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. C., Kwon Y. K., Joh S. J., Lindberg A. M., Kwon J. H., Kim J. H., Kim S. J. (2006). Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae. J Gen Virol 87, 3307–3316 10.1099/vir.0.81804-0 [DOI] [PubMed] [Google Scholar]
- Knowles N. J., Hovi T., Hyypiä T., King A. M. Q., Lindberg A. M., Pallansch M. A., Palmenberg A. C., Simmonds P., Skern T. & other authors (2012). Picornaviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, pp. 855–880 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. San Diego, CA: Elsevier [Google Scholar]
- Kofstad T., Jonassen C. M. (2011). Screening of feral and wood pigeons for viruses harbouring a conserved mobile viral element: characterization of novel Astroviruses and Picornaviruses. PLoS ONE 6, e25964 10.1371/journal.pone.0025964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wimmer E., Paul A. V. (2009). Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta 1789, 495–517 10.1016/j.bbagrm.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev A. V., Liebhaber S. A. (2002). The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8, 265–278 10.1017/S1355838202024627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C. & other authors (2011). cdd: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39 (database issue), D225–D229 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvil P., Knowles N. J., Mockett A. P., Britton P., Brown T. D. K., Cavanagh D. (1999). Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J Gen Virol 80, 653–662 [DOI] [PubMed] [Google Scholar]
- Nicholas K. B., Nicholas H. B. (1997). GeneDoc: a tool for editing and annotating multiple sequence alignments. National Resource for Biomedical Supercomputing, accessed 2 December 2011. http://www.psc.edu/biomed/genedoc
- Pankovics P., Boros Á., Reuter G. (2012). Novel picornavirus in domesticated common quail (Coturnix coturnix) in Hungary. Arch Virol 157, 525–530 10.1007/s00705-011-1192-8 [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M. J., Spackman E., Day J. M., Rives D. (2007). Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms. Avian Dis 51, 674–680 10.1637/0005-2086(2007)51[674:PMOCTF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M. J., Day J. M., Jackwood M. W., Spackman E. (2008). Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis 52, 235–244 10.1637/8174-111507-Reg.1 [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Poperechny K. V., Maslova S. V., Melchers W. J., Slot H. J., Agol V. I. (1996). Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO J 15, 5428–5436 [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. (2007). Picornaviridae: the viruses and their replication. In Fields Virology, fifth edn, pp. 795–838 Edited by Knipe D. M., Howley P. M. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Reuter G., Farkas T., Berke T., Jiang X., Matson D. O., Szücs G. (2002). Molecular epidemiology of human calicivirus gastroenteritis outbreaks in Hungary, 1998 to 2000. J Med Virol 68, 390–398 10.1002/jmv.10216 [DOI] [PubMed] [Google Scholar]
- Rohll J. B., Moon D. H., Evans D. J., Almond J. W. (1995). The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol 69, 7835–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K., Bushell M., Spriggs K. A., Willis A. E. (2008). Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans 36, 641–647 10.1042/BST0360641 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C. H., Tsai H. J. (2007). Sequence analysis of a duck picornavirus isolate indicates that it together with porcine enterovirus type 8 and simian picornavirus type 2 should be assigned to a new picornavirus genus. Virus Res 129, 104–114 10.1016/j.virusres.2007.06.023 [DOI] [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651 10.1128/JVI.02301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., Lau S. K., Yuen K. Y. (2006). Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr Opin Infect Dis 19, 401–407 10.1097/01.qco.0000244043.08264.fc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., Lau S. K., Huang Y., Lam C. S., Poon R. W., Tsoi H. W., Lee P., Tse H., Chan A. S. & other authors (2010). Comparative analysis of six genome sequences of three novel picornaviruses, turdiviruses 1, 2 and 3, in dead wild birds, and proposal of two novel genera, Orthoturdivirus and Paraturdivirus, in the family Picornaviridae. J Gen Virol 91, 2433–2448 10.1099/vir.0.021717-0 [DOI] [PubMed] [Google Scholar]
- Yamashita T., Sakae K., Tsuzuki H., Suzuki Y., Ishikawa N., Takeda N., Miyamura T., Yamazaki S. (1998). Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol 72, 8408–8412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Abaeva I. S., Marintchev A., Pestova T. V., Hellen C. U. T. (2011). Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res 39, 4851–4865 10.1093/nar/gkr045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31, 3406–3415 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]