Abstract
Chlamydia trachomatis is a major cause of bacterial sexually transmitted infections worldwide. In 2006, a new variant of C. trachomatis (nvCT), carrying a 377 bp deletion within the plasmid, was reported in Sweden. This deletion included the targets used by the commercial diagnostic systems from Roche and Abbott. The nvCT is clonal (serovar/genovar E) and it spread rapidly in Sweden, undiagnosed by these systems. The degree of spread may also indicate an increased biological fitness of nvCT. The aims of this study were to describe the genome of nvCT, to compare the nvCT genome to all available C. trachomatis genome sequences and to investigate the biological properties of nvCT. An early nvCT isolate (Sweden2) was analysed by genome sequencing, growth kinetics, microscopy, cell tropism assay and antimicrobial susceptibility testing. It was compared with relevant C. trachomatis isolates, including a similar serovar E C. trachomatis wild-type strain that circulated in Sweden prior to the initially undetected expansion of nvCT. The nvCT genome does not contain any major genetic polymorphisms – the genes for central metabolism, development cycle and virulence are conserved – or phenotypic characteristics that indicate any altered biological fitness. This is supported by the observations that the nvCT and wild-type C. trachomatis infections are very similar in terms of epidemiological distribution, and that differences in clinical signs are only described, in one study, in women. In conclusion, the nvCT does not appear to have any altered biological fitness. Therefore, the rapid transmission of nvCT in Sweden was due to the strong diagnostic selective advantage and its introduction into a high-frequency transmitting population.
INTRODUCTION
Chlamydia trachomatis is the major cause of bacterial sexually transmitted infections worldwide and it can give rise to severe complications such as pelvic inflammatory disease, ectopic pregnancy and infertility. Genital tract infections are caused by a limited number of C. trachomatis serovars (genovars) that appear to represent a stable pool of pathogens. However, C. trachomatis strains resistant to several antimicrobials have been induced in vitro (Wang et al., 2005), raising concerns that C. trachomatis strains resistant to antimicrobials used in routine treatment of chlamydial infection or C. trachomatis strains with enhanced pathogenic properties may emerge naturally, as observed with Chlamydia suis (Lenart et al., 2001). In October 2006, a new variant of C. trachomatis (nvCT) was first reported in Sweden (Ripa [amp ] Nilsson, 2006); it has spread rapidly in Sweden and has also appeared in other European countries (de Barbeyrac et al., 2007; Health Protection Agency, 2008; Herrmann et al., 2008; Hoffmann [amp ] Jensen, 2007; Lynagh et al., 2007; Moghaddam [amp ] Reinton, 2007; Reinton et al., 2009; Unemo et al., 2007; Westh [amp ] Jensen, 2008).
C. trachomatis has a unique developmental cycle which takes place within a specialized cytoplasmic compartment known as an inclusion (Ward, 1983). The obligate intracellular nature of chlamydial development places severe restrictions on studying the biology of these micro-organisms. Most C. trachomatis isolates carry a small plasmid which encodes eight proteins; the role of the plasmid has recently come under close scrutiny since it was demonstrated to be a transcriptional regulator of chromosomal genes and a virulence factor (Carlson et al., 2008). Attempts to cure C. trachomatis of its plasmid have been unsuccessful (Pickett et al., 2005). However, live, naturally occurring, plasmid-free isolates of C. trachomatis do exist but are exceedingly rare, and they do not spread in the population (Farencena et al., 1997; Peterson et al., 1990; Stothard et al., 1998), which may indicate a reduced biological fitness. Plasmid-free strains are unable to accumulate glycogen in their inclusions, which is a unique characteristic of C. trachomatis that can be visualized using iodine staining (Matsumoto et al., 1998). Thus, this unique property is closely linked to the presence of the plasmid, which is now taken as an indicator of virulence (Carlson et al., 2008). Other potential virulence factors have been described, e.g. a type III secretion system that delivers chlamydial effector proteins such as Incs (involved in controlling interactions with the host cell, inclusion development and avoidance of lysosomal targeting), Pmps (autotransporters that interact with the host cell e.g. in attachment) and Tarp (potentially involved in invasion) into the inclusion vacuole and membrane or into the host cytoplasm, which then subvert the host cell for the benefit of the bacterium (Kleba [amp ] Stephens, 2008; Rockey et al., 2000).
Preliminary analyses showed that the Swedish nvCT carries a plasmid with a 377 bp deletion within coding sequence (CDS) 1 (Ripa [amp ] Nilsson, 2007). Subsequent full sequence characterization of the plasmid showed that it additionally carries a 44 bp duplication immediately upstream of both CDS2 and CDS3 (Seth-Smith et al., 2009). The 377 bp deletion covers a region including the target sequences for the independently produced single-target nucleic acid amplification tests (NAATs) manufactured by Roche Diagnostics and Abbott Laboratories, which were used in two-thirds of the counties in Sweden. Failure to detect nvCT in samples submitted for routine diagnosis resulted in false-negative reports with a consequent huge impact on the overall national detection rates in Sweden, especially in counties where these tests were used (Söderblom et al., 2006; Velicko et al., 2007).
Screening for nvCT showed that its proportion, compared with wild-type C. trachomatis (wtCT) strains, varied widely from 7 to 64 % between different counties across Sweden (Herrmann et al., 2008; Unemo et al., 2007). The lowest proportions were found in the one-third of Swedish counties that were using a commercial NAAT (from Becton Dickinson) which targets a plasmid sequence outside the deletion in nvCT. Abbott and Roche designed new dual-target assays that simultaneously target the affected sequence of the nvCT plasmid and another sequence of the plasmid and the chromosomal DNA (ompA); these are Abbott RealTime CT/NG and the Roche Cobas TaqMan CT v2.0, respectively (Hadad et al., 2008).
Several sequence-based typing systems are now available for application to C. trachomatis (Pedersen et al., 2009). Use of variable number tandem repeats (VNTR; Pedersen et al., 2008) and multilocus sequence typing (MLST; Klint et al., 2007) typing systems with the highest resolving power showed that nvCT is clonal; it belongs to serovar E and the ompA sequence is identical to the prototype C. trachomatis reference strain E/Bour. The nvCT was assigned to MLST sequence type 21 (hctB), 19 (CT058), 1 (CT144), 2 (CT172) and 1 (pbpB) (Herrmann et al., 2008; Unemo et al., 2007) and VNTR type 8.7.1 (CT1335, CT1299 and CT1291, respectively) (Pedersen et al., 2008).
To determine when nvCT first appeared in the Swedish population, the only two archival collections of C. trachomatis available, one from Örebro county in central Sweden (1999–2000) and the other from Malmö (2000–2001) in southern Sweden, were screened but no nvCT was found. This result suggested that the nvCT likely arose after 2000. The subsequent rapid and wide transmission of nvCT across Sweden and the initial observation that the prevalence of nvCT remained high despite the introduction of the Cobas TaqMan CT v2.0 in some counties using the Roche NAATs (Hadad et al., 2008) suggested that nvCT, once established and in addition to the diagnostic selective advantage, may have novel biological properties that give it further selective advantage by conferring an increased biological fitness (Hadad et al., 2008; Herrmann et al., 2008; Unemo et al., 2007). A comprehensive case control study involving 1878 patients compared nvCT with wtCT infections; this study showed a significant reduction of symptomatic urethral infection and abdominal pain amongst nvCT-infected women (Bjartling et al., 2009). This difference in symptoms would confer a selective advantage on nvCT, as patients would be less likely to seek diagnosis allowing greater opportunities for transmission. These data taken together suggest that nvCT is a strain which has recently emerged as a result of a rare genetic event from amongst the established population of highly conserved C. trachomatis strains, and hints at potential new biological properties of nvCT.
The aims of our study were to characterize the nvCT strain from Sweden. Here, we present its complete genome sequence (which is the first for a serovar E strain), comparisons with genome sequences of other C. trachomatis serovars (A, B, D and L2) and we investigate the biological properties of nvCT in terms of morphology, developmental cycle and cell tropism.
METHODS
C. trachomatis isolates.
All C. trachomatis isolates examined in this study are described in Table 1. Briefly, the clinical nvCT isolate (Sweden2) was recovered in McCoy cells in October 2006 from a urethral sample from a Swedish man, suffering from symptomatic chlamydial urethritis. The similar clinical ‘wild-type’ serovar E strain (Sweden3) was isolated in McCoy cells in 2001 in the same area of Sweden, i.e. circulated in Sweden prior to the undetected expansion of nvCT. Compared with nvCT, this strain that we refer to as wtCT has an identical ompA sequence and only differs by one nucleotide in VNTR typing (Pedersen et al., 2008). Furthermore, the prototype reference strain E/Bour (also with an identical ompA sequence to nvCT), one plasmid-free E strain (C599) and one lymphogranuloma venereum (LGV) strain (L2/434/BU) were included for phenotypic comparisons.
Table 1.
C. trachomatis strains examined in this study
| Strain | Origin | Site | Date | Serovar | Plasmid | Reference |
|---|---|---|---|---|---|---|
| nvCT (Sweden2) | Malmö, Sweden | Male urethra | 2006 | E | pSW2 | Seth-Smith et al. (2009) |
| Sweden3 (wtCT) | Malmö, Sweden | Female cervix | 2001 | E | pSW3 | Seth-Smith et al. (2009) |
| Bour (wtCT) | USA | Conjunctiva | 1959 | E | pBour | Hanna et al. (1959) |
| C599 (plasmid-free) | Indianapolis, USA | Male urethra | 1996 | E | Plasmid-free | Stothard et al. (1998) |
| L2/434/BU (wtCT) | San Francisco, USA | Bubo | 1969 | L2 | pL2 | Schachter [amp ] Meyer (1969) |
Preparation of DNA for genome sequencing.
A large quantity of the nvCT isolate was prepared in mycoplasma-free McCoy cell cultures by expansion of the culture from the original swab; elementary bodies (EBs) were harvested and gradient-purified prior to storage at −80 °C, as described previously (Skipp et al., 2005). Genomic DNA was extracted and purified from the EBs using the wizard genomic purification kit (Promega) according to the manufacturer's instructions. The only modification was that purified EBs were incubated with proteinase K for 1 h at 60 °C prior to addition of the nuclei lysis solution. The quality of the genomic DNA recovered by using this method was assessed by agarose gel electrophoresis and by spectrophotometric analysis. An accurate determination of plasmid and genome copies for each DNA preparation was obtained by performing 5′-exonuclease (TaqMan) assays, as described previously (Pickett et al., 2005).
Genome sequencing.
The genome of the nvCT isolate was sequenced using paired-end library preparation and shotgun sequencing LR70 on Genome Sequencer FLX (GS FLX; Roche Diagnostics) in two lanes. Using GS FLX Newbler Assembler (Roche), a de novo assembly produced three contigs representing the chromosome, including one contig representing assembled rRNA operons and a single contig for the plasmid. These chromosomal contigs comprised 1 037 359 bp and 510 844 reads, representing a coverage of 119×. For confirmation, the nvCT isolate was also sequenced using Illumina Genome Analyser (Illumina). The assembly of these sequences produced six contigs covering the chromosome, totalling 1 054 452 bp, with the largest contig representing 458 705 bp. The coverage for these contigs ranged from 165× to 498×. Again, the de novo assembly was confounded by repeats, with reads covering the two rRNA operons assembled as a single contig. Other gaps between the chromosomal contigs occurred at homopolymeric tracts (13Cs, 12As, 12Ts) and repeats associated with the tarp gene. The plasmid was present as a separate contig. Integration of the datasets was performed using ICORN (http://icorn.sourceforge.net/), with the ordered GS FLX contigs as a scaffold upon which the Illumina Genome Analyser reads were mapped, and to which corrections were performed. Only one iteration was required to find all of the discrepancies: 90 % of the reads (26 845 892/29 752 262) mapped uniquely, covering 99.9994 % of the genome and representing a genome coverage of greater than 40×, allowing a very high confidence in the final sequence. The contigs were joined using single mapping reads spanning contig boundaries. The nvCT genome was annotated, analysed and compared with all previously sequenced C. trachomatis genomes (Table 2) using the Artemis (Berriman [amp ] Rutherford, 2003) and ACT (Carver et al., 2005) software packages. The genome sequence and annotation have been deposited in the EMBL database under accession number FN652779.
Table 2.
General features of the genomes of the nvCT and all other available genome sequences of C. trachomatis strains
All genome sequences have a G+C content of 41.3 %, two rRNA operons and 37 tRNA operons.
| Strain | Serovar | Biovar | Origin | EMBL accession no. | Chromosome (bp) | No. predicted CDSs | Coding density (%) | Average gene size (bp) | No. pseudogenes* | Plasmid size (bp) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| nvCT (Sweden2) | E | Trachoma (genitotropic) | Sweden | FN652779 | 1 042 839 | 889 | 89 | 1056 | 14 | 7169 | This study |
| UW-3/CX† | D | Trachoma (genitotropic) | USA | AE001273 | 1 042 519 | 894 | 90 | 1050 | 5 | 7493 | Stephens et al. (1998) |
| TZ1A828/OT | B | Trachoma (oculotropic) | Tanzania | FM872307 | 1 044 282 | 879 | 89 | 1051 | 14 | 7502 | Seth-Smith et al. (2009) |
| Jali20 | B | Trachoma (oculotropic) | The Gambia | FM872307 | 1 044 352 | 875 | 89 | 1056 | 18 | 7506 | Seth-Smith et al. (2009) |
| HAR-13† | A | Trachoma (oculotropic) | Saudi Arabia | CP000051 | 1 044 459 | 920 | 90 | 1032 | 8 | 7510 | Carlson et al. (2005) |
| 434/BU | L2 | LGV | USA | AM884176 | 1 038 842 | 889 | 89 | 1052 | 15 | 7499 | Thomson et al. (2008) |
| UCH-1 | L2b | LGV | England | AM884177 | 1 038 869 | 889 | 89 | 1052 | 15 | 7500 | Thomson et al. (2008) |
*Pseudogene nos do not include cytotoxin gene fragments.
†These genomes have been annotated elsewhere, thus predicted CDSs and pseudogene numbers may not be comparable.
Growth characteristics.
Phase-contrast microscopy of the nvCT isolate, Sweden3 (wtCT), prototype reference strain E/Bour and L2/434/BU strain were used to follow the development cycle. Quantitative real-time PCR was used to compare the growth characteristics of nvCT and Sweden3 (wtCT). To accurately compare the characteristics of these two isolates during early and exponential phases of growth, confluent monolayers of mycoplasma-free McCoy cells were infected at an m.o.i. of 1.0 and cultured at 37 °C in 5 % CO2 for 54 h, as described previously (Skilton et al., 2009). For each time point, cells were infected in triplicate and the infection was stopped at 6, 18, 30, 42 and 54 h post-infection. Samples were then stored at −80 °C after snap-freezing. Subsequently, genomic and plasmid DNA was extracted in a microplate format, the residue was resuspended in 100 μl nuclease-free water, and the samples were diluted 1 in 100 prior to quantitative real-time PCR analysis. The quantitative real-time PCR protocol to determine the absolute number of chlamydial plasmids and genomes in samples using 5′-exonuclease (TaqMan) assays with unlabelled primers and carboxyfluorescein/carboxytetramethylrhodamine (FAM/TAMRA) dual-labelled probes has been described previously (Pickett et al., 2005).
Iodine staining.
The nvCT isolate, Sweden3 (wtCT), prototype reference strain E/Bour and plasmid-free E strain (C599; as a staining control) were cultured on coverslips as described above. Inclusions were examined by phase-contrast microscopy then washed several times with PBS and fixed with methanol. Coverslips were stained with 5 % iodine stain (containing both potassium iodide and iodine in 50 % ethanol) for 10 min. The stain was then changed for 2.5 % iodine stain for 10 min and mounted in 5 % iodine stain in glycerol (1 : 1) for microscopy.
High quality digital time lapse video photomicroscopy.
The nvCT isolate and prototype reference strain E/Bour were cultured as described above. Time lapse video microscopy was performed on these strains in McCoy cells as described previously (Skilton et al., 2009).
Cell tropism study.
Confluent monolayers of Hep2, McCoy, BGMK, Vero and 293A cell lines were infected at an m.o.i. of 1.0 with the nvCT isolate, Sweden3 (wtCT), prototype reference strain E/Bour and L2/434/BU (the inocula were prepared in McCoy cells) and cultured at 37 °C in 5 % CO2. At the end of the developmental cycle, the chlamydial growth (inclusion formation) was scored. EBs were harvested and used to infect a fresh culture of the same cells. This was repeated four times.
Transmission electron microscopy (TEM).
The nvCT isolate, Sweden3 (wtCT), and prototype reference strain E/Bour were cultured in BGMK cells at 37 °C in 5 % CO2 for 48 h, as described previously (Skilton et al., 2009).
Antimicrobial susceptibility.
The MIC of tetracycline, erythromycin and ciprofloxacin was determined for the nvCT isolate and Sweden3 (wtCT) in McCoy cells. Briefly, the strains were propagated in McCoy cell cultures on coverslips in plastic vials. When the cultures showed at least 10 inclusions in each high power field (magnification of ×200), a 24-well plate was inoculated. After centrifugation, the inoculum was removed and the wells were replenished with 1.5 ml RPMI medium containing 1 % fetal calf serum and 1 μg cycloheximide ml−1. A two-step dilution series of each antibiotic was also added, starting from 0.05 and then 0.1, 0.25, 0.5 and 1.0 μg ml−1. After fixation in methanol, the inclusions were stained using a fluorochrome-labelled C. trachomatis-specific monoclonal antibody and were examined by using fluorescence microscopy.
RESULTS
The nvCT (Sweden2) genome sequence and genomic comparisons
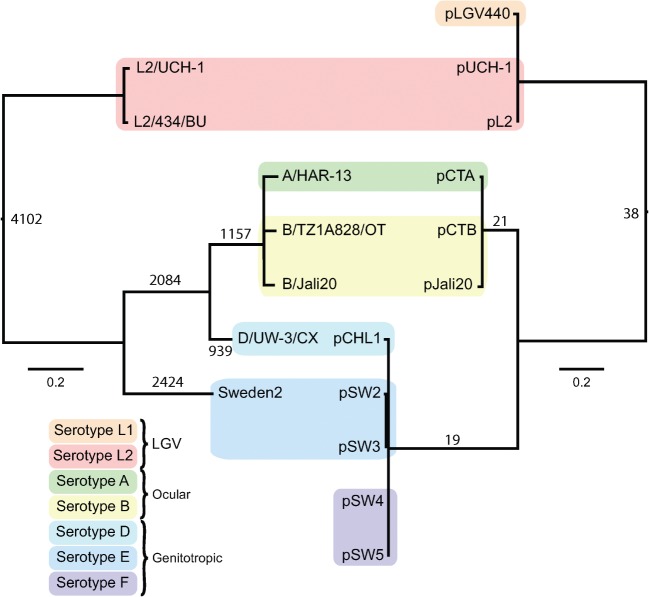
The genome of the unique and currently circulating nvCT (Sweden2) is the first genome sequence of a C. trachomatis serovar/genovar E strain. Genomic DNA was sequenced on both Genome Sequencer FLX and Illumina Genome Analyser, with the data integrated using ICORN software to produce a single contig. The general features of the nvCT genome are summarized in Table 2, with all the complete C. trachomatis genomes included for comparison. The nvCT chromosome consists of 1 042 839 bp (Table 2); it has a high level of sequence identity to, and is syntenic with, all other available genomes from C. trachomatis strains. The plasmid (7169 bp) sequence was as described previously (Seth-Smith et al., 2009). A comparative phylogeny of the genomes and their plasmids confirmed the previous observation (Seth-Smith et al., 2009) that the plasmids have co-evolved with their cognate chromosome (Fig. 1). Compared with the most closely related C. trachomatis genome (D/UW-3/CX), the nvCT has 5896 single nucleotide polymorphisms (SNPs) (Table 3), and no whole functional gene differences were observed.
Fig. 1.
Phylogenetic relationships within C. trachomatis. Comparison of tree topologies produced by maximum-likelihood analyses of plasmid (right) and full genome (left) sequences. The serotypes (serovars/genovars) are indicated. Bars, number of substitutions per SNP site. Numbers on nodes represent numbers of SNPs occurring on that branch.
Table 3.
Pairwise comparison of SNP numbers between complete genome sequences of all available genome-sequenced C. trachomatis strains
| L2b/UCH-1 | L2/434/BU | A/HAR-13 | B/TZ1A828/OT | B/Jali20 | D/UW-3/CX | |
|---|---|---|---|---|---|---|
| L2/434/BU | 487 | |||||
| A/HAR-13 | 8688 | 8693 | ||||
| B/TZ1A828/OT | 8603 | 8588 | 1335 | |||
| B/Jali20 | 8565 | 8554 | 1258 | 1148 | ||
| D/UW-3/CX | 8138 | 8102 | 3652 | 3623 | 3534 | |
| nvCT (Sweden2; E) | 7705 | 7670 | 7115 | 7023 | 7000 | 5896 |
The nvCT genome is closest in size to that of D/UW-3/CX, with the genomes of serovars A and B containing an additional 1–2 kb, and the genomes of serovars L2 and L2b strains containing approximately 4 kb less than the nvCT (Table 2). This difference is accounted for by two large indels in the plasticity zone (PZ), including one within the cytotoxin locus, which is 3745 bp.
This trend in genome size is not reflected in the number of CDS, with the nvCT predicted to encode 889 genes, the same as the L2/L2b strains, whereas the serovar B strains TZ1A828/OT and Jali20 are predicted to carry fewer genes, 879 and 875, respectively. However, the numbers of CDSs present in the genomes of A/HAR-13 and D/UW-3/CX may reflect alternative annotation methods rather than genuine whole gene differences (Table 2).
Within the genomes of serovar E (nvCT), B, L2 and L2b strains, 14–18 pseudogenes have been identified (Table 2), with B/Jali20 harbouring the most. Of the 14 pseudogenes found within the nvCT genome, those that are unique to nvCT include ΨSW2_6011, encoding succinate dehydrogenase subunit B; all genomes sequenced to date contain at least one defunct component within this gene cluster. The nvCT also carries other pseudogenes; these include ΨSW2_8191, which would have encoded a putative exported protease, and four conserved hypothetical/membrane/exported proteins of unknown function: ΨSW2_0111, ΨSW2_1021, ΨSW2_4821 and ΨSW2_8891 (Supplementary Table S1, available with the online version of this paper).
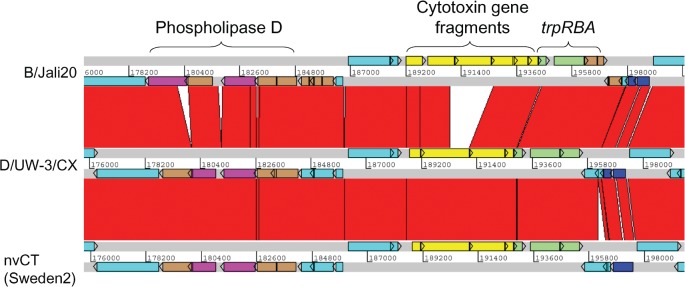
Within the PZ, which is the region in which the greatest sequence variation amongst chlamydiae has been identified (Thomson et al., 2008), the sequence of nvCT resembles most closely that of D/UW-3/CX (Fig. 2). Both of these strains have the same disruptions to the phospholipase D operon, with SW2_1551, SW2_1561 and SW2_1571 intact and SW2_1581 carrying the same mutation as the D/UW-3/CX homologue. The nvCT and D/UW-3/CX also encode complete trp operons, which are usually intact in genitotropic strains, but non-functional in oculotropic (e.g. B/Jali20) and LGV strains (Caldwell et al., 2003). In addition, nvCT displays a cytotoxin locus nearly identical to that of D/UW-3/CX, which produces an active cytotoxin (Belland et al., 2001), indicating that the nvCT may also express this activity. One difference in this operon is the absence of the start codon for CT_165 (D/UW-3/CX notation); thus, for expression of the homologous gene in nvCT, SW2_1641, the protein product would need to be translated from a start codon 35 aa downstream. However, no defined functional domain is affected by this alteration.
Fig. 2.
Comparison of the PZ in C. trachomatis nvCT (Sweden2, serovar E), Jali20 (serovar B) and D/UW-3/CX (serovar D). The grey lines indicate forward and reverse reading frames of sequenced genomes, with predicted CDSs superimposed. The red bars indicate regions of 95–100 % nucleotide identity. Brown CDSs denote pseudogenes. The phospholipase D locus (purple) contains pseudogenes in all strains. The cytotoxin locus (yellow) in D/UW-3/CX produces an active cytotoxin (Belland et al., 2001). This locus is identical in the nvCT, except for an SNP causing a putative 35 aa N-terminal truncation in the leftmost CDS. The trp operon (green) is complete in strains D/UW-3/CX and the nvCT, but has pseudogene trpA in Jali20. The nvCT carries an insertion of 307 bp in a CDS encoding a hypothetical protein downstream of the trp operon, causing an N-terminal extension of 127 aa. Additionally, an SNP in the nvCT relative to D/UW-3/CX causes read-through of CDSs encoding two conserved hypothetical proteins to create one CDS of 247 aa (dark blue). Other CDSs are turquoise.
Additional sequence variations within CDSs of the nvCT genome were observed (these are summarized in Supplementary Table S2), including in hctB, tarp and ompA; the higher diversity in these genes is consistent with previous observations. There are also examples of sequence variation creating alternate C and N termini in several CDSs. The CDS SW2_1731 contains a large insertion of 307 bp relative to other strains, forming a hypothetical protein of 290 aa. Orthologues of this gene appear to be truncated or otherwise disrupted in other C. trachomatis strains. Unlike the situation in strain A/HAR-13, the secF gene, SW2_4571, is present as a full-length copy as described for the serovar B strains (Seth-Smith et al., 2009).
Accordingly, the nvCT genome does not contain any major genetic polymorphisms and the genes for central metabolism, development cycle and virulence, etc. are conserved, giving no indication of altered biological fitness. Furthermore, no new genes or genetic polymorphisms were found that would confer the nvCT with antimicrobial resistance. This is also consistent with our phenotypic antimicrobial susceptibility testing, which showed that the nvCT does not have reduced susceptibility to the antimicrobials used routinely to treat C. trachomatis infections, i.e. the MICs of tetracycline (0.25 mg l−1), erythromycin (0.25 mg l−1) and ciprofloxacin (0.25 mg l−1) were low.
Inclusion formation and growth characteristics of nvCT (Sweden2)
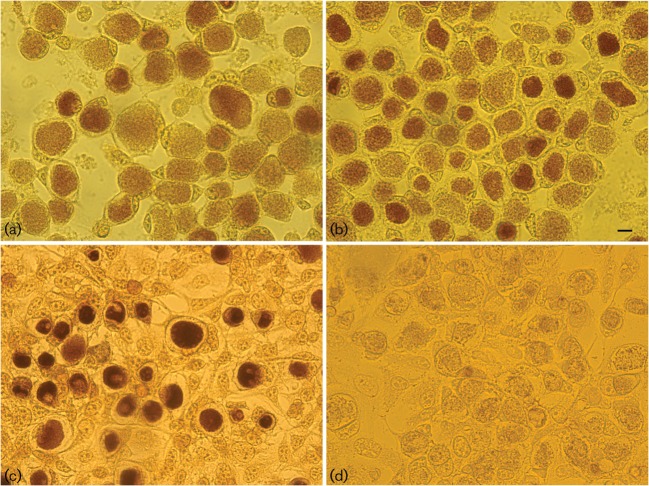
The accumulation of glycogen in mature inclusions was originally used to distinguish C. trachomatis from Chlamydophila psittaci by staining inclusions with iodine. Attempts to reveal the molecular mechanism by which glycogen synthesis is regulated by the plasmid have been unsuccessful, although it is clear that the plasmid itself does not encode the genes for glycogen synthesis. The lack of a system for manipulating the C. trachomatis genome makes natural variants or mutant strains such as nvCT potentially valuable for determining structure–function relationships; whilst the nvCT genome is essentially conserved when compared with other chlamydial isolates, this strain was primarily characterized by a significant deletion within CDS1 of the plasmid. CDS1 encodes a replication protein that is thought to regulate plasmid replication through an iteron binding mechanism and thus is potentially a critical factor in plasmid maintenance (Thomas et al., 1997). Thus, it was of interest to investigate whether the naturally occurring CDS1 deletion within the nvCT plasmid has an effect on the ability of this strain to accumulate glycogen, which might reflect altered biological fitness and virulence. Fig. 3 shows that mature nvCT inclusions appear normal when compared with the reference strain E/Bour and Sweden3 (wtCT).
Fig. 3.
Iodine-stained inclusions of the C. trachomatis strains nvCT [Sweden2 (a)], Sweden3 [wtCT (b)], E/Bour [prototype reference E strain (c)] and C599 [plasmid-free E strain (d)] in McCoy cells. Bar, 4 μm. Stained inclusions containing glycogen are dark brown, as seen in (a)–(c).
The growth characteristics of nvCT and Sweden3 (wtCT) were analysed by quantitative real-time PCR and phase-contrast microscopy in McCoy cells; both strains exhibited similar growth kinetics (Supplementary Fig. S1). They also grew at a similar rate to the E/Bour strain, but as expected, grew more slowly than L2, as judged by phase-contrast microscopy. Cell infection by the nvCT and E/Bour was analysed by high-quality digital time lapse video photomicroscopy (Supplementary Fig. S2); the video showed that, for both E/Bour and nvCT, the inclusion development is similar and that nvCT has a typical C. trachomatis phenotype.
Co-cultivation of nvCT and Sweden3 (wtCT) showed that these strains can be co-propagated and coincide and, at least in vitro, seemed to be equally transmissible, i.e. they did not out-compete each other during many passages. In several Swedish sample collections, rare co-infections of nvCT and wtCT strains have also been identified, which supports the hypothesis that the nvCT does not out-compete the wtCT strains in vivo (data not shown).
Cell tropism
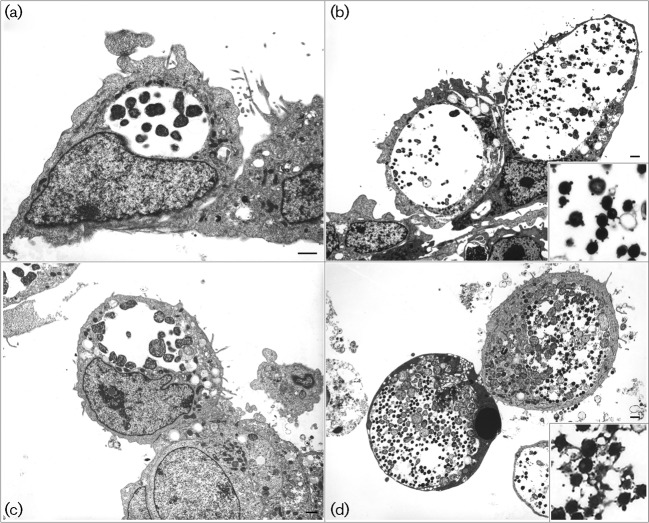
The cell tropism experiments, using the commonly available Hep2, McCoy, BGMK, Vero and 293A cell lines, are summarized in Supplementary Table S3. These data suggest that there are only some subtle differences in the cell tropism of the nvCT and Sweden3 (wtCT). Briefly, the reference C. trachomatis E/Bour strain (also a serovar E isolate) grew well in all cells except Vero cells. The nvCT and Sweden3 also displayed the same cell tropism towards Vero cells, i.e. they could initially infect Vero cells but only at a very low level and then could not be passaged or expanded further. In contrast, E/Bour grew well and expanded rapidly in BGMK cells (a monkey cell line as Vero cells), whereas both the nvCT and Sweden3 could only initially infect BGMK cells and complete a single developmental cycle; further attempts at passage of Sweden3 were unsuccessful whilst the nvCT rapidly adapted to grow in BGMK cells following the second passage. Therefore, we looked closely at the developmental cycles of nvCT, Sweden3 and E/Bour by TEM to see if there were any major differences. However, these strains looked similar both at the mid stage of the developmental cycle (Fig. 4a and c) and when mature inclusions had formed (Fig. 4b and d). We noticed that some inclusions contained EBs with membrane blebs; this was not an artefact of the fixation process because adjacent inclusions in the same preparation displayed this property and the E/Bour strain also had some inclusions, with EBs carrying blebs which occurred regardless of cell type. Furthermore, TEM preparations performed independently in two different laboratories made the same observations.
Fig. 4.
TEM of the C. trachomatis strains nvCT [Sweden2 (a) and (b)] and E/Bour [prototype reference E strain (c) and (d)] in BGMK cells. Electron micrographs were taken at 24 h [mid stage of developmental cycle (a) and (c)] and 48 h [mature inclusions (b) and (d)], and showed no major differences between the strains. The enlarged images in the lower right corner of (b) and (d) show EBs with membrane blebs.
DISCUSSION
The results of this study show that the emergence and rapid and nationwide transmission of the nvCT in Sweden was due to the strong diagnostic selective advantage. This is supported by our data which show that the nvCT does not have any obvious biological advantage, or even disadvantage, i.e. the nvCT does not have any altered biological fitness. The nvCT belongs to the most commonly reported genitotropic serovar (serovar E) worldwide; in Sweden, serovar E strains constitute about 40–50 % of the C. trachomatis population (Fredlund et al., 2004; Jurstrand et al., 2001; Lysén et al., 2004; Pedersen et al., 2009; Suchland et al., 2003). Comparisons of the nvCT genome, which is also the first genome sequence for serovar E, were made with all the complete genome sequences available (serovars A, B, D and L2). Compared with these genomes and several other relevant wtCT isolates, nvCT does not display any genetic polymorphisms that appear to result in an altered biological fitness; the genes for central metabolism, development cycle and virulence were conserved. Phenotypic characteristics, such as growth kinetics, development cycle, morphology, glycogen accumulation, plasmid copy number (Seth-Smith et al., 2009) and antimicrobial susceptibility were unchanged. In addition, co-cultivation of nvCT and Sweden3 (wtCT serovar E) showed that these strains can coincide in vitro; additionally, co-infections of nvCT and wtCT strains have been identified and it has previously been shown that in Örebro county in Sweden, after excluding all the nvCT infections, the serovar distributions before and after the introduction of the nvCT were similar (Jurstrand et al., 2010). These data further support the notion that the nvCT does not out-compete the wtCT serovar E strains. All this is also supported by the observations that the nvCT and wtCT infections are very similar in terms of epidemiological distribution and that differences in clinical signs have been rare (Anagrius [amp ] Loré, 2008; Marions et al., 2008; Unemo et al., 2007); these have only been described in women in one large and well-designed study (Bjartling et al., 2009). Nevertheless, the effect of small-scale gene variation such as single amino acid alterations, and alterations in timing and level of gene transcription and translation cannot be completely excluded based on the present genomic and phenotypic data.
The transmission of the nvCT has had a huge impact on the national detection rates of C. trachomatis in Sweden (Söderblom et al., 2006; Velicko et al., 2007), with higher proportions of nvCT in the counties using Roche/Abbott diagnostics systems than those using the BD system, which was able to detect the nvCT throughout the period that it arose. However, the recent national Swedish data show that the increase in the C. trachomatis incidence, i.e. after ‘catch-up’ of the previously missed nvCT-positive cases in early 2007 (Velicko et al., 2007), followed a similar trend to that observed from the mid-1990s (Swedish Institute for Infectious Disease Control, 2009), and the proportion of the nvCT strain compared with wtCT has now started to decline after the selective diagnostic advantage for nvCT was removed (Klint et al., 2009). Single cases of nvCT have been described in a few countries beyond Scandinavia; these include Ireland, France and Scotland (de Barbeyrac et al., 2007; Health Protection Agency, 2008; Herrmann et al., 2008; Lynagh et al., 2007; Unemo et al., 2009). However, current knowledge on the presence and prevalence of nvCT beyond Sweden is limited due to the small number of recent studies and the fact that many laboratories in European countries still cannot detect the nvCT (Reischl et al., 2009; Unemo et al., 2009). Furthermore, the commercial NAATs that can detect nvCT do not allow its discrimination from wtCT, and nvCT-discriminatory in-house NAATs are rarely used. Now that nvCT infections are being routinely diagnosed and treated, and because nvCT has similar biological fitness to the wtCT, presumably the nvCT will not be eradicated but will reach equilibrium with the wtCT strains in the C. trachomatis population. The proportions of nvCT infections from different Swedish counties have started to converge towards an equilibrium, following removal of the selective diagnostic advantage for nvCT (Klint et al., 2009). The nvCT is already spreading in other countries, and it could be in early or even late transmission in several. Laboratories using commercial NAATs or in-house NAATs that do not detect the nvCT are encouraged to (i) monitor their C. trachomatis incidence rate to identify unexplained significant declines, (ii) frequently participate in effective internal and external quality assurance and control schemes and (iii) consider changing to a testing system that detects the nvCT.
In conclusion, the most plausible explanation for the emergence of the nvCT is that it was due to a recent single genetic event, which appears to be neutral with regard to biological fitness. It is likely to have occurred within a single bacterial cell that clonally expanded and initially existed as a co-infection together with the wild-type parent/progenitor C. trachomatis. The nvCT and progenitor were initially transmitted simultaneously; however, at some time, the nvCT was able to separate from the progenitor and cause a single clonal infection, by chance rather than by out-competition of the progenitor due to increased biological fitness. The nvCT was then rapidly and widely transmitted due to the strong diagnostic selective advantage. Accordingly, nvCT escaped detection and thus could spread rapidly, especially in high-frequency transmitting populations. Effective control measures (treatment and contact tracing) that were only applied to the wild-type allowed nvCT to proliferate. This may also be a scenario and a model for expansion and wide transmission of antimicrobial-resistant C. trachomatis strains that may possibly emerge in the future, but the only difference is, that if this occurs, the selection will be biological and not diagnostic.
Acknowledgments
The authors are grateful to the core sequencing facilities and especially Kicki Holmberg and Joakim Lundeberg, KTH Genome Center, Royal Institute of Technology, Stockholm, Sweden, and Thomas Dan Otto, The Wellcome Trust Sanger Institute, Cambridge, UK, for help with ICORN. The authors are also grateful to Dr B. Van der Pol for the generous gift of the plasmid-free C. trachomatis strain C599. This study was supported by grants from the Örebro County Council Research Committee, the Foundation for Medical Research at Örebro University Hospital, Örebro, Sweden, and the Wellcome Trust grant no 080348 to I. N. C.
Abbreviations
CDS, coding sequence
EBs, elementary bodies
LGV, lymphogranuloma venereum
MLST, multilocus sequence typing
NAAT, nucleic acid amplification test
nvCT, new variant of C. trachomatis
PZ, plasticity zone
TEM, transmission electron microscopy
SNPs, single nucleotide polymorphisms
VNTR, variable number tandem repeats
wtCT, wild-type C. trachomatis
Footnotes
The GenBank/EMBL/DDBJ accession number for the genome sequence of the nvCT isolate reported in this paper is FN652779.
Three supplementary tables and two supplementary figures (including one showing high quality digital time lapse video photomicroscopy of infection of McCoy cells with various C. trachomatis strains) are available with the online version of this paper.
References
- Anagrius, C. [amp ] Loré, B. (2008). Clinical experiences of new variant Chlamydia trachomatis in a Swedish high prevalence county. In 24th International Union against Sexually Transmitted Infections – Europe conference, Milan, abstract FOC4.12.
- Belland, R. J., Scidmore, M. A., Crane, D. D., Hogan, D. M., Whitmire, W., McClarty, G. [amp ] Caldwell, H. D. (2001). Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc Natl Acad Sci U S A 98, 13984–13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman, M. [amp ] Rutherford, K. (2003). Viewing and annotating sequence data with Artemis. Brief Bioinform 4, 124–132. [DOI] [PubMed] [Google Scholar]
- Bjartling, C., Osser, S., Johnson, A. [amp ] Persson, K. (2009). Clinical manifestations and epidemiology of the new genetic variant of Chlamydia trachomatis. Sex Transm Dis 36, 529–535. [DOI] [PubMed] [Google Scholar]
- Caldwell, H. D., Wood, H., Crane, D., Bailey, R., Jones, R. B., Mabey, D., Maclean, I., Mohammed, Z., Peeling, R. [amp ] other authors (2003). Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111, 1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, J. H., Porcella, S. F., McClarty, G. [amp ] Caldwell, H. D. (2005). Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun 73, 6407–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, J. H., Whitmire, W. M., Crane, D. D., Wicke, L., Virtaneva, K., Sturdevant, D. E., Kupko, J. J., III, Porcella, S. F., Martinez-Orengo, N. [amp ] other authors (2008). The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76, 2273–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver, T. J., Rutherford, K. M., Berriman, M., Rajandream, M. A., Barrell, B. G. [amp ] Parkhill, J. (2005). ACT: the Artemis comparison tool. Bioinformatics 21, 3422–3423. [DOI] [PubMed] [Google Scholar]
- de Barbeyrac, B., Raherison, S., Cado, S., Normandin, F., Clerc, M., Clairet, V., Bébéar, C. [amp ] Goulet, V. (2007). French situation concerning the Swedish Chlamydia trachomatis variant. Euro Surveill 12, E11–E12. [DOI] [PubMed] [Google Scholar]
- Farencena, A., Comanducci, M., Donati, M., Ratti, G. [amp ] Cevenini, R. (1997). Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect Immun 65, 2965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund, H., Falk, L., Jurstrand, M. [amp ] Unemo, M. (2004). Molecular genetic methods for diagnosis and characterisation of Chlamydia trachomatis and Neisseria gonorrhoeae: impact on epidemiological surveillance and interventions. APMIS 112, 771–784. [DOI] [PubMed] [Google Scholar]
- Hadad, R., Fredlund, H. [amp ] Unemo, M. (2008). Evaluation of the new COBAS TaqMan CT Test v2.0 and impact on the proportion of new variant Chlamydia trachomatis by the introduction of diagnostics detecting new variant C. trachomatis in Örebro county, Sweden. Sex Transm Infect 85, 190–193. [DOI] [PubMed] [Google Scholar]
- Hanna, L., Thygeson, P. [amp ] Jawetz, E. (1959). Elementary-body virus isolated from clinical trachoma in California. Science 130, 1339–1340. [DOI] [PubMed] [Google Scholar]
- Health Protection Agency (2008). New variant Chlamydia trachomatis in Scotland. HPS Weekly Report 42 (2008/39). http://www.hps.scot.nhs.uk/ewr/redirect.aspx?id=39092. Accession date: 10th January 2010.
- Herrmann, B., Törner, A., Low, N., Klint, M., Nilsson, A., Velicko, I., Söderblom, T. [amp ] Blaxhult, A. (2008). Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerg Infect Dis 14, 1462–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, S. [amp ] Jensen, J. S. (2007). Mutant Chlamydia trachomatis in Denmark. Euro Surveill 12, E7–E8. [DOI] [PubMed] [Google Scholar]
- Jurstrand, M., Falk, L., Fredlund, H., Lindberg, M., Olcén, P., Andersson, S., Persson, K., Albert, J. [amp ] Bäckman, A. (2001). Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J Clin Microbiol 39, 3915–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurstrand, M., Christerson, L., Klint, M., Fredlund, H., Unemo, M. [amp ] Herrmann, B. (2010). Characterization of Chlamydia trachomatis by ompA sequencing and multilocus sequence typing in a Swedish county before and after identification of the new variant. Sex Transm Infect 86, 56–60. [DOI] [PubMed] [Google Scholar]
- Kleba, B. [amp ] Stephens, R. S. (2008). Chlamydial effector proteins localized to the host cell cytoplasmic compartment. Infect Immun 76, 4842–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klint, M., Fuxelius, H. H., Goldkuhl, R. R., Skarin, H., Rutemark, C., Andersson, S. G., Persson, K. [amp ] Herrmann, B. (2007). High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J Clin Microbiol 45, 1410–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klint, M., Hadad, R., Christerson, L., Loré, B., Anagrius, C., Österlund, A., Larsson, I., Sylvan, S., Hallén, A. [amp ] other authors (2009). Spread of the new variant of Chlamydia trachomatis within Sweden over time. In 18th International Society for Sexually Transmitted Disease Research conference, London, United Kingdom, abstract P3.33.
- Lenart, J., Andersen, A. A. [amp ] Rockey, D. D. (2001). Growth and development of tetracycline-resistant Chlamydia suis. Antimicrob Agents Chemother 45, 2198–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynagh, Y., Walsh, A. [amp ] Crowley, B. (2007). First report of the new variant strain of Chlamydia trachomatis in Ireland. EPI-Insight 8, 4. http://www.ndsc.ie/hpsc/EPI-Insight/Volume82007/File,2424,en.PDF. Accession date: 10th January 2010.
- Lysén, M., Österlund, A., Rubin, C.-J., Persson, T., Persson, I. [amp ] Herrmann, B. (2004). Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J Clin Microbiol 42, 1641–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marions, L., Rotzen-Östlund, M., Grillner, L., Edgardh, K., Tiveljung-Lindell, A., Wikström, A. [amp ] Lidbrink, P. (2008). High occurrence of a new variant of Chlamydia trachomatis escaping diagnostic tests among STI clinic patients in Stockholm, Sweden. Sex Transm Dis 35, 61–64. [DOI] [PubMed] [Google Scholar]
- Matsumoto, A., Izutsu, H., Miyashita, N. [amp ] Ohuchi, M. (1998). Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol 36, 3013–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam, A. [amp ] Reinton, N. (2007). Identification of the Swedish Chlamydia trachomatis variant among patients attending a STI clinic in Oslo, Norway. Euro Surveill 12, E070301.3. [DOI] [PubMed] [Google Scholar]
- Pedersen, L. N., Pødenphant, L. [amp ] Møller, J. K. (2008). Highly discriminative genotyping of Chlamydia trachomatis using omp1 and a set of variable number tandem repeats. Clin Microbiol Infect 14, 644–652. [DOI] [PubMed] [Google Scholar]
- Pedersen, L. N., Herrmann, B. [amp ] Møller, J. K. (2009). Typing Chlamydia trachomatis: from egg yolk to nanotechnology. FEMS Immunol Med Microbiol 55, 120–130. [DOI] [PubMed] [Google Scholar]
- Peterson, E. M., Markoff, B. A., Schachter, J. [amp ] de la Maza, L. M. (1990). The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 23, 144–148. [DOI] [PubMed] [Google Scholar]
- Pickett, M. A., Everson, J. S., Pead, P. J. [amp ] Clarke, I. N. (2005). The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151, 893–903. [DOI] [PubMed] [Google Scholar]
- Reinton, N., Moi, H., Bjerner, J. [amp ] Moghaddam, A. (2009). The Chlamydia mutant nvC. trachomatis in Norway. In 18th International Society for Sexually Transmitted Disease Research conference, London, United Kingdom, abstract P3.34.
- Reischl, U., Straube, E. [amp ] Unemo, M. (2009). The Swedish new variant of Chlamydia trachomatis (nvCT) remains undetected by many European laboratories as revealed in the recent PCR/NAT ring trial organised by INSTAND e.V., Germany. Euro Surveill 14, pii: 19302.. [PubMed] [Google Scholar]
- Ripa, T. [amp ] Nilsson, P. A. (2006). A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro Surveill 11, E061109.2.. [DOI] [PubMed] [Google Scholar]
- Ripa, T. [amp ] Nilsson, P. A. (2007). A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic amplification tests. Sex Transm Dis 34, 255–256. [DOI] [PubMed] [Google Scholar]
- Rockey, D. D., Lenart, J. [amp ] Stephens, R. S. (2000). Genome sequencing and our understanding of chlamydiae. Infect Immun 68, 5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter, J. [amp ] Meyer, K. F. (1969). Lymphogranuloma venereum. II. Characterization of some recently isolated strains. J Bacteriol 99, 636–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith, H. M. B., Harris, S. R., Persson, K., Marsh, P., Barron, A., Bignell, A., Bjartling, C., Clark, L., Cutcliffe, L. T. [amp ] other authors (2009). Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skilton, R. J., Cutcliffe, L. T., Barlow, D., Wang, Y., Salim, O., Lambden, P. R. [amp ] Clarke, I. N. (2009). Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PloS One 6, e7723. [DOI] [PMC free article] [PubMed]
- Skipp, P., Robinson, J., O'Connor, C. D. [amp ] Clarke, I. N. (2005). Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics 5, 1558–1573. [DOI] [PubMed] [Google Scholar]
- Söderblom, T., Blaxhult, A., Fredlund, H. [amp ] Herrmann, B. (2006). Impact of a genetic variant of Chlamydia trachomatis on national detection rates in Sweden. Euro Surveill 11, E061207.1. [DOI] [PubMed] [Google Scholar]
- Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R. L. [amp ] other authors (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. [DOI] [PubMed] [Google Scholar]
- Stothard, D. R., Williams, J. A., Van der Pol, B. [amp ] Jones, R. B. (1998). Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect Immun 66, 6010–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland, R. J., Eckert, L. O., Hawes, S. E. [amp ] Stamm, W. E. (2003). Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988–1996. Sex Transm Dis 30, 357–361. [DOI] [PubMed] [Google Scholar]
- Swedish Institute for Infectious Disease Control (SMI) (2009). Data and Statistics. http://www.smittskyddsinstitutet.se/in-english/statistics/. Accession date: 10th January 2010.
- Thomas, N. S., Lusher, M., Storey, C. C. [amp ] Clarke, I. N. (1997). Plasmid diversity in Chlamydia. Microbiology 143, 1847–1854. [DOI] [PubMed] [Google Scholar]
- Thomson, N. R., Holden, M. T., Carder, C., Lennard, N., Lockey, S. J., Marsh, P., Skipp, P., O'Connor, C. D., Goodhead, I. [amp ] other authors (2008). Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res 18, 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M., Olcén, P., Agné-Stadling, I., Feldt, A., Jurstrand, M., Herrmann, B., Persson, K., Nilsson, P., Ripa, T. [amp ] Fredlund, H. (2007). Experiences with the new genetic variant of Chlamydia trachomatis in Örebro county, Sweden – proportion, characteristics and effective diagnostic solution in an emergent situation. Euro Surveill 12, E5–E6. [DOI] [PubMed] [Google Scholar]
- Unemo, M., Rossouw, A., James, V. [amp ] Jenkins, C. (2009). Can the Swedish new variant of Chlamydia trachomatis (nvCT) be detected by UK NEQAS participants from seventeen European countries and five additional countries/regions in 2009? Euro Surveill 14, pii: 19206. [DOI] [PubMed] [Google Scholar]
- Velicko, I., Kühlmann-Berenzon, S. [amp ] Blaxhult, A. (2007). Reasons for the sharp increase of genital chlamydia infections reported in the first months of 2007 in Sweden. Euro Surveill 12, E5–E6. [DOI] [PubMed] [Google Scholar]
- Wang, S. A., Papp, J. R., Stamm, W. E., Peeling, S. W., Martin, D. H. [amp ] Holmes, K. K. (2005). Evaluation of antimicrobial resistance and treatment failures for Chlamydia trachomatis: a meeting report. J Infect Dis 191, 917–923. [DOI] [PubMed] [Google Scholar]
- Ward, M. E. (1983). Chlamydial classification, development and structure. Br Med Bull 39, 109–115. [DOI] [PubMed] [Google Scholar]
- Westh, H. [amp ] Jensen, J. S. (2008). Low prevalence of the new variant of Chlamydia trachomatis in Denmark. Sex Transm Infect 84, 546–547. [DOI] [PubMed] [Google Scholar]