Abstract
As the only prion disease affecting free-ranging animals, ante-mortem identification of affected cervids has become paramount in understanding chronic wasting disease (CWD) pathogenesis, prevalence and control of horizontal or vertical transmission. To seek maximal sensitivity in ante-mortem detection of CWD infection, this study used paired tonsil biopsy samples collected at various time points from 48 CWD-exposed cervids to compare blinded serial protein misfolding cyclic amplification (sPMCA) with the assay long considered the ‘gold standard’ for CWD detection, immunohistochemistry (IHC). sPMCA-negative controls (34 % of the samples evaluated) included tissues from mock-inoculated animals and unspiked negative controls, all of which tested negative throughout the course of the study. It was found that sPMCA on tonsil biopsies detected CWD infection significantly earlier (2.78 months, 95 % confidence interval 2.40–3.15) than conventional IHC. Interestingly, a correlation was observed between early detection by sPMCA and host PRNP genotype. These findings demonstrate that in vitro-amplification assays provide enhanced sensitivity and advanced detection of CWD infection in the peripheral tissues of cervids, with a potential role for spike or substrate genotype in sPMCA amplification efficiency.
Introduction
Chronic wasting disease (CWD) is an efficiently transmitted prion disease or transmissible spongiform encephalopathy (TSE) of cervids (e.g. deer, elk and moose), and is the only known prion disease affecting free-ranging, non-domestic animals. Whilst the origins of CWD are uncertain, the disease has been present in wild cervid populations of northern Colorado and southern Wyoming for over 40 years (Williams & Young, 1980, 1982) and has now been identified in both captive and free-ranging cervids in 20 states, two Canadian provinces and the Republic of Korea (Sohn et al., 2002; Sigurdson, 2008; National Wildlife Health Centre, 2011). With intensified national surveillance efforts, CWD continues to be identified in areas previously thought to be free of infection, including recent discoveries in Missouri and Maryland in the USA (International Society for Infectious Diseases, 2010, 2011). The prevalence of CWD varies across North America but can be as high as 30 % in endemic areas and up to 100 % in captive populations (Miller et al., 1998; Williams, 2005; Keane et al., 2008a).
Definitive identification of infected individuals, and thus accurate estimation of prevalence, relies on the selection of an appropriate diagnostic specimen. The obex, a region of the caudal medulla, is generally considered the most appropriate tissue for confirmation of TSE infection by conventional diagnostic assays in deer and other species (Peters et al., 2000; OIE, 2010; USDA, 2011). In some species – whitetail and mule deer, for example – immunohistochemical (IHC) analysis of the retropharyngeal lymph nodes (RLNs) may more readily detect CWD protease-resistant prion protein (PrPres), as it has been shown that the RLNs and other alimentary tract-associated lymphoid centres are early sites of PrPres accumulation following oral exposure in deer (Sigurdson et al., 1999; Keane et al., 2008b). Unfortunately, obex and RLN samples are only accessible post-mortem; thus, efforts over the past decade have been made to identify peripherally accessible sites from which an ante-mortem diagnosis may be made. To this end, several investigators have evaluated peripheral lymphoid tissues of sheep, deer and elk, such as the third eyelid, tonsil and recto-anal mucosal associated lymphoid tissue by IHC and found sensitivities (when compared with IHC of the obex) approaching 100 % in both pre-clinical and clinical individuals (van Keulen et al., 1996; Hill et al., 1997; Sigurdson et al., 1999; O’Rourke et al., 2000; González et al., 2006; Spraker et al., 2006; Dennis et al., 2009). Other groups have retrospectively analysed human tonsil biopsies by either conventional IHC or Western blotting to estimate the prevalence of Creutzfeldt–Jakob disease (CJD), although to date there have been no reports of subclinical CJD cases in those populations evaluated (Hill et al., 1999; Frosh et al., 2004; Clewley et al., 2009). Despite the limited success when assessing human peripheral tissues, the positive identification of PrPres accumulation in the peripheral tissues of naturally infected sheep and cervids, often months prior to the onset of clinical signs, demonstrates the utility of this approach in the ante-mortem detection of prion infection in these and potentially in other species.
Also imperative to accurate estimation of TSE prevalence rates is a sensitive and specific ‘gold standard’ diagnostic assay. Despite the continued development and implementation of ELISA-based assays (e.g. TeSeE ELISA, Bio-Rad; HerdCheck EIA, IDEXX Laboratories) for rapid screening of cattle, sheep and cervids, IHC is currently considered the test of choice for confirmation of prion infection in cervids and domestic animals (Spraker et al., 2002; OIE, 2010; USDA, 2011). The true sensitivity and specificity of IHC in the detection of infected individuals is unknown, although it is generally acknowledged that the assay is not infallible, potentially because of either low levels of PrPres in the tissues of subclinical animals (Haley et al., 2009a) or excessively harsh pre-treatment steps to abolish cellular PrPC cross-reactivity (Safar et al., 2005). This, in turn, has led to the development of a variety of assays that involve either amplification of PrPres such as serial protein misfolding cyclic amplification (sPMCA) (Saborio et al., 2001; Soto et al., 2005), fluorometric quantification of seeding activity such as real-time quaking-induced conversion (RT-QuIC) (Atarashi et al., 2008; Wilham et al., 2010) and other approaches that avoid harsh proteolytic treatments such as a conformation-dependent immunoassay (CDI) (Safar et al., 2002; Thackray et al., 2007).
In the present study, we carried out a comparative analysis of paired tonsil biopsies from 48 CWD-exposed or naïve whitetail deer using IHC of three rounds of blinded sPMCA, which, in our experience, restricts the appearance of false-positive results occasionally observed in further rounds of amplification (Haley et al., 2009a, 2011). We hypothesized that sPMCA would permit identification of earlier time points of PrPres accumulation in tonsil biopsies, offering enhanced diagnostic utility over IHC.
Results
To assist in the identification of early time points of peripheral (tonsilar) accumulation of PrPres in infected deer, and perhaps to shed light on the sensitivity of IHC in clinical samples, we compared IHC evaluation of tonsil biopsies from 48 CWD-exposed or naïve deer with a retrospective, blinded analysis using sPMCA.
IHC detection of CWD PrPres in tonsil biopsies
Analysis of serially collected tonsil biopsies by IHC demonstrated PrPres in deer exposed to CWD through various inocula (e.g. CWD-positive brain, blood and blood components, environmental fomites) via selected routes (intracranial, intravenous, intraperitoneal, oral and through environmental exposure). These results have been presented elsewhere (Mathiason et al., 2006, 2009, 2010) and are summarized in Table 1.
Table 1. Summary of source animals with tonsil biopsies evaluated by sPMCA.
IV, Intravenous; IP, intraperitoneal; IC, intracranial; PO, per os; WBCs, white blood cells; NPD, no prion detected; (+) inoculum source from CWD-positive donor; (−) inoculum source from CWD-negative donor.
| Animal ID | PrP genotype (position 96) | Inoculum | Exposure route | Initial IHC+ tonsil biopsy (months) | Observation period (months) | Obex IHC results at necropsy |
| 104 | G/S | (+) Brain | PO | 12 | 19 | Positive |
| 135 | G/S | (+) Brain | IC | 17 | 17 | Positive |
| 138 | G/G | (+) Brain | IC | 6 | 23 | Positive |
| 148 | G/G | (+) Brain | IC | 12 | 19 | Positive |
| 103 | G/G | (−) Brain | PO/IC | NPD | 19 | NPD |
| 123 | G/G | (−) Brain | PO/IC | NPD | 19 | NPD |
| 4488 | G/S | (−) Brain | PO/IC | NPD | 19 | NPD |
| 4516 | G/G | (−) Brain | PO/IC | NPD | 19 | NPD |
| 114 | G/G | (+) Blood | IP | 19 | 19 | Positive |
| 133 | G/G | (+) Blood | IV | 12 | 26 | Positive |
| 137 | G/S | (+) Blood | IV | 19 | 19 | Positive |
| 302 | G/S | (+) Blood | IV | 12 | 27 | Positive |
| 347 | G/S | (+) Blood | IV | 6 | 27 | Positive |
| 372 | G/G | (+) Blood | IV | 12 | 26 | Positive |
| 374 | G/G | (+) Blood | IV | 6 | 24 | Positive |
| 4116 | G/G | (+) Blood | IV | 12 | 19 | Positive |
| 4119 | G/S | (+) Blood | IV | 12 | 24 | Positive |
| 4502 | G/G | (+) Blood | IV | 6 | 24 | Positive |
| 4513 | G/G | (+) Blood | IV | 6 | 19 | Positive |
| 4128 | G/G | (+) WBCs | IV | 12 | 19 | Positive |
| 4506 | G/S | (+) WBCs | IV | 19 | 19 | Positive |
| 4512 | G/G | (+) WBCs | IV | 15 | 18 | Positive |
| 4521 | G/S | (+) WBCs | IV | 19 | 19 | Positive |
| 307 | G/G | (+) Monocytes | IV | NPD | 19 | NPD |
| 311 | G/S | (+) Monocytes | IV | NPD | 19 | NPD |
| 360 | G/G | (+) Monocytes | IV | NPD | 19 | NPD |
| 373 | G/G | (+) Monocytes | IV | NPD | 19 | NPD |
| 323 | G/S | (+) Platelets | IV | NPD | 19 | NPD |
| 324 | G/G | (+) Platelets | IV | 19 | 19 | Positive |
| 325 | G/G | (+) Platelets | IV | 12 | 19 | Positive |
| 357 | G/G | (+) Platelets | IV | 19 | 19 | Positive |
| 4117 | G/G | (+) Plasma | IV | NPD | 19 | NPD |
| 4410 | G/S | (+) Plasma | IV | NPD | 19 | NPD |
| 4515 | G/G | (+) Plasma | IV | NPD | 19 | NPD |
| 303 | G/S | (+) B-cells | IV | 12 | 19 | Positive |
| 322 | G/G | (+) B-cells | IV | 19 | 19 | Positive |
| 346 | G/G | (+) B-cells | IV | 12 | 19 | Positive |
| 371 | G/G | (+) B-cells | IV | 19 | 19 | Positive |
| 113 | G/G | (+) Saliva | PO | 12 | 19 | Positive |
| 132 | G/G | (+) Saliva | PO | 19 | 19 | Positive |
| 147 | G/G | (+) Saliva | PO | NPD | 19 | NPD |
| 111 | G/S | (+) Urine/faeces | PO | NPD | 27 | NPD |
| 124 | G/S | (+) Urine/faeces | PO | NPD | 27 | NPD |
| 134 | G/G | (+) Urine/faeces | PO | NPD | 27 | NPD |
| 141 | G/G | (+) Urine/faeces | PO | NPD | 27 | NPD |
| 150 | G/G | (+) Urine/faeces | PO | NPD | 27 | NPD |
| 4129 | G/S | (+) Environment | Unknown | 19 | 19 | Positive |
| 4461 | G/S | (+) Environment | Unknown | 19 | 19 | Positive |
sPMCA successfully amplifies PrPres in tonsil biopsies
Following sPMCA analysis of tonsil biopsy specimens, sample identities were revealed and the results tabulated (Figs 1 and 2). Of 238 samples analysed, 43 (18 %) were from mock-inoculated negative controls, which, along with an additional 37 unspiked controls [i.e. negative Tg(CerPrP) brain homogenate only; 16 % of total samples evaluated; see Methods], remained negative in blinded, duplicate sPMCA analysis. In 17/48 cases (35 %), sPMCA-amplified PrPres from biopsy specimens at a time point prior to definitive identification by IHC, including five animals that remained IHC negative at necropsy. In 3/46 cases (6.5 %), sPMCA failed to amplify PrPres in terminal samples that were IHC positive for PrPres; in another individual, IHC was positive 6 months prior to sPMCA detection. sPMCA findings were congruous with IHC results from 27/48 deer evaluated throughout the time points examined in each case; 13 of these deer were negative at all time points by both assays, whilst the remaining 14 had consistent results (i.e. IHC−/sPMCA− or IHC+/sPMCA+) in both assays at each collection time point.
Fig. 1.
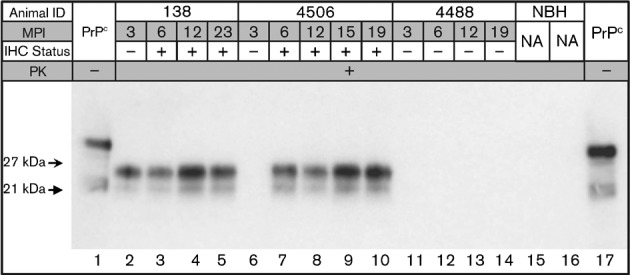
Western blot analysis of tonsil biopsy sPMCA from deer numbers 138 (inoculated intracranially with CWD-positive brain tissue), 4506 (inoculated intravenously with CWD-positive white blood cells) and 4488 (inoculated intracranially and per os with CWD-negative brain tissue) (Table 1). Biopsy dates, in months post-inoculation (MPI), as well as IHC results, are given for each individual and tonsil biopsy, respectively. Non-proteinase K-treated brain homogenate is included in lanes 1 and 17 for band size reference, and unspiked normal brain homogenate (NBH) controls are shown in lanes 15 and 16. na, Not applicable.
Fig. 2.
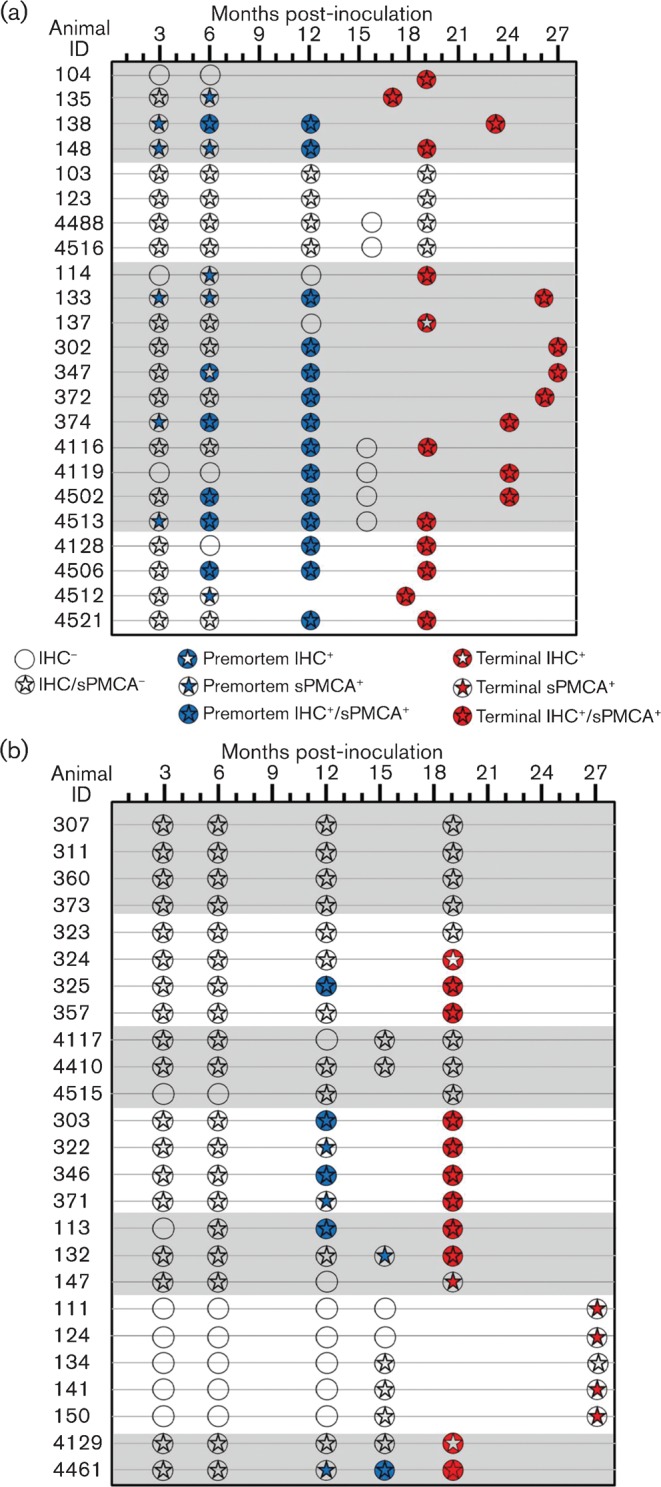
Summary of the tonsil biopsy results for sPMCA versus IHC. Of the 48 deer examined, sPMCA findings were consistent with IHC results in 27 individuals, 13 of which remained negative by both assays throughout the collection period. In 17/48 deer (35 %), sPMCA identified CWD infection at time points prior to conventional IHC, including four IHC-negative deer identified at necropsy. In 4/48 deer (8 %), IHC identified CWD infections in samples that did not amplify PrPres using sPMCA, including three deer that were negative by sPMCA at the termination of the study. Statistical analysis showed that sPMCA could identify CWD PrPres significantly earlier (2.78 months) than IHC. Deer are grouped by inoculum type (shown by shading or no shading) corresponding to the groupings in Table 1.
sPMCA amplifies PrPres at time points significantly earlier than IHC detection
Calculating differences in the rate of CWD detection.
On average, CWD PrPres was amplified by sPMCA in tonsil biopsies 2.78 months prior to IHC detection [maximum-likelihood 95 % confidence interval (CI) 2.40–3.15 months]. When examining individual deer genotypes, this difference was more pronounced for G/G genotype deer (3.72 months earlier than IHC, maximum-likelihood 95 % CI, 3.26–4.18 months). Genotype G/S deer were slightly more likely to have CWD detected earlier by sPMCA (0.89 months earlier than IHC, maximum-likelihood 95 % CI, 0.24–1.54 months), but this effect was not significant (as shown by the above statistics). Overall, our analyses demonstrated sPMCA to be a more sensitive technique for detecting CWD infection in deer than conventional IHC. sPMCA was a more sensitive technique for deer of genotype G/G, but not genotype G/S. It is possible that with a larger number of genotype G/S deer we may have detected a difference between IHC and sPMCA.
Comparing IHC and sPMCA and host genotype for timing of CWD detection.
Overall, in our analysis of all 48 individuals, CWD was detected significantly earlier by sPMCA than by IHC (sign test, S = 17, P = 0.007). This difference was driven by detection of CWD in genotype G/G deer (expressing G/G at position 96 of the cervid PrP, n = 30 individuals; sign test, S = 13, P = 0.002) and was not observed in genotype G/S deer (expressing G/S at position 96, n = 18 individuals; sign test, S = 4, P>0.05). When those individuals testing negative at necropsy by either method were removed from the analysis, sPMCA detected CWD earlier in 12/27 deer (44 %), IHC in one individual (4 %), and there was no difference in 14 deer (52 %). Using this approach, CWD was again detected significantly earlier by sPMCA than by IHC (sign test, S = 12, P = 0.003). Similarly, this difference was driven by detection of CWD in G/G genotype deer (n = 18 individuals; sign test, S = 10, P = 0.002) and was not observed in G/S genotype deer (n = 9 individuals; sign test, S = 2, P>0.05).
Determining detection probabilities for IHC and sPMCA over time.
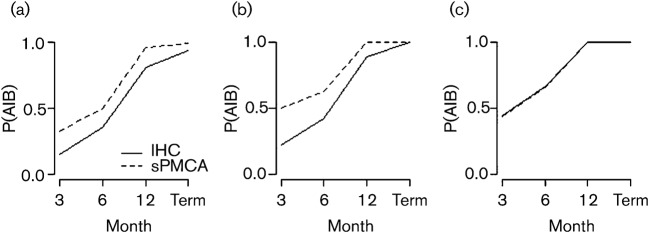
Using Bayes’ rule, we were also able to account for the influence of false positives and false negatives on the probability of detecting CWD given that the deer is infected [P(A|B); see Methods]. Using this conditional probability, our results demonstrated that the probability of detecting an infected deer increased over time for both IHC and sPMCA (Fig. 3a–c). However, overall, sPMCA had a greater probability of detecting a CWD-infected deer than IHC across all time intervals (Fig. 3a). Similar to the above findings, this difference was observable for genotype G/G deer (Fig. 3b), but not for genotype G/S deer (Fig. 3c).
Fig. 3.
Probability of detecting CWD PrPres by IHC or sPMCA over time, given that the deer is infected [P(A|B)]. P(A|B) was calculated for all deer (a), genotype G/G (b) and genotype G/S (c) individuals. Term, Termination of experiment.
sPMCA of tonsil biopsies demonstrates a high level of intra-run agreement
Because tonsil biopsies were run in duplicate by sPMCA on two different sonicators (designated A and B), we sought to determine the level of agreement between the replicate experiments using a commercially available calculator to determine unweighted Cohen’s κ value. A summary of the sPMCA results are presented in Table 2. The observed κ value for the sum of sPMCA experiments was 0.80 (sem 0.046; 95 % CI 0.71–0.89), which is considered substantial to near-perfect agreement under the guidelines proposed by Landis & Koch (1977).
Table 2. Summary of results in duplicate sPMCA experiments.
Tonsil biopsies were evaluated in duplicate on two different sonicators, designated A and B, and scored as positive or negative based on WB results. Corresponding results from duplicated experiments were tallied and evaluated categorically to derive a value for Cohen’s κ (0.80±0.046).
| Sonicator B | Sonicator A | |
| + | − | |
| + | 48 | 10 |
| − | 7 | 169 |
Discussion
Among the TSEs, CWD is unique in its high level of both transmission, with basic reproductive value (R0) estimates ranging from 1.3 to >10 (Miller et al., 2006; Almberg et al., 2011), and prevalence in wild populations, ranging from 1 to 30 % in endemic areas (Williams, 2005). Determination of CWD prevalence in wild cervids relies on rapid screening using ELISA-based techniques, such as the Bio-Rad TeSeE ELISA (Hibler et al., 2003), followed by confirmatory diagnosis using the ‘gold standard’ diagnostic assay – IHC of either obex or RLNs collected post-mortem (Spraker et al., 2002, 2004; Keane et al., 2008b; USDA, 2011). Experimentally, peripheral tissue samples collected ante-mortem (third eyelid, tonsil and rectal-associated lymphoid tissue) have been shown to approach near-perfect agreement with obex samples collected at necropsy (O’Rourke et al., 2000, 2003; Dennis et al., 2009), offering hope for a sensitive, pre-clinical and ante-mortem test for mammalian and human prion diseases.
The potential for low levels of PrPres, beyond the detection limits of conventional assays, in peripherally accessible tissues or body fluids as a result of either pre- or subclinical infection (Haley et al., 2009a) or sensitivity to harsh pre-treatments (Safar et al., 2005) has prompted the development of alternative assays, seeking greater sensitivity than conventional diagnostics, including sPMCA (Saborio et al., 2001; Castilla et al., 2005), RT-QuIC (Atarashi et al., 2007; Orrú et al., 2009) and CDI (Safar et al., 2005). Here, we reported that amplification of PrPres in tonsil biopsies from CWD-exposed deer by sPMCA allowed significantly earlier detection of infection than IHC, by a mean of 2.78 months. The blinded evaluation of samples, inclusion of appropriate negative controls (~34 % of the samples evaluated) and intra-experimental agreement of sPMCA results additionally raises our confidence in these findings.
The results reported here clearly demonstrate the enhanced diagnostic sensitivity of sPMCA over conventional IHC, which should raise concern that the continued reliance on confirmatory IHC could underestimate prion disease prevalence in natural settings. Future analyses of tonsil biopsies – notably in human populations where the identification of subclinical infection has so far proven difficult – using sPMCA or other developing detection systems may allow more sensitive estimation of prevalence rates. Without bioassay of samples analysed in the current study, it remains to be shown whether sPMCA-positive biopsies truly harbour infectious CWD prions. Our previous work, however, provides support for sPMCA amplification of bona fide infectious prions (Meyerett et al., 2008), the correlation of sPMCA positivity with infectivity and the presence of true infectivity in conventional IHC-negative, sPMCA-positive samples (Haley et al., 2009a, b; Gough et al., 2012).
Whilst the sPMCA findings may point to a significant improvement over IHC in detection sensitivity, individual genotypes may play a role in the likelihood of advanced detection of PrPres in tonsil biopsies. Deer homozygous for an allele encoding glycine at position 96 of the cervid PrP were significantly more likely to demonstrate advanced sPMCA amplification; conversely, deer heterozygous at position 96, with alleles encoding glycine and serine, seemed more likely to exhibit congruous sPMCA and IHC results, or in some cases a failure of amplification in the face of an IHC-positive result. This apparent variability in sensitivity could reflect a number of vagaries of the sPMCA assay, including – importantly – the substrate genotype used for amplification. Host genotype has been shown to play an important role in susceptibility to TSE infection in vivo (Windl et al., 1996; Hunter et al., 1997; Johnson et al., 2006), whilst sPMCA substrate genotype has likewise been found to affect amplification efficiency in vitro (Bucalossi et al., 2011). Although the sPMCA protocol in the present study has been standardized and used successfully to analyse numerous tissues with various genotypic backgrounds in previous studies (Haley et al., 2009a, 2011), further evaluation of the effect of substrate genotype in sPMCA from CWD cases is warranted. Future sPMCA studies utilizing peripheral tissues may benefit from enhanced sensitivity with (i) attention to the NBH genotypic background, (ii) additional sample replicates, (iii) analysis of a number of biopsies from each time point or target tissue and (iv) increased frequency of biopsy collection.
Whilst advanced identification of PrPres in extraneural tissues is certainly important for improved TSE surveillance, early peripheral accumulation of prions may also serve as a harbinger of shedding in bodily fluids. In line with this, recent reports have identified infectious prions in excreta from pre-clinical, asymptomatic deer, 7 months or more prior to clinical signs in some individuals (Mathiason et al., 2009; Tamgüney et al., 2009), a time frame consistent with early detection of PrPres in tonsil biopsies of cervids (Mathiason et al., 2009, 2010). Studies currently under way in our laboratory are seeking to determine the relationship between the peripheral accumulation of CWD PrPres (e.g. in tonsilar tissues or rectal biopsies), as indicated by sPMCA versus IHC, and the onset of shedding in excreta.
In summary, we have reported the use of sPMCA to identify time points of peripheral accumulation of CWD PrPres in tonsilar tissues, on average 2.78 months (95 % CI 2.40–3.15) prior to conventional IHC detection. The identification of amplifiable PrPres in tonsil biopsies that were negative by conventional IHC suggests that conventional assays may understate the prevalence of CWD infection. The role of substrate genotype and the relationship between peripheral accumulation of CWD prions and the onset of shedding in bodily fluids are areas of future and ongoing research that we are currently pursuing.
Methods
Infected cervids.
Forty-eight white-tailed deer (Odocoileus virginianus) were exposed to CWD from positive and negative sources in various forms (urine and faeces, saliva, environmental fomites, blood or blood components and brain tissue) and by various routes (orally, intravenously, intracranially, intraperitoneally and through environmental exposure) (Mathiason et al., 2006, 2009, 2010). All animals were inoculated and maintained in dedicated indoor, restricted-access CWD research facilities in accord with Colorado State University Institutional Animal Care and Use Committee guidelines. The sources of inoculum was terminally ill mule deer (Odocoileus hemionus) of unknown PrP genotype (courtesy of Dr M. Miller, Colorado Division of Wildlife, CO, USA) and white-tailed deer utilized in previous and concurrent studies of either of two genotypes: homozygous for glycine (G/G) at cervid PrP position 96, or heterozygous at that position with alleles for both glycine and serine (G/S). Details of the study deer are summarized in Table 1, and deer were selected for this study based on the availability of tonsil biopsies for retrospective analysis by sPMCA.
Study samples.
At various time points (typically 3, 6, 12 and occasionally 15 months post-inoculation) following CWD exposure, as well as at necropsy (17–27 months post-inoculation), animals were sedated and palatine tonsils were biopsied using sterile biopsy equipment. As the sampling regime varied across the duration of the original deer studies (Mathiason et al., 2006, 2009, 2010), with samples occasionally exhausted for repeat IHC analyses, pre-terminal collections were sporadically unavailable for sPMCA in some deer. Biopsy specimens were divided after collection, with half of the sample fixed in 10 % formalin prior to IHC analysis and half frozen at −80 °C for sPMCA. Equal amounts of tissue (~20–50 mg) were used for each analysis.
IHC.
IHC was performed as described by Spraker et al. (1997). Briefly, 3–5 mm sections of formalin-fixed, formic acid-treated tissues were deparaffinized at 60–70 °C for 1 h, rehydrated through a series of xylene/ethanol baths and treated in formic acid a second time for 5 min, prior to a 20 min antigen retrieval cycle (Target Retrieval Solution; Dako) in a 2100 Retriever (PickCell Laboratories). Slides were further processed with the aid of a Ventana Discovery autostainer utilizing a Ventana Red Map stain kit, a PrPCWD-specific primary mAb (clone BAR224; Cayman Chemical) and a biotinylated secondary goat anti-mouse antibody (Ventana). After autostaining, the slides were rinsed quickly in a warm water and detergent solution, passed through a series of dehydration baths and mounted under a coverslip (Mathiason et al., 2006, 2009, 2010).
Preparation of NBH for sPMCA.
NBH, the substrate for prion conversion in vitro, was prepared from naïve Tg(CerPrP)5037 mice in a room that had not previously been used for prion research (Haley et al., 2009a, b, 2011; Kurt et al., 2009). Transgenic Tg(CerPrP)5037 mice express the elk variant of the cervid PrP protein, encoding glycine at position 96 and glutamine at position 226. Following euthanasia and perfusion with 5 mM EDTA in PBS, whole brain was collected and placed on ice. Brain homogenates were prepared as a 10 % (w/v) solution in PMCA buffer [1 % (v/v) Triton X-100, 5 mM EDTA, 150 mM NaCl in PBS (pH 7.2)] with the addition of Complete Protease Inhibitor (Roche Pharmaceuticals) using a dounce homogenizer. Homogenates were then centrifuged for 1 min at 2000 r.p.m. in a bench microfuge and the supernatant frozen in single-experiment aliquots at –80 °C in a ‘prion-free’ room until use in sPMCA. Each preparation was composed of brain from four to six mice to minimize the potential influence of expression variation in cervid PrP or other co-factors (Browning et al., 2004; Angers et al., 2009). Multiple batches of NBH were utilized over the course of the experiments.
sPMCA of tonsil biopsies.
Tonsil biopsies were thawed briefly and ~20 mg of each was trimmed individually and homogenized as a 1 % solution (w/v) in PMCA buffer using a FastPrep tissue homogenizer for 60 s at power setting 6.5. All biopsies were prepared in individual microcentrifuge tubes and homogenized, in parallel and in the same machine, concurrently with positive and negative controls. Prepared homogenates were coded to allow blinded evaluation by sPMCA.
Ten microlitres of test or control tissue homogenate was added to 50 µl NBH and evaluated in duplicate and in parallel with positive and negative tissue-matched controls, in adjacent wells of a 96-well plate (USA Scientific), along with NBH prepared from unexposed Tg(CerPrP)5037 mice as additional, unseeded negative controls. Plates were then sonicated using an ultrasonic processor (Misonix) and incubated at 37 °C. Sonication parameters were set at 40 s bursts at power level 7.0, followed by 30 min of incubation. Ninety-two cycles of sonication were performed over 48 h, with a 10 µl aliquot of sonicated material transferred to 50 µl fresh NBH for serial amplification. Following the third round of amplification, samples were evaluated by Western blotting, as described below, for the presence of PrPres. A sample was considered positive when at least one of the two replicates showed amplification of PrPres. In each experimental run, between 20 and 30 % of samples evaluated were tissue-matched negative controls, and an additional 10–20 % of the samples were unspiked, NBH-negative controls.
Western blotting of sPMCA-amplified tonsil biopsies.
Following the third and final round of sPMCA amplification, an aliquot of each sonicated sample was subjected to Western blotting for evaluation of the PrPres signal. Fifteen microlitres of sample homogenate was mixed with 7 µl sample buffer [0.1 % (v/v) Triton X-100, 4 % (w/v) SDS in PBS] and digested with 3 µl proteinase K (final concentration 60 µg ml−1) for 20 min at 37 °C followed by 10 min at 45 °C. Twenty microlitres of this preparation was run on a pre-cast SDS-PAGE gel (12 % acrylamide; Invitrogen) in a Bio-Rad electrophoresis apparatus for 1 h at 160 mV. Samples were then transferred to a PVDF membrane (Millipore) for 1 h at 115 mV in a Bio-Rad transfer apparatus. PVDF membranes were subsequently probed with a PrP-specific mAb (clone BAR224) conjugated in house to HRP, diluted 1 : 20 000 in 5 % (w/v) powdered milk in 0.2 % Tween 20 in Tris-buffered saline (TBST) for 1 h. Following washing with TBST, immunoreactivity was detected using an enhanced chemiluminescent detection system (ECL-Plus; Amersham Biosciences) in an LAS 3000 imaging system. (Fuji Photo Film; Fuji Inc,)
Statistical analysis of data.
A number of important factors potentially influenced the timing of CWD detection in this study and were critical to consider in our analyses. First, individual deer varied in the rate at which CWD was detected (see Results). Secondly, sampling intervals were not equal or even for all individual deer (Fig. 2). For some individual deer, CWD could not be detected by IHC or sPMCA at the time the terminal sample was collected (see Results and Fig. 2) and, as such, the timing of first identification is unknown for these animals. The study included two genotypes of deer, and this may have affected CWD accumulation profiles and detection sensitivities between IHC and sPMCA. Finally, it was important to consider the influence of false positives and false negatives on detection probabilities for either method.
Calculating differences in the rate of CWD detection.
For individuals in whom CWD was detected by both methods, we determined the mean difference (in months) in the rate at which CWD was detected. This was calculated using maximum-likelihood estimation, which enabled us to profile more accurate estimates of the 95 % CI, given the data around this mean, than sds. Differences in the rate of CWD detection were determined for all deer and for each genotype individually.
Comparing IHC and sPMCA and host genotype for timing of CWD detection.
To account for individuals in whom CWD was not detected by either detection method at the time of necropsy, we evaluated how timing of CWD detection differed between IHC and sPMCA for: (i) individuals in whom CWD was ultimately detected by both methods, and (ii) for all individuals, with the conservative prediction that individuals who were CWD negative by either method at the terminal sample collection would be positive at a following hypothetical sampling date (30 months). We made these comparisons for all deer and for each genotype individually. We analysed whether timing of CWD detection differed between IHC and sPMCA using a sign test. Because the sign test is a ranking test, it enabled unbiased quantification of how timing in detection differed, enabling us to account for the first four of the five factors outlined above.
Sign tests explicitly evaluated the direction/rank of difference (earlier, later or equivalent timing of detection) between IHC and sPMCA, rather than the magnitude of that difference. Thus, our hypothetical allocation of samples to be positive at 30 months had no impact on analytical outcomes. Additionally, our allocation of individuals in whom CWD had not been detected to be positive by both tests at 30 months made our analysis slightly more conservative towards detecting no difference between IHC and sPMCA. On the basis of negative controls being truly negative (see Results), these individuals were logically removed prior to these analyses.
Determining detection probabilities for IHC and sPMCA over time.
To account for the influence of false positives and false negatives on detection of CWD, we utilized Bayes’ rule to calculate the conditional probability:
P(A|B) is the probability of detecting CWD given that the deer is infected, P(B|A) is the probability of being infected given a positive test, P(A) is the probability of detecting CWD, Pr(pos) and Pr(neg) are the true proportion of the study population infected and uninfected (negative controls), respectively, Pr(iden pos) and Pr(iden neg) are the proportion of deer correctly identified as infected and uninfected, respectively, and P(B) is the probability of being infected with CWD [equivalent to Pr(pos)]. The parameter values and sources are given in Table 3.
Table 3. Parameter values used for determining the probability of detecting CWD using IHC or sPMCA over time, given that the deer is infected [P(A|B)].
| Parameter | Month | All deer | Genotype 1 | Genotype 2 | |||
| IHC | sPMCA | IHC | sPMCA | IHC | sPMCA | ||
| P(B|A) | Constant | 1.000* | 0.992† | 1.000* | 0.992† | 1.000* | 0.992† |
| P(A)‡ | 3 | 0.129 | 0.289 | 0.182 | 0.408 | 0.308 | 0.305 |
| 6 | 0.313 | 0.436 | 0.348 | 0.520 | 0.462 | 0.459 | |
| 12 | 0.710 | 0.838 | 0.727 | 0.862 | 0.769 | 0.844 | |
| Term | 0.846 | 0.897 | 0.846 | 0.922 | 0.882 | 0.880 | |
| P(B)§ | 3 | 0.871 (27) | 0.871 (27) | 0.818 (18) | 0.818 (18) | 0.692 (9) | 0.692 (9) |
| 6 | 0.875 (28) | 0.875 (28) | 0.826 (19) | 0.826 (19) | 0.692 (9) | 0.692 (9) | |
| 12 | 0.871 (27) | 0.871 (27) | 0.818 (18) | 0.818 (18) | 0.692 (9) | 0.692 (9) | |
| Term | 0.897 (35) | 0.897 (35) | 0.846 (22) | 0.846 (22) | 0.765 (13) | 0.765 (13) | |
Assumed value, based on IHC being the gold standard diagnostic test.
Conservative value derived from Haley et al. (2011). If based on the present study, this value would have been 1.
Pr(pos), Pr(neg), Pr(iden pos) and Pr(iden neg) values were determined from data collected in the present study.
Values vary due to fluctuations in the sample size of infected deer at monthly intervals, shown in parentheses. The number of negative controls (n = 4) was invariant among months.
We calculated the probability of detecting CWD given that the deer is infected [P(A|B)] for IHC and sPMCA across all deer and for both genotypes. Adequate samples enabled us to undertake this analysis for data from months 3, 6, 12 and the experimental termination (17–27 months; Table 3). The samples analysed were restricted to individuals in whom CWD was only detected by one or both methods by the termination of the experiment.
All analyses were undertaken using program r version 12.13.0 (http://www.r-project.org/, packages bsda and Stats4).
Evaluation of intra-run variability in duplicate experiments.
As all samples were run in duplicate on two different sonicating machines, we sought to compare the inter-run variability in experimental results. A commercially available calculator for the categorical Cohen’s κ value was used to define the agreement of results between the two different experiments (http://faculty.vassar.edu/lowry/kappa.html). Because sPMCA results were bimodal (i.e. either positive or negative), a non-weighted approach was used to calculate Cohen’s κ value. The significance of correlation was determined using the method described by Landis & Koch (1977).
Acknowledgements
This work was supported by grants from the National Institutes of Health (K01RR026270, R01NS061902). We sincerely thank all of those who have made important contributions to this manuscript, including Karl Miller, Bob Warren, David Osborn and Sallie Dahmes, without whom the primary experiments in cervids could not have been performed, and Dr Michael Miller, who provided initial samples of CWD-positive brain and bodily fluids. Other notable contributors who provided assistance with assay development and interpretation include Timothy Kurt and Amy Nalls. Davis Seelig assisted with the conceptual development of figures in the manuscript. For long-term care and sample collection from source deer and transgenic mice, we sincerely thank Sheila Hays, Kelly Anderson and Jeanette Hayes-Klug. Without each of their contributions and unique skills, this work could not have been completed.
References
- Almberg E. S., Cross P. C., Johnson C. J., Heisey D. M., Richards B. J. (2011). Modeling routes of chronic wasting disease transmission: environmental prion persistence promotes deer population decline and extinction. PLoS ONE 6, e19896 10.1371/journal.pone.0019896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers R. C., Seward T. S., Napier D., Green M., Hoover E., Spraker T., O’Rourke K., Balachandran A., Telling G. C. (2009). Chronic wasting disease prions in elk antler velvet. Emerg Infect Dis 15, 696–703 10.3201/eid1505.081458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi R., Moore R. A., Sim V. L., Hughson A. G., Dorward D. W., Onwubiko H. A., Priola S. A., Caughey B. (2007). Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods 4, 645–650 10.1038/nmeth1066 [DOI] [PubMed] [Google Scholar]
- Atarashi R., Wilham J. M., Christensen L., Hughson A. G., Moore R. A., Johnson L. M., Onwubiko H. A., Priola S. A., Caughey B. (2008). Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat Methods 5, 211–212 10.1038/nmeth0308-211 [DOI] [PubMed] [Google Scholar]
- Browning S. R., Mason G. L., Seward T., Green M., Eliason G. A., Mathiason C., Miller M. W., Williams E. S., Hoover E., Telling G. C. (2004). Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J Virol 78, 13345–13350 10.1128/JVI.78.23.13345-13350.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucalossi C., Cosseddu G., D’Agostino C., Di Bari M. A., Chiappini B., Conte M., Rosone F., De Grossi L., Scavia G. & other authors (2011). Assessment of the genetic susceptibility of sheep to scrapie by protein misfolding cyclic amplification and comparison with experimental scrapie transmission studies. J Virol 85, 8386–8392 10.1128/JVI.00241-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla J., Saá P., Soto C. (2005). Detection of prions in blood. Nat Med 11, 982–985 [DOI] [PubMed] [Google Scholar]
- Clewley J. P., Kelly C. M., Andrews N., Vogliqi K., Mallinson G., Kaisar M., Hilton D. A., Ironside J. W., Edwards P. & other authors (2009). Prevalence of disease related prion protein in anonymous tonsil specimens in Britain: cross sectional opportunistic survey. BMJ 338, b1442 10.1136/bmj.b1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M. M., Thomsen B. V., Marshall K. L., Hall S. M., Wagner B. A., Salman M. D., Norden D. K., Gaiser C., Sutton D. L. (2009). Evaluation of immunohistochemical detection of prion protein in rectoanal mucosa-associated lymphoid tissue for diagnosis of scrapie in sheep. Am J Vet Res 70, 63–72 10.2460/ajvr.70.1.63 [DOI] [PubMed] [Google Scholar]
- Frosh A., Smith L. C., Jackson C. J., Linehan J. M., Brandner S., Wadsworth J. D., Collinge J. (2004). Analysis of 2000 consecutive UK tonsillectomy specimens for disease-related prion protein. Lancet 364, 1260–1262 10.1016/S0140-6736(04)17143-2 [DOI] [PubMed] [Google Scholar]
- González L., Dagleish M. P., Bellworthy S. J., Sisó S., Stack M. J., Chaplin M. J., Davis L. A., Hawkins S. A., Hughes J., Jeffrey M. (2006). Postmortem diagnosis of preclinical and clinical scrapie in sheep by the detection of disease-associated PrP in their rectal mucosa. Vet Rec 158, 325–331 10.1136/vr.158.10.325 [DOI] [PubMed] [Google Scholar]
- Gough K. C., Baker C. A., Rees H. C., Terry L. A., Spiropoulos J., Thorne L., Maddison B. C. (2012). The oral secretion of infectious scrapie prions occurs in preclinical sheep with a range of PRNP genotypes. J Virol 86, 566–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley N. J., Mathiason C. K., Zabel M. D., Telling G. C., Hoover E. A. (2009a). Detection of sub-clinical CWD infection in conventional test-negative deer long after oral exposure to urine and feces from CWD+ deer. PLoS ONE 4, e7990 10.1371/journal.pone.0007990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley N. J., Seelig D. M., Zabel M. D., Telling G. C., Hoover E. A. (2009b). Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS ONE 4, e4848 10.1371/journal.pone.0004848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley N. J., Mathiason C. K., Carver S., Zabel M., Telling G. C., Hoover E. A. (2011). Detection of chronic wasting disease prions in salivary, urinary, and intestinal tissues of deer: potential mechanisms of prion shedding and transmission. J Virol 85, 6309–6318 10.1128/JVI.00425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibler C. P., Wilson K. L., Spraker T. R., Miller M. W., Zink R. R., DeBuse L. L., Andersen E., Schweitzer D., Kennedy J. A. & other authors (2003). Field validation and assessment of an enzyme-linked immunosorbent assay for detecting chronic wasting disease in mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), and Rocky Mountain elk (Cervus elaphus nelsoni). J Vet Diagn Invest 15, 311–319 10.1177/104063870301500402 [DOI] [PubMed] [Google Scholar]
- Hill A. F., Zeidler M., Ironside J., Collinge J. (1997). Diagnosis of new variant Creutzfeldt–Jakob disease by tonsil biopsy. Lancet 349, 99–100 10.1016/S0140-6736(97)24002-X [DOI] [PubMed] [Google Scholar]
- Hill A. F., Butterworth R. J., Joiner S., Jackson G., Rossor M. N., Thomas D. J., Frosh A., Tolley N., Bell J. E. & other authors (1999). Investigation of variant Creutzfeldt–Jakob disease and other human prion diseases with tonsil biopsy samples. Lancet 353, 183–189 10.1016/S0140-6736(98)12075-5 [DOI] [PubMed] [Google Scholar]
- Hunter N., Goldmann W., Foster J. D., Cairns D., Smith G. (1997). Natural scrapie and PrP genotype: case–control studies in British sheep. Vet Rec 141, 137–140 10.1136/vr.141.6.137 [DOI] [PubMed] [Google Scholar]
- International Society for Infectious Diseases (2010). Chronic wasting disease, cervids – USA (03): (Missouri). http://www.promedmail.org/direct.php?id=20100303.0697
- International Society for Infectious Diseases (2011). Chronic wasting disease, cervid - USA (05): (Maryland). http://www.promedmail.org/direct.php?id=20110212.0486
- Johnson C., Johnson J., Vanderloo J. P., Keane D., Aiken J. M., McKenzie D. (2006). Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol 87, 2109–2114 10.1099/vir.0.81615-0 [DOI] [PubMed] [Google Scholar]
- Keane D. P., Barr D. J., Bochsler P. N., Hall S. M., Gidlewski T., O’Rourke K. I., Spraker T. R., Samuel M. D. (2008a). Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Invest 20, 698–703 10.1177/104063870802000534 [DOI] [PubMed] [Google Scholar]
- Keane D. P., Barr D. J., Keller J. E., Hall S. M., Langenberg J. A., Bochsler P. N. (2008b). Comparison of retropharyngeal lymph node and obex region of the brainstem in detection of chronic wasting disease in white-tailed deer (Odocoileus virginianus). J Vet Diagn Invest 20, 58–60 10.1177/104063870802000110 [DOI] [PubMed] [Google Scholar]
- Kurt T. D., Telling G. C., Zabel M. D., Hoover E. A. (2009). Trans-species amplification of PrPCWD and correlation with rigid loop 170N. Virology 387, 235–243 10.1016/j.virol.2009.02.025 [DOI] [PubMed] [Google Scholar]
- Landis J. R., Koch G. G. (1977). The measurement of observer agreement for categorical data. Biometrics 33, 159–174 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- Mathiason C. K., Powers J. G., Dahmes S. J., Osborn D. A., Miller K. V., Warren R. J., Mason G. L., Hays S. A., Hayes-Klug J. & other authors (2006). Infectious prions in the saliva and blood of deer with chronic wasting disease. Science 314, 133–136 10.1126/science.1132661 [DOI] [PubMed] [Google Scholar]
- Mathiason C. K., Hays S. A., Powers J., Hayes-Klug J., Langenberg J., Dahmes S. J., Osborn D. A., Miller K. V., Warren R. J. & other authors (2009). Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS ONE 4, e5916 10.1371/journal.pone.0005916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiason C. K., Hayes-Klug J., Hays S. A., Powers J., Osborn D. A., Dahmes S. J., Miller K. V., Warren R. J., Mason G. L. & other authors (2010). B cells and platelets harbor prion infectivity in the blood of deer infected with chronic wasting disease. J Virol 84, 5097–5107 10.1128/JVI.02169-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerett C., Michel B., Pulford B., Spraker T. R., Nichols T. A., Johnson T., Kurt T., Hoover E. A., Telling G. C., Zabel M. D. (2008). In vitro strain adaptation of CWD prions by serial protein misfolding cyclic amplification. Virology 382, 267–276 10.1016/j.virol.2008.09.023 [DOI] [PubMed] [Google Scholar]
- Miller M. W., Wild M. A., Williams E. S. (1998). Epidemiology of chronic wasting disease in captive Rocky Mountain elk. J Wildl Dis 34, 532–538 [DOI] [PubMed] [Google Scholar]
- Miller M. W., Hobbs N. T., Tavener S. J. (2006). Dynamics of prion disease transmission in mule deer. Ecol Appl 16, 2208–2214 10.1890/1051-0761(2006)016[2208:DOPDTI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- National Wildlife Health Centre (2011). Chronic wasting disease (CWD). http://www.nwhc.usgs.gov/disease_information/chronic_wasting_disease/
- O’Rourke K. I., Baszler T. V., Besser T. E., Miller J. M., Cutlip R. C., Wells G. A., Ryder S. J., Parish S. M., Hamir A. N. & other authors (2000). Preclinical diagnosis of scrapie by immunohistochemistry of third eyelid lymphoid tissue. J Clin Microbiol 38, 3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke K. I., Zhuang D., Lyda A., Gomez G., Williams E. S., Tuo W., Miller M. W. (2003). Abundant PrP(CWD) in tonsil from mule deer with preclinical chronic wasting disease. J Vet Diagn Invest 15, 320–323 10.1177/104063870301500403 [DOI] [PubMed] [Google Scholar]
- OIE (2010). Chapter 2.4.6. Bovine spongiform encephalopathy. In OIE Terrestrial Manual 2010, pp. 1–12 http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.04.06_BSE.pdf
- Orrú C. D., Wilham J. M., Hughson A. G., Raymond L. D., McNally K. L., Bossers A., Ligios C., Caughey B. (2009). Human variant Creutzfeldt–Jakob disease and sheep scrapie PrPres detection using seeded conversion of recombinant prion protein. Protein Eng Des Sel 22, 515–521 10.1093/protein/gzp031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J., Miller J. M., Jenny A. L., Peterson T. L., Carmichael K. P. (2000). Immunohistochemical diagnosis of chronic wasting disease in preclinically affected elk from a captive herd. J Vet Diagn Invest 12, 579–582 10.1177/104063870001200618 [DOI] [PubMed] [Google Scholar]
- Saborio G. P., Permanne B., Soto C. (2001). Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 10.1038/35081095 [DOI] [PubMed] [Google Scholar]
- Safar J. G., Scott M., Monaghan J., Deering C., Didorenko S., Vergara J., Ball H., Legname G., Leclerc E. & other authors (2002). Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat Biotechnol 20, 1147–1150 10.1038/nbt748 [DOI] [PubMed] [Google Scholar]
- Safar J. G., Geschwind M. D., Deering C., Didorenko S., Sattavat M., Sanchez H., Serban A., Vey M., Baron H. & other authors (2005). Diagnosis of human prion disease. Proc Natl Acad Sci U S A 102, 3501–3506 10.1073/pnas.0409651102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson C. J. (2008). A prion disease of cervids: chronic wasting disease. Vet Res 39, 41 10.1051/vetres:2008018 [DOI] [PubMed] [Google Scholar]
- Sigurdson C. J., Williams E. S., Miller M. W., Spraker T. R., O’Rourke K. I., Hoover E. A. (1999). Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). J Gen Virol 80, 2757–2764 [DOI] [PubMed] [Google Scholar]
- Sohn H.-J., Kim J.-H., Choi K.-S., Nah J.-J., Joo Y.-S., Jean Y.-H., Ahn S.-W., Kim O.-K., Kim D.-Y., Balachandran A. (2002). A case of chronic wasting disease in an elk imported to Korea from Canada. J Vet Med Sci 64, 855–858 10.1292/jvms.64.855 [DOI] [PubMed] [Google Scholar]
- Soto C., Anderes L., Suardi S., Cardone F., Castilla J., Frossard M. J., Peano S., Saa P., Limido L., Carbonatto M. (2005). Pre-symptomatic detection of prions by cyclic amplification of protein misfolding. FEBS Lett 579, 638–642 10.1016/j.febslet.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Spraker T. R., Miller M. W., Williams E. S., Getzy D. M., Adrian W. J., Schoonveld G. G., Spowart R. A., O’Rourke K. I., Miller J. M., Merz P. A. (1997). Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J Wildl Dis 33, 1–6 [DOI] [PubMed] [Google Scholar]
- Spraker T. R., O’Rourke K. I., Balachandran A., Zink R. R., Cummings B. A., Miller M. W., Powers B. E. (2002). Validation of monoclonal antibody F99/97.6.1 for immunohistochemical staining of brain and tonsil in mule deer (Odocoileus hemionus) with chronic wasting disease. J Vet Diagn Invest 14, 3–7 10.1177/104063870201400102 [DOI] [PubMed] [Google Scholar]
- Spraker T. R., Balachandran A., Zhuang D., O’Rourke K. I. (2004). Variable patterns of distribution of PrPCWD in the obex and cranial lymphoid tissues of Rocky Mountain elk (Cervus elaphus nelsoni) with subclinical chronic wasting disease. Vet Rec 155, 295–302 10.1136/vr.155.10.295 [DOI] [PubMed] [Google Scholar]
- Spraker T. R., Gidlewski T. L., Balachandran A., VerCauteren K. C., Creekmore L., Munger R. D. (2006). Detection of PrPCWD in postmortem rectal lymphoid tissues in Rocky Mountain elk (Cervus elaphus nelsoni) infected with chronic wasting disease. J Vet Diagn Invest 18, 553–557 10.1177/104063870601800605 [DOI] [PubMed] [Google Scholar]
- Tamgüney G., Miller M. W., Wolfe L. L., Sirochman T. M., Glidden D. V., Palmer C., Lemus A., DeArmond S. J., Prusiner S. B. (2009). Asymptomatic deer excrete infectious prions in faeces. Nature 461, 529–532 Corrigendum Nature 466, 653 10.1038/nature08289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray A. M., Hopkins L., Bujdoso R. (2007). Proteinase K-sensitive disease-associated ovine prion protein revealed by conformation-dependent immunoassay. Biochem J 401, 475–483 10.1042/BJ20061264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA (2011). Chronic wasting disease. http://www.aphis.usda.gov/animal_health/animal_diseases/cwd/
- van Keulen L. J., Schreuder B. E., Meloen R. H., Mooij-Harkes G., Vromans M. E., Langeveld J. P. (1996). Immunohistochemical detection of prion protein in lymphoid tissues of sheep with natural scrapie. J Clin Microbiol 34, 1228–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilham J. M., Orrú C. D., Bessen R. A., Atarashi R., Sano K., Race B., Meade-White K. D., Taubner L. M., Timmes A., Caughey B. (2010). Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 6, e1001217 10.1371/journal.ppat.1001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. S. (2005). Chronic wasting disease. Vet Pathol 42, 530–549 10.1354/vp.42-5-530 [DOI] [PubMed] [Google Scholar]
- Williams E. S., Young S. (1980). Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis 16, 89–98 [DOI] [PubMed] [Google Scholar]
- Williams E. S., Young S. (1982). Spongiform encephalopathy of Rocky Mountain elk. J Wildl Dis 18, 465–471 [DOI] [PubMed] [Google Scholar]
- Windl O., Dempster M., Estibeiro J. P., Lathe R., de Silva R., Esmonde T., Will R., Springbett A., Campbell T. A. & other authors (1996). Genetic basis of Creutzfeldt–Jakob disease in the United Kingdom: a systematic analysis of predisposing mutations and allelic variation in the PRNP gene. Hum Genet 98, 259–264 10.1007/s004390050204 [DOI] [PubMed] [Google Scholar]