Abstract
Chromatin immunoprecipitation followed by sequencing analysis (ChIP-Seq) is a powerful method to investigate genome-wide distributions of chromatin-binding proteins and histone modifications in any genome with a known sequence. Application of this technique to a variety of developmental and differentiation systems has provided global views of cis regulatory elements, transcription factor function, and epigenetic processes involved in the control of gene transcription. Here, we describe several technical aspects of the ChIP-Seq assay to reduce bias and background noise, and to consistently generate high quality data.
INTRODUCTION
Immune function involves the tight control of transcription by the interplay of cis-regulatory elements and trans-acting transcription factors, which is largely influenced by the epigenetic landscape of immune cells. Previous studies have identified numerous transcription factors and chromatin modifiers implicated in innate and adaptive immunity. However, traditionally, only one or a few individual genes or regulatory regions have been studied using techniques such as ChIP-PCR assays. Recent technical advances in global techniques such as DNA microarrays and next-generation sequencing allow analysis of entire genomes. Identifying and characterizing genome-wide locations of transcription factors, chromatin-modifying enzymes, and the modification status of histones is imperative to comprehensively understand transcriptional regulation of the immune system under diverse biological conditions. Recent applications of ChIP-Seq to several transcription factors and epigenetic modifications have propelled efforts to characterize their global cistromes and to understand immune memory [1,2,3,4]
In this review we highlight several technical aspects of ChIP-Seq that should be considered to obtain high-quality genome-wide data, including consideration of antibodies, controls, library construction, and statistical analysis.
Antibodies
The quality of antibodies used for ChIP-Seq experiments is one of the most important factors that contribute to the quality of the data generated from these studies. Antibodies that offer high sensitivity and specificity are necessary for ChIP-Seq assays because they allow for the detection of enrichment peaks without substantial background noise. Many commercial antibodies that have been tested for their use in ChIP studies are available. However, results from various groups have shown that not all commercial antibodies that are designated as ChIP “grade” or “qualified” can be successfully used to interrogate genome-wide protein-DNA interactions. Certain antibodies that are sufficient for detecting locus specific enrichment using ChIP-PCR may not be suitable for ChIP-Seq studies. As a general rule, if an antibody shows ≥ 5-fold enrichment in ChIP-PCR assays at several positive-control regions compared to negative control regions, it usually works well for ChIP-Seq. Because enrichment may vary from target to target, multiple genomic loci should be tested for their enrichment following ChIP.
It is also important to consider the potential cross-reactivity of antibodies with closely related family members that may serve unique or redundant roles in the immune system. The specificity of an antibody can be directly addressed by performing a western blot for a protein of interest using an RNAi knockdown or knockout model. In these cases, because expression of the protein should be reduced to background levels, any protein that is detected by western blotting can be assumed to be non-specific. Performing ChIP using a higher concentration of antibodies, which can be acquired upon request from several companies, or pooled monoclonal antibodies, may also be considered to enrich for factor-occupied DNA sequences.
In cases where specific antibodies are unavailable, epitope-tagged proteins can be expressed, and then ChIP is performed using a tag specific antibody [5,6]. The most frequently used tags include HA, Flag, Myc, and V5. Although this method has been successful in certain applications, their efficiency in ChIP varies depending on the specific protein it is fused to and its location in the protein (N- or C-terminus). In addition to epitope antibodies, the target protein can also be tagged with a biotin acceptor sequence, which can be labeled with biotin via biotin ligase either in vivo or in vitro. The high affinity of biotin-streptavidin interaction can withstand stringent wash conditions and thus significantly reduce background noise [6,7]. This is particularly advantageous when partially denaturing conditions are required to expose epitopes, such as components of large protein complexes. One caveat to this approach is that overexpression of proteins may lead to altered genomic binding profiles due to excess protein in the cell. Therefore, it is important to ensure that protein expression levels do not exceed the endogenous levels.
The clonality, or heterogeneity, of the antibody should also be considered when choosing an antibody. Monoclonal antibodies recognize a single epitope on an antigen, which may be beneficial for reducing background noise in ChIP studies. However, the use of monoclonal antibodies may result in a decreased signal if the epitope is masked by surrounding chromatin components or if the protein lies within a larger protein complex. While epitope recognition may be problematic for any antibody in ChIP studies regardless of its clonality, polyclonal antibodies offer the flexibility of recognizing multiple epitopes, which may boost signal levels in cases where epitopes are masked by surrounding material. Because there is not a definitive rule for choosing the appropriate clonality of an antibody for ChIP studies, it is best to test several antibodies if they are available. This will provide greater confidence that identified peaks are true positives.
Cell number
The abundance of the protein or histone modification to be investigated, and the quality of the antibody, should be considered when determining a starting cell number for ChIP-Seq analysis. As the signal-to-noise ratio is directly correlated with the cell number, using a greater number of cells tends to give higher signal-to-noise ratios. Therefore, it is important to empirically determine the minimum number of cells, whenever possible. ChIP-Seq experiments typically require one to ten million cells resulting in 10–100 ng of ChIP DNA. One million cells is usually sufficient to analyze abundant proteins such as RNA polymerase II and localized histone modifications such as H3K4me3, while ten million cells may be required to assay less abundant proteins or diffuse histone modifications. However, several alternative protocols have been designed using smaller numbers of cells (104–105) to profile genome-wide distributions of histone modifications [8], although these methods have not been demonstrated yet to work well for transcription factors. The advantage of this approach lies is the ability to use 10–100 fold less cells relative to conventional ChIP-seq protocols, which may be beneficial for studying rare cell types.
Controls
An important part of designing ChIP-Seq experiments is determining what controls to use for the experiment. Artifacts may arise from the following steps of experimentation: (1) chromatin fragmentation: open chromatin regions are easier to shear than closed chromatin regions and thus may be associated with higher background signals [9]; (2) unrecognized cross reactivity of antibodies; (3) variable sequencing efficiency of DNA regions with different base compositions. While both non-specific IgGs and chromatin inputs have been used as controls, IgGs may be less desirable in certain circumstances due to the following reasons: (1) Most IgGs are not the true pre-immune serum from the same animal from which the specific antibody was raised, and (2) IgGs usually pull down much less DNA than a specific antibody and thus limited genomic regions from the control may be over-amplified during the library construction step. In this case the resulting sequence reads will not sufficiently cover the genome as a background model for peak identification. Therefore, chromatin inputs serve as better controls for bias in chromatin fragmentation and variations in sequencing efficiency; additionally, they provide greater and more evenly distributed coverage of the genome. However, normal IgGs and chromatin inputs are not the appropriate controls for addressing cross reactivity of antibodies, which can be controlled for using true pre-immune serum or a different specific antibody for the same factor that recognizes a different epitope. Additional controls for antibody specificity include targeted deletion or RNAi knockdown of the factor of interest. There is an abundance of knockout mice available for transcription factors that have important regulatory functions in the immune system (e.g. Gata3, Stat1/2/6, etc), which would serve as ideal controls to test antibody specificity. In these cases, because expression of the protein should be reduced to background levels, any potential DNA binding events can be assumed to be non-specific.
Replicates
High quality ChIP-Seq datasets are valuable resources for the community. Many factors, including cell culture conditions, ChIP, and library construction, may contribute to variability between datasets. To ensure reliability of the data, it is necessary to perform biological replicate experiments. While there is not a consensus as to the correct number of replicates that should be used, at least duplicate biological experiments should be performed. Although only one ChIP grade antibody is available for most histone modifications and transcription factors, it is recommended to validate ChIP-Seq data using a different antibody whenever possible to control for potential antibody cross-reactivity.
Chromatin fragmentation
Chromatin must be fragmented into manageable sizes (~200–300 bp) before ChIP by sonication or enzymatic means (usually with micrococcal nuclease - MNase treatment) with or without cross-linking depending on the purpose of the experiment. For histone modifications, MNase digestion of native chromatin into mononucleosome-sized particles may be the preferred method because it generates high-resolution data for nucleosome modifications and eliminates artifactual signals caused by cross-linking with other genomic regions. However, this method may potentially suffer from a loss of signal due to unstable nucleosomes. For mapping binding sites of transcription factors, sonication of formaldehyde cross-linked chromatin may be the preferred method because MNase degrades linker DNA, where transcription factors tend to bind. Although different sizes of chromatin fragments may work well for ChIP-PCR assays, the optimal size range of chromatin for ChIP-Seq analysis should be between 150 and 300 bp. DNA fragments within this size range, which are equivalent to mono- and dinucleosome chromatin fragments, provide high resolution of binding sites, and they work well for next generation sequencing platforms. The conditions used to sonicate chromatin need to be optimized for each cell type because they are highly variable and depend on the cell type, the number of cells used, fixation conditions, type of sonicator, and sonicator settings. It is important to avoid oversonication of chromatin when transcription factors are to be evaluated in ChIP studies, while oversonication may not be as problematic for histone modifications. Preparing nuclei prior to fixation may also help to reduce background which may be observed with whole cell chromatin extracts.
Sonication buffers may also influence ChIP-Seq results. Sonication in SDS containing buffers may disrupt protein-protein and protein-DNA interactions and therefore may expose antibody epitopes buried inside a protein complex and improve specific signals. For example, efficient mapping of methylation of H3K79, which is located in the nucleosome core, requires sonication of chromatin in SDS-containing buffers [10]. SDS containing buffers also increase sonication efficiency, and may be appropriate for evaluating transcription factors, which are tightly bound to DNA. However, addition of SDS to sonication buffers may result in a loss of signal for proteins not directly bound to DNA, such as epigenetic regulators.
Library Construction and sequencing
Library construction from ChIP DNA may be performed using standard protocols specific to the sequencing platform. Typically, library construction includes end-repair, single A-addition, adapter-ligation, size selection and gel purification, and PCR using primers specific to the sequencing platform. During the size selection step it is important to melt the agarose at room temperature (~22°C) rather than at 50°C, which may otherwise result in a G+C bias due to a loss of A+T-rich sequences[11]. During the PCR amplification step it is important not to over amplify adapter-ligated DNA products, which may result in a loss of specific signal, bias, or redundancy in the number of sequencing tags. Overamplification can typically be avoided by reducing the number of PCR cycles or decreasing the amount of template DNA used for PCR. One way to determine whether overamplification has occurred is to compare the size of the adapter ligated product to the PCR product. Overamplified PCR products will generally exhibit an increased shift in the size relative to adapter ligated products (e.g. a 200–400 bp adapter ligated product may shift to >300–500 bp).
Although the exact modifications of adapters and primers sold by Illumina and other NGS sequencing reagent providers are not publicly available, adapters and PCR primers can be custom ordered from other companies to reduce costs. For example, custom PCR primers with a phosphorothioate between the two bases at the end have produced similar results for some investigators [11].
The quantity of PCR products is usually sufficient for sequencing as long as the product bands are visible on an agarose gel with ethidium bromide staining. Libraries can be sequenced 25 bp from one end of the DNA templates, which typically provides relatively good coverage (66 %) [12] of uniquely mappable sequences in the human genome (build hg19). Longer sequencing reads of 30–35 bp improve the mappability to 70.9 and 74.1 % [12], respectively, and may be preferred if cost is not an issue.
Libraries can be sequenced using either single-end (which generates short sequence reads from one end of the DNA template) or paired-end (which generates short sequence reads from both ends) sequencing strategies. While both methods have been successful, there are several advantages to using paired-end sequencing: (1) increased sequencing coverage, (2) improved alignment efficiency to repetitive regions because more sequence information is obtained from each DNA template, and (3) increased ability to detect fragment sizes. In cases where ChIP-enriched DNA fragments partially overlap or contain repetitive sequences, sequencing both ends may allow for more accurate mapping to the genome compared with single-end sequencing, which may otherwise result in a loss of repetitive sequences during the analysis.
The number of sequencing reads required to reach a reasonable genomic coverage is contingent upon several factors including antibody affinity and the number of target sites in the genome. For H3K4me3 distributions, five million sequence reads are sufficient to reach saturation of its target sites, while 20 million reads may be required to achieve a reasonable coverage for H3K27me3 profiles. A more quantitative approach to determine the appropriate depth of sequencing involves evaluating the saturation point, or the number of reads after which additional sequencing does not identify new binding or enrichment sites [13]. One lane of sequencing on a Genome Analyzer IIA, which typically generates around 20 million mapped unique reads, is usually sufficient for most modifications and transcription factors. It is also possible to pool several libraries, such as for H3K4me3 modifications, using indexing adaptors to reduce the cost of sequencing.
Data Analysis
The vendor-supplied analysis package includes an image analysis tool that transforms the pixel values into intensities and a base-calling tool to convert the intensities into sequences. Because the output sequences are short in length and high in error rates, third-party base-calling tools have been developed to increase base-call accuracy and yield [14]. Alternative tools correct for potential errors after base-calling [15], and facilitate genome alignment and de novo assembly [16]. However, the potential benefit to peak calling for ChIP-Seq data remains unexplored. Short sequence reads, with or without error corrections, are then mapped to a reference genome using a variety of alignment programs. Due to the recent development of alignment tools, short read alignment is no longer a bottleneck in the data analysis process; for a comprehensive summary of various alignment tools, we refer readers to a recent review article [17].
The quality of ChIP-Seq data can be inspected using a combination of methods. First, it is important to evaluate the summary report generated by the vendor-supplied analysis pipeline. For example, the “Summary.html” from CASAVA contains a set of comprehensive performance measures for data generated from Illumina GA platforms. The next step involves converting the sequence alignments to an appropriate format, uploading them to a Genome Browser display, and examining several genomic regions of interest (e.g., known targets of a transcription factor). Another qualitative measure for determining the quality of ChIP-Seq data involves searching for sequence motifs within tag-enriched regions or peaks [18]. In addition, it may be useful to examine the distribution of tag profiles around certain genomic features (e.g., transcriptional start sites). We also suggest to parallelize the inspection using input or IgG controls, and to minimize the bias using specific tools [19]. As mentioned previously, it is also important to validate selected ChIP-Seq peaks using quantitative PCR.
After the initial quality inspection, peak calling is performed to identify tag-enriched regions from the ChIP-Seq data. Multiple algorithms are available and their comparisons constitutes the themes of several publications [18]. When considering which tool to choose, it is important to recognize that there are two fundamental types of peaks, sharp and broad. The inspection of tag distributions in a genome browser together with prior knowledge helps to reach an initial idea about what the peaks look like. Algorithms like MACS [20] work well for identifying sharp peaks of most sequence-specific transcription factors, while programs like SICER [21] and CCAT [22] are appropriate for identifying broad peaks of most histone modifications and chromatin binding proteins. Our experience is that CCAT has greater sensitivity for identifying peaks, while SICER has greater specificity. However, because CCAT requires negative controls to estimate noise rates, this algorithm may not be applicable to datasets where negative controls are absent, such as for FAIRE-Seq. Another method, ZINBA, has recently been developed to identify both sharp and broad peaks [23]. Because these tools are designed for different purposes, a performance comparison between them may not be fair. On the other hand, efforts made to evaluate these methods have been limited largely due to the absence of objective benchmark standards [18].
Reads mapped to multiple sites (multi-reads) are discarded during "normal" analysis. Consequently, peaks in highly repetitive regions are overlooked. However, repetitive regions have been linked to important biology functions such as disease susceptibility, immunity and defense. A new method has recently been proposed to incorporate multi-reads with a weighted alignment scheme into peak detection [24]. Since most of the novel peaks reside in repetitive regions, this method will be of particular interest to the analysis of ChIP-Seq data from proteins that selectively bind to repetitive regions.
Another important issue in data analysis is how to compare the levels of histone modifications or transcription factor binding between two different cell types or under different conditions. Due to variations in ChIP conditions, the level of noise may vary significantly between different samples even for the same antibody. Because scaling the data to sequence depth does not eliminate systematic errors, normalization algorithms are needed to enable comparisons across samples. A recent tool, DIME, has been developed to classify significantly enriched regions between two ChIP-Seq samples based on an estimation of multivariate mixture models [25]. Because the method partitions the genome into bins that are larger in size than a typical TF binding site, it may serve as a first-pass algorithm to identify candidate differential binding regions, especially in cases of low sequencing depth. A sub-module of SICER is also able to identify differentially enriched regions between two conditions when regions of interest are specified (e.g., changes in the levels of histone modifications at promoters between two samples).
Because genome-wide data are being generated at a rapid rate, it is important that analytical tools are developed at a similar rate to support the storage and analysis needs that users encounter. We expect a flurry of software with user friendly interfaces (GUI) to be released in the near future and adopted by biologists with diverse backgrounds.
Summary
Global surveys of sequence-specific transcription factor binding and chromatin structure in a variety of cell types and species provides comprehensive information to understand dynamic processes such as stem cell differentiation, immune memory formation, disease progression, and responses to external environmental stimuli. ChIP-Seq is a powerful technology to evaluate protein-DNA interactions on a global scale. Findings from studies utilizing ChIP-Seq have improved our knowledge of epigenetic landscapes that regulate chromatin structure and transcriptional profiles. Because datasets generated from ChIP-Seq studies are valuable resources for the community, it would be beneficial if datasets generated from different laboratories could be directly compared. For this purpose, experimental conditions must be optimized and standardized.
In this review, we discussed several important technical considerations for performing ChIP-seq experiments and data analysis, as summarized in Figures 1–2, that may help to yield reliable and reproducible datasets. As ChIP-Seq protocols are further improved and data analysis platforms become more manageable to experimental biologists, application of this technology will help to build comprehensive global views of immune processes, thus aiding our understanding of disease states.
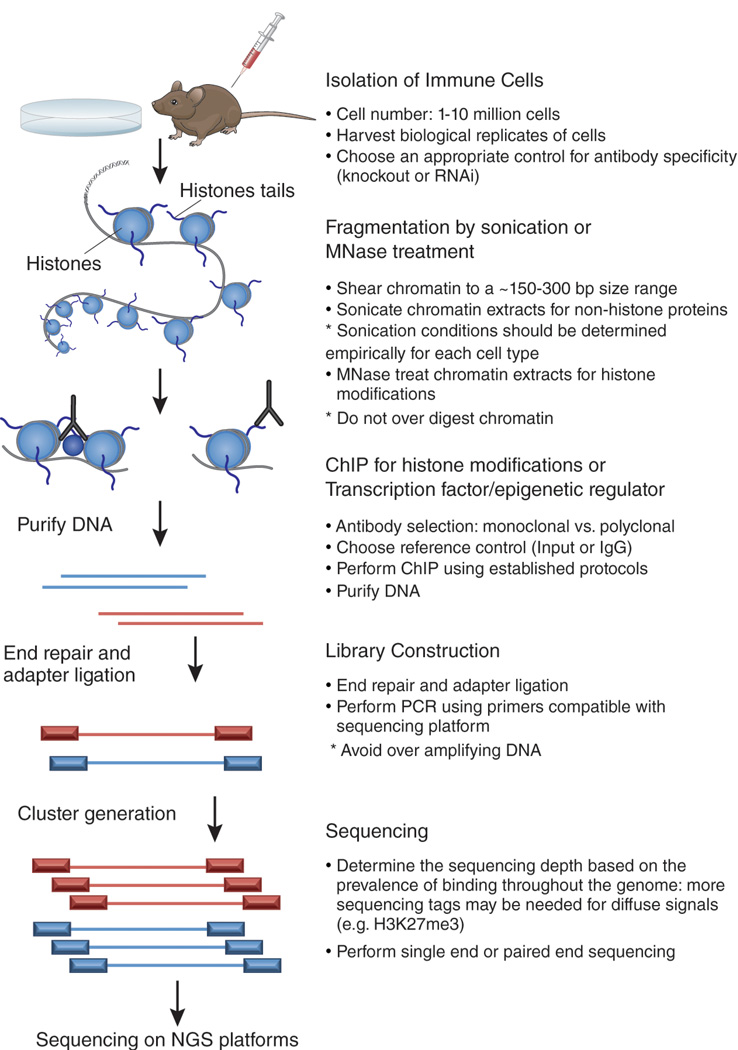
Figure 1. ChIP-seq experimental design.
Chromatin immunoprecipitation followed by next-generation sequencing (NGS) is a powerful tool to investigate protein-DNA interactions on a global scale. Before performing ChIP-seq it is important to determine the appropriate controls for antibody specificity. After isolation of an ideal number of cells, chromatin is sheared into an ideal size range by sonication or enzymatic means (MNase). Next, ChIP is performed using high-quality antibodies to enrich for factor-occupied DNA sequences. A reference genome should also be included in this step to control for the ChIP experiment. Following purification of ChIP-enriched DNA, library construction is performed to allow for sequencing on next-generation sequencing platforms. Library construction typically includes end-repair, single A addition, adapter ligation, and PCR using primers compatible with the sequencing platform. Following cluster generation, single or paired-end sequencing is performed on NGS platforms.
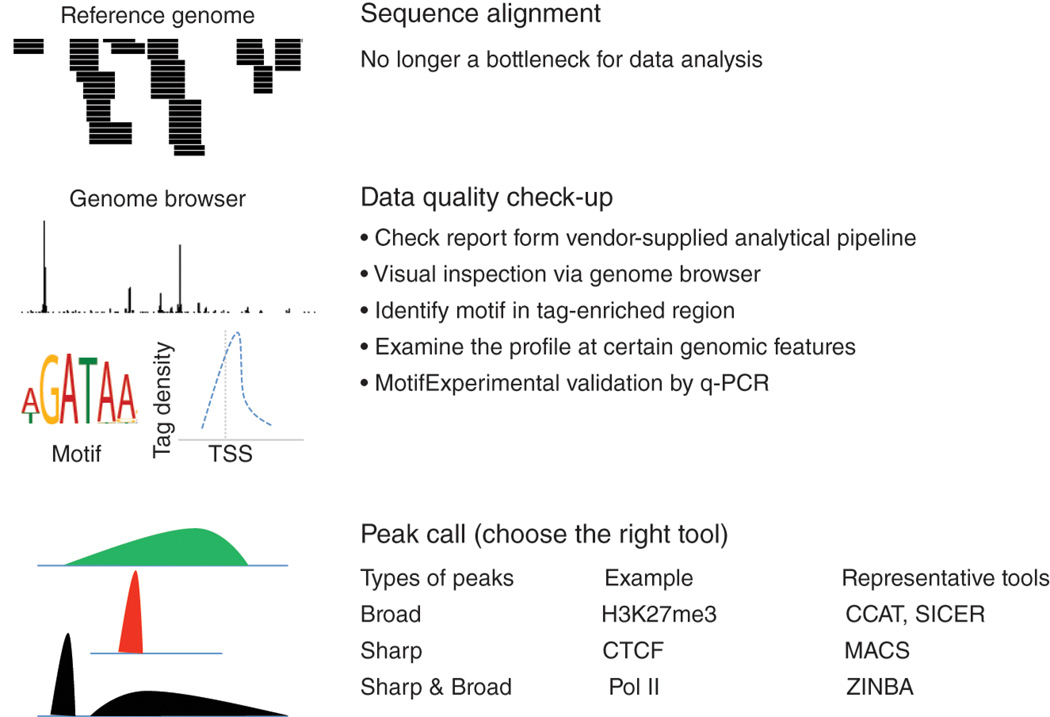
Figure 2. Common procedures for ChIP-seq data analysis.
After base-calling, short read sequences are aligned to a reference genome. Data quality is inspected by a combination of various strategies such as visual inspection using a genome browser, motif identification and Q-PCR validation. The initial inspection or prior knowledge provides information about whether the peaks are broad, sharp or both. Different algorithms have been developed to identify peaks from these three groups, of which several representatives are shown in the figure.
ACKNOWLEDGEMENTS
Research in the author’s laboratory is supported by the Division of Intramural Research, National Heart, Lung and Blood Institute. We thank Brian Abraham and Daniel Northrup for critically reviewing the manuscript and Kairong Cui for helpful discussions.
REFERENCES
- 1.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Northrup DL, Zhao K. Application of ChIP-Seq and Related Techniques to the Study of Immune Function. Immunity. 2011;34:830–842. doi: 10.1016/j.immuni.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodziej KE, Pourfarzad F, de Boer E, Krpic S, Grosveld F, et al. Optimal use of tandem biotin and V5 tags in ChIP assays. BMC Mol Biol. 2009;10:6. doi: 10.1186/1471-2199-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, et al. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, et al. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehler R, Issac H, Cloonan N, Grimmond SM. The uniqueome: a mappability resource for short-tag sequencing. Bioinformatics. 2011;27:272–274. doi: 10.1093/bioinformatics/btq640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Brief Bioinform. 2011 doi: 10.1093/bib/bbq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao WC, Chan AH, Song YS. ECHO: A reference-free short-read error correction algorithm. Genome Res. 2011 doi: 10.1101/gr.111351.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley DR, Schatz MC, Salzberg SL. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szalkowski AM, Schmid CD. Rapid innovation in ChIP-seq peak-calling algorithms is outdistancing benchmarking efforts. Brief Bioinform. 2010 doi: 10.1093/bib/bbq068. [DOI] [PubMed] [Google Scholar]
- 19.Cheung MS, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zang C, Schones DE, Zeng C, Cui K, Zhao K, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Handoko L, Wei X, Ye C, Sheng J, et al. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics. 2010;26:1199–1204. doi: 10.1093/bioinformatics/btq128. [DOI] [PubMed] [Google Scholar]
- 23.Rashid N, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung D, Kuan PF, Li B, Sanalkumar R, Liang K, et al. Discovering Transcription Factor Binding Sites in Highly Repetitive Regions of Genomes with Multi-Read Analysis of ChIP-Seq Data. PLoS Comput Biol. 2011;7:e1002111. doi: 10.1371/journal.pcbi.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taslim C, Huang T, Lin S. DIME: R-package for identifying differential ChIP-seq based on an ensemble of mixture models. Bioinformatics. 2011;27:1569–1570. doi: 10.1093/bioinformatics/btr165. [DOI] [PMC free article] [PubMed] [Google Scholar]