Abstract
p53 is a pivotal tumor suppressor which induces apoptosis, cell-cycle arrest and senescence in response to stress signals. Although p53 transcriptional activation is important for these responses, the mechanisms underlying tumor suppression have been elusive. To date, no single or compound mouse knockout of specific p53 target genes has recapitulated the dramatic tumor predisposition that characterizes p53-null mice. Recently, however, analysis of knock-in mice expressing p53 transactivation domain mutants has revealed a group of primarily novel direct p53 target genes that may mediate tumor suppression in vivo. Here, we present an overview of well-known p53 target genes and the tumor phenotypes of the cognate knockout mice, then segue into the recent identification of new p53 transcriptional targets and how they enhance our understanding of p53 transcriptional networks central for tumor suppression.
p53: complexity at a molecular and network scale
p53 has been studied extensively due to its paramount importance in tumor suppression. The significance of p53 in tumor suppression in humans is highlighted by its inactivation in over half of all human cancers and by the dramatic cancer predisposition of individuals with Li-Fraumeni syndrome, who inherit a mutant p53 allele. In addition, mice deficient for p53 develop cancer with 100% penetrance [1, 2]. Although we have some understanding of the molecular mechanisms by which p53 functions in tumor suppression, it is increasingly evident that our current knowledge is incomplete. The discovery of vast and varied transcriptional targets controlled by p53 raises new questions about how these networks coordinate to promote tumor suppression. Mouse models have been instrumental in beginning to decipher the networks through which p53 functions in vivo, and the insights gained from these studies will be the subject of this review.
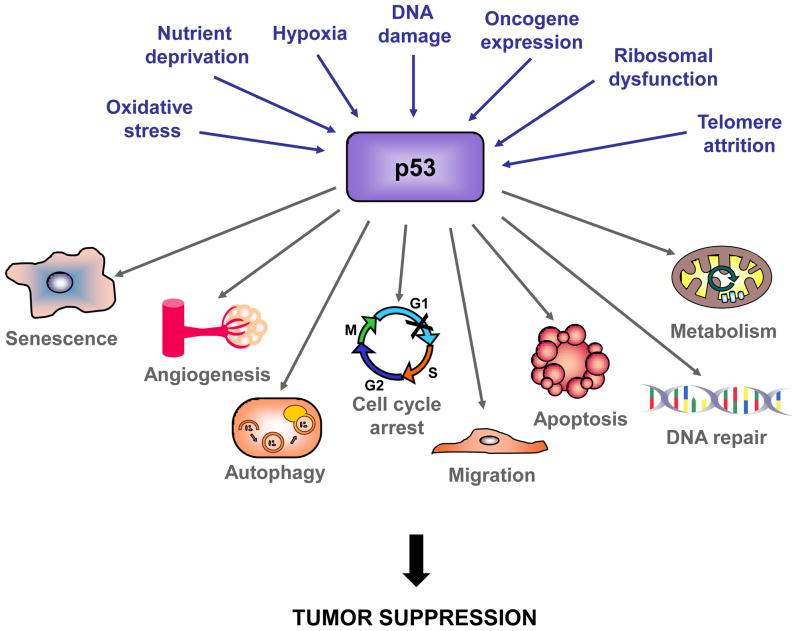
p53 plays a fundamental role in the response to cellular stress, which can, at least in part, explain its tumor suppression function (Figure 1). For example, p53 responds to hyperproliferative signals caused by oncogene expression by inducing apoptosis or cellular senescence as safeguards against tumorigenesis (Text Box 1). In the absence of cellular stress, p53 is bound by its negative regulator, Mdm2, an E3 ubiquitin ligase that promotes its degradation. Oncogene activation can trigger expression of Arf, which disrupts the p53-Mdm2 interaction, leading to stabilization and activation of p53 [1]. p53 also responds to acute DNA damage signals by inducing apoptosis or cell cycle arrest to prevent the genomic instability and increased risk for carcinogenesis associated with propagation of damaged cells [3]. As with the response to oncogenic signaling, the response of p53 to DNA damage may also have a role in tumor suppression, as nascent human and mouse tumors display activation of DNA damage pathway components, including p53 (See Text Box 2, [4]). Studies using mouse lymphoma and fibrosarcoma models, however, suggest that p53-mediated responses to acute DNA damage are dispensable for tumor suppression, instead highlighting the importance of Arf in p53-mediated tumor suppression [5–7]. Whether the molecular trigger for p53-mediated tumor suppression is oncogene signaling through Arf or DNA damage is an area of active debate and investigation, and both are likely to be important (Text Box 2). Defining the triggers for p53 activation in tumor suppression in different settings will be key for fully elaborating the functional p53 tumor suppressor network.
Figure 1. p53 responds to a plethora of stress signals and regulates diverse responses.
Myriad cellular stress signals can activate p53, and p53 can respond by regulating many cellular processes that could contribute to tumor suppression.
Text Box 1. Mouse models reveal key roles for apoptosis and cell cycle arrest in p53-mediated tumor suppression.
The discovery that p53-null mice are highly susceptible to spontaneous tumors not only provides compelling evidence for the importance of p53 in tumor suppression, but also underscores the utility of mouse models for studying this process in vivo [83]. The ability of p53 to trigger apoptosis and cell cycle arrest in response to DNA damage and oncogene activation suggests obvious cellular mechanisms for tumor suppression by p53, and mouse models confirm roles for both of these effector functions. The first evidence for the importance of apoptosis in p53-mediated tumor suppression in vivo came from a mouse model of brain cancer driven by expression of an SV40 large T-antigen mutant, T121, which inactivates the Retinoblastoma (Rb) family of tumor suppressors. In the presence of p53, tumors grow slowly and are characterized by high levels of apoptosis, but without p53, apoptosis is minimal and tumors grow rapidly [84]. In addition, in Eμ-Myc transgenic mice, a model for B-cell lymphoma, disruption of apoptosis through expression of Bcl-2 or dominant-negative caspase 9 confers a tumor growth advantage similar to p53 loss, specifically highlighting the importance of p53- mediated apoptosis in suppressing cancer development [18]. Mouse models have also shown that p53-dependent growth arrest and senescence contribute to its role in tumor suppression. Knock-in mice expressing p53R172P (also called Trp53515C) – a tumor-derived p53 mutant that promotes cell cycle arrest but not apoptosis – are significantly more cancer-resistant than p53- null mice [75]. Furthermore, in telomerase-deficient mice, expression of p53R172P inhibits spontaneous tumorigenesis through senescence [85]. The pivotal role of senescence in p53- mediated tumor suppression was further demonstrated in mouse lung and prostate cancer models driven by activated BRAF [86] and Pten loss, respectively [87]. The relative importance of p53-induced apoptosis and senescence in tumor suppression is likely cell-type specific, as illustrated by studies in which restoration of p53 function in p53-deficient tumors triggered apoptosis in lymphomas and senescence in sarcomas [88]. The question of how p53 drives different responses in different settings is of great interest, and numerous mechanisms implicating different co-factors for p53 have been proposed to explain the context-dependence of these responses.
Text Box 2. DNA damage responses in p53-mediated tumor suppression.
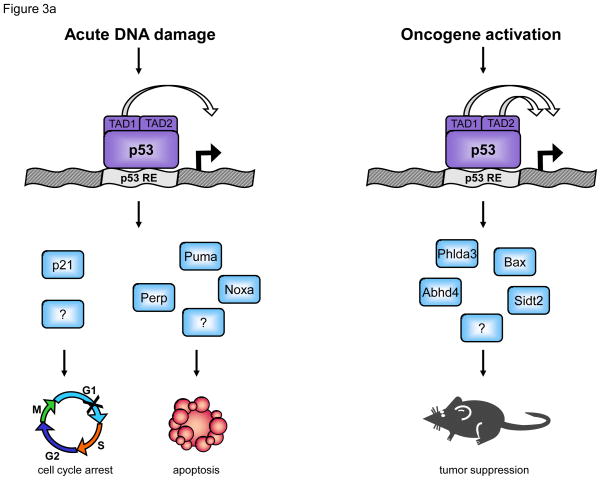
The role for the DNA damage response in triggering p53 tumor suppressor function has been a topic of some debate. A model has been proposed whereby oncogene-induced proliferation triggers replication fork collapse, the formation of double strand DNA breaks, and the activation of ATM and ATR kinases. This cascade then ultimately impinges upon p53 to promote a tumor suppressive response. The model is supported by evidence of markers for DNA damage pathway activation, including p53 activation, in human and mouse precancerous lesions [4]. Studies in mouse lymphoma and fibrosarcoma models, however, suggest that p53-mediated tumor suppression requires oncogene-triggered induction of the Arf tumor suppressor but not the acute damage response [5–7]. This notion is supported by recent studies of knock-in mice expressing a p53 TAD1 mutant, p5325,26. The failure of the p5325,26 mutant to efficiently activate classical p53 target genes essential for apoptosis and cell cycle arrest, such as Puma and p21, and to mediate cell cycle arrest and apoptosis in response to acute DNA damage [14, 15] supports the notion that robust transactivation of these target genes is required for p53 effector function downstream of acute DNA damage. However, the ability of p5325,26 to suppress tumor formation in vivo in mouse models for diverse cancers including NSCLC, fibrosarcoma, medulloblastoma, and B cell lymphoma [15, 77] suggests not only that full transactivation is dispensable for tumor suppression, but also that p53 responses to acute DNA damage are not required for tumor suppression in various settings. These different observations may be reconciled by invoking the possibility that DNA damage induced in incipient tumors is a low level stress that provokes a mechanistically distinct pathway from the acute DNA damage response [4]. It is possible that the p5325,26 mutant may selectively respond to chronic, low-level DNA damage but not acute DNA damage. Further experimentation will determine whether the mechanisms of p53 action downstream of acute and chronic DNA damage are distinct. This notion provides an explanation for how both the oncogene-Arf and DNA damage signaling pathways could be important for tumor suppression.
The best-characterized molecular function of p53 in driving apoptosis, cell cycle arrest, or senescence is as a transcriptional activator, although p53 has other biochemical activities including the ability to repress transcription and to promote apoptosis through direct interaction with apoptotic regulators in the cytosol [1, 2]. Like most transcription factors, p53 contains distinct domains responsible for sequence-specific DNA binding and transcriptional activation. The DNA binding domain comprises residues 100–300 and directs p53 to p53-response elements (p53 RE). The DNA binding domain is the most common site for mutations in cancer [8], underscoring the importance of p53 DNA binding function for tumor suppression. Two distinct transcriptional activation domains (TADs), spanning residues 1–40 and 40–83, cooperate for full p53 transactivation capacity (Figure 2). These domains were defined initially by their ability to confer activation potential on a Gal4 DNA binding domain in reporter assays, and residues within these domains critical for transactivation were pinpointed through additional reporter assays [9–11]. While both TADs are present in full-length p53, an amino-terminally truncated form of p53 lacking the first TAD, ΔN40, generated either through alternative splicing or translational initiation, has been described [12]. Apart from the observation that ΔN40 p53 can cause premature aging in mice when overexpressed [13], there has been very limited insight into the respective roles of these two TADs gleaned from cell culture studies. Recently, however, the specific contributions of each domain to various p53 functions in vivo have been clarified through the generation of TAD mutant knock-in mice, and we now appreciate that the TADs are differentially required for the activation of distinct sets of p53 target genes and for different biological functions ([14, 15]; see below). This knowledge can now be harnessed to map the transcriptional networks critical for p53 function in tumor suppression.
Figure 2. Transactivation domains mediate interactions between p53 and cofactors.
The domain organization of p53 includes two N-terminal transcriptional activation domains (TADs), a proline rich domain, a DNA binding domain, a tetramerization (Tet) domain, and a basic region. The sequence alignment of mouse and human p53 is shown, with the asterisks indicating the location of the mutations in the TAD mutants. The LW residues mutated in p5325,26 knock-in mouse strains correspond to amino acids 25 and 26 in mouse p53 and 22 and 23 in human p53, and the FF residues mutated in the p5353,54 knock-in mouse correspond to amino acids 53 and 54 in both mouse and human p53 (marked in red). A variety of protein cofactors that regulate chromatin remodeling and/or transcriptional initiation interact with p53 via one or both TADs, including the transcriptional regulator proteins TAF6, TAF9, TBP, TRAP80, TFIIH, and histone acetyltransferases p300, CBP, and GCN5, a component of the human STAGA (STP3-TAF(II)31-GCN5-L acetylase) complex. Please note that the exact boundaries of the interaction sites are not precise.
p53 directly regulates over 125 gene targets [2, 16], and genome-wide chromatin immunoprecipitation (ChIP) studies have revealed that p53 binds to thousands of genes [17]. Although some p53 targets have clear links to p53’s cellular functions in apoptosis and cell cycle arrest, some are implicated in other cellular processes in which p53 is also involved, including DNA repair, metabolism, and cell migration, that could also contribute to tumor suppression [1] (Figure 1). In this review, we will discuss our current knowledge of the p53 transcriptional networks involved in tumor suppression. We will highlight those p53 target genes most thoroughly studied in mouse tumor models, many of which were identified because of their robust induction in response to DNA damage signals, and describe what is known about their cellular functions and roles in tumor suppression. We will then describe a new tumor suppression-associated transcriptional program identified by dissection of p53 TADs, which augments our understanding of the networks involved in p53-mediated tumor suppression. Our discussion will reveal that we are just beginning to scratch the surface of the transcriptional circuitry employed by p53 in suppressing cancer development.
Elucidating the functions of p53 target genes in vivo
Revealing in vivo roles of p53 apoptosis target genes
Mouse models have demonstrated the importance of p53 apoptotic function in tumor suppression (Text Box 1, [18]). Thus investigating p53 apoptotic target genes may be important for understanding the molecular mechanisms of p53-mediated tumor suppression. p53 can trigger apoptosis via the intrinsic or extrinsic signaling pathways, which converge at the level of caspase activation, but differ in upstream stimuli. The intrinsic apoptotic pathway is regulated by the ratio of pro-apoptotic to pro-survival Bcl-2 family members. Pro-apoptotic Bcl-2 effector proteins such as Bax and Bak oligomerize at the mitochondrial outer membrane, resulting in membrane permeabilization (MOMP) and release of cytochrome c, eventually activating effector caspases. The pro-survival Bcl-2 family members, including Bcl-2, Bcl-XL, and Mcl-1 bind directly to Bax and Bak, inhibiting MOMP. BH3 (Bcl-2 Homology Domain 3)-only pro-apoptotic Bcl-2 family members, including Puma and Noxa, perturb these interactions, liberating Bax and Bak to promote apoptosis. p53 can engage the intrinsic cell death pathway through induction of Bax, Puma, and Noxa. In contrast, the extrinsic apoptotic pathway is activated by engagement of transmembrane death domain proteins at the cell surface, and p53 participates in this pathway by inducing transcription of death receptors and ligands such as Fas, Killer/DR5, and Pidd.
Bax is one of the earliest studied p53 transcriptional targets, and genetic experiments revealed its importance for DNA damage-induced apoptosis in neurons [19–21] and oncogene-expressing mouse embryo fibroblasts (MEFs) [22], but not thymocytes or intestinal crypt cells [23, 24]. While Bax-deficient mice are characterized by lymphoid hyperplasia, loss of Bax alone does not lead to tumor development in mouse models [24, 25]. In the context of oncogene activation, however, Bax does display tumor suppressor activity, as Bax deficiency accelerates tumorigenesis in mouse models of mammary, brain, and pancreatic beta cell cancer, as well as in B-cell lymphoma driven by Eμ-Myc, [26–29] (Table 1). It is noteworthy that Bax also participates in a transcription-independent pro-apoptotic function of p53. p53 protein interacts directly with pro-apoptotic and/or pro-survival Bcl2 family members to induce MOMP [30]. Thus, the requirement for Bax in p53-dependent apoptosis and tumor suppression may not reflect an exclusive role as a transcriptional target.
Table 1.
Phenotypes of p53 apoptosis target gene mouse knockout strains
| p53-dependent apoptotic phenotype of null cells | Knockout mouse phenotype | Knockout mouse tumor models | Refs | |
|---|---|---|---|---|
|
Bax Bcl-2 - associated X protein |
Apoptosis deficient:
Apoptosis competent:
|
|
Enhanced tumorigenesis:
|
[19–22, 24–29] |
|
Puma (Bbc3) p53 up- regulated modulator of apoptosis (Bcl- 2 binding component 3) |
Apoptosis deficient:
Apoptosis competent:
|
|
Enhanced tumorigenesis:
|
[33–38, 43] |
|
Noxa (Pmaip) Phorbol-12- myristate-13- acetate-induced protein 1 |
Apoptosis deficient:
Apoptosis competent:
|
|
No effect:
|
[34, 36, 43] |
|
Perp p53 apoptosis effector related to PMP-22 |
Apoptosis deficient:
Apoptosis competent:
|
|
Enhanced tumorigenesis:
|
[47, 48, 89] |
|
Dr5 (Tnfrsf10b, Killer, TRAIL- R2) Death receptor 5 (Tumor necrosis factor receptor superfamily, member 10b) |
Apoptosis deficient:
Apoptosis competent:
|
|
Enhanced tumorigenesis:
|
[49, 50] |
|
Fas (CD95) TNF receptor superfamily member 6 |
Apoptosis competent:
|
|
Enhanced tumorigenesis:
Decreased tumorigenesis:
|
[52–54, 90] |
|
Pidd p53-induced protein with a death domain |
Apoptosis competent:
|
|
Not determined | [51] |
The p53 target genes Pmaip1 and Bbc3 genes encode the BH3-only proteins Noxa and Puma, respectively, [31, 32] and are essential mediators of the apoptotic arm of the p53 pathway. Puma deficiency nearly or completely abolishes DNA damage-induced, p53-dependent apoptosis in lymphocytes and neurons [33, 34], suggesting that it is a central p53 apoptosis mediator. Given the critical role of Puma in apoptosis in diverse cell types [33–35], it is surprising that Puma−/− mice are not prone to spontaneous tumor development [33, 34]. This finding may be explained by Puma playing a role specifically downstream of potent stress signals, such as acute DNA damage, that may not be relevant for p53 action in tumor suppression. However, like Bax loss, Puma deficiency accelerates tumorigenesis in the context of oncogene activation in the Eμ-Myc lymphoma model [36–38].
Noxa displays more limited pro-apoptotic potential than Puma, which may be explained by its specific affinity for Mcl1, which contrasts the ability of Puma to bind all pro-survival Bcl-2 family members equivalently [39]. A specific requirement for Noxa in DNA damage-induced apoptosis is seen in certain cell types (Table 1) [34, 40–42]. Since Noxa deficiency compromises apoptosis less than Puma loss, it is not surprising that Noxa−/− mice are not predisposed to spontaneous tumorigenesis [34] and that Noxa deficiency fails to accelerate lymphomagenesis in the Eμ-Myc model [36]. Analysis of mice deficient for both Puma and Noxa has reinforced the dominant role for Puma in p53-mediated apoptosis. With a few exceptions, levels of apoptosis in most cell types from Noxa−/−Puma−/− mice are indistinguishable from those in Puma−/− mice after various stimuli [43]. Loss of both Noxa and Puma is also insufficient to initiate tumor formation, as no spontaneous tumors are observed in double homozygous mutant mice [43]. Moreover, Eμ-Myc/Noxa−/−Puma−/− mice develop tumors with similar kinetics to Eμ-Myc/Puma−/− mice, suggesting a minimal role for Noxa in tumor suppression [36]. Importantly, tumorigenesis in Eμ-Myc/Puma−/− mice is substantially delayed relative to Eμ-Myc/p53+/− mice [36], suggesting that other target genes, possibly those with non-apoptotic function(s), contribute to p53-mediated tumor suppression in this model.
Mapping the p53 networks involved in tumor suppression can be informed by identifying target genes selective to specific cellular responses. Interestingly, while Bax, Noxa, and Puma are induced by p53 to similar levels in the contexts of apoptosis and cell cycle arrest and therefore do not specify the apoptotic cell fate [40, 44, 45], the p53 target gene Perp is upregulated to high levels during p53-mediated apoptosis compared to cell cycle arrest. Furthermore, Perp overexpression is sufficient to induce apoptosis [46]. Perp encodes a tetraspan membrane protein that localizes to desmosomes, intercellular adhesion junctions, in keratinocytes [47]. Analysis of Perp in mouse models has uncovered a role in adhesion, apoptosis and tumor suppression in vivo. As Perp−/− mice display dramatic lethal blistering in the epidermis and other stratified epithelia, tumor studies have relied on conditional knockout strategies [47]. In a mouse model for squamous cell carcinoma (SCC) in which mice lacking Perp in the epidermis were exposed to chronic UVB radiation [48], Perp-deficient mice developed tumors with reduced latency, and tumors were more advanced, than in controls, indicating that Perp loss promotes both tumor initiation and progression. [48].
Additional mouse studies have queried the role of the extrinsic apoptotic pathway in p53 function in vivo. Mice deficient for the p53 target genes Dr5 (also known as Killer and Trail receptor 2), Pidd (p53 Induced Death Domain), and Fas/CD95 are not prone to developing spontaneous tumors and have variable tumor phenotypes in chemical or genetic mouse cancer models (Table 1) [49–54]. Collectively, studies of p53 apoptosis target genes in mouse models demonstrate roles for many of these genes in tumor suppression, although their roles are not sufficient to explain full p53 function. To broadly understand the role of p53 transactivation in tumor suppression, we must also consider those p53 targets that promote cell cycle arrest and senescence.
Revealing in vivo roles of p53 cell cycle arrest and senescence target genes
The cyclin-dependent kinase inhibitor p21 (Cdkn1a) was the first p53 target gene to be identified [55, 56]. p21 is important for the G1 checkpoint response, as p21 loss compromises p53-mediated G1 arrest in response to DNA damage [57, 58]. Unlike p53−/− MEFs, however, p21−/− MEFs are not immortal, undergoing senescence similarly to wild-type MEFs [59]. Although initial reports suggested that p21-null mice do not display an enhanced tumor predisposition [57, 58], a subsequent study suggested a slightly accelerated mean latency of spontaneous tumor onset compared to controls [60], but far from mirroring the phenotype of p53-null mice [57]. In most, but not all cancer models driven by chemical carcinogens, irradiation, or oncogene expression, p21 nullizygosity promotes some aspect of tumorigenesis, including tumor initiation, progression, or metastasis (Table 2 [61–64]). Collectively, these studies suggest that p21 deficiency can enable tumorigenesis in select settings.
Table 2.
Phenotypes of p53 cell cycle arrest and senescence target gene mouse knockout strains
| p53-dependent cell cycle arrest phenotype of null cells | Knockout mouse phenotype | Knockout mouse tumor models | Refs | |
|---|---|---|---|---|
|
p21 Cyclin dependent kinase inhibitor 1A |
Deficient:
Competent:
|
Multiple reports:
|
No effect:
Enhanced tumorigenesis:
|
[57–60, 62–64, 91–94] |
|
Gadd45a Growth arrest and DNA- damage- inducible 45 alpha |
Deficient:
|
|
Enhanced tumorigenesis:
Decreased tumorigenesis:
|
[65–68, 95] |
|
Pml Promyelocytic leukemia |
Deficient:
|
|
Enhanced tumorigenesis:
No effect:
|
[69–73] |
|
Ptprv Protein tyrosine phosphatase, receptor type, V |
Deficient:
|
|
Enhanced tumorigenesis:
|
[74] |
Other p53 target genes involved in senescence and cell cycle arrest also demonstrate context-specific tumor suppressor activity (Table 2). Gadd45a is a p53 target gene with a demonstrated role in controlling G2/M progression. Gadd45a−/− MEFs display chromosomal defects, including aneuploidy and gene amplification, similar to p53−/− MEFs [65], suggesting that Gadd45a is critical for maintenance of genomic stability by p53. While Gadd45a deficiency alone does not predispose mice to spontaneous tumor development, mice exhibit increased γ- and UV- radiation-induced carcinogenesis [65, 66]. In the context of oncogene activation, loss of Gadd45a both accelerates and inhibits tumor onset, revealing that Gadd45a function is highly context-dependent [67, 68]. Pml is a direct p53 target gene [69], essential for senescence triggered by oncogenic Ras expression [70, 71]. Pml-deficient mice do not exhibit a spontaneous tumor predisposition, but are highly susceptible to infection, confounding tumor analyses [72]. Nonetheless, Pml loss can promote tumorigenesis in specific models [72, 73]. Finally, the p53-inducible Ptprv gene encodes a transmembrane tyrosine phosphatase [74]. While Ptprv−/− MEFs are deficient in cell cycle arrest responses to acute DNA damage, Ptprv−/− mice do not develop spontaneous tumors within the first year of life, but do develop more papillomas than wild-type mice after DMBA treatment [74]. Exactly how Ptprv functions in tumor suppression remains unclear.
Perspectives on individual target gene knockouts
While the analysis of individual p53 transcriptional targets through mouse knockout strategies has revealed the importance of these genes for tumor suppression in vivo in specific settings, mice deficient for any single target gene fail to recapitulate the dramatic, completely penetrant spontaneous tumor phenotype of p53-null mice (Tables 1 and 2). In fact, many of these knockout mice do not display any spontaneous tumor phenotype, although loss of these targets can contribute to tumor development in the face of hyperproliferative signals or DNA damage. Given that many p53 target genes have p53-independent functions in apoptosis, growth arrest, DNA repair, and other cellular processes, it is also possible that the tumor phenotypes of mice deficient for these genes are unrelated to their being direct effectors of p53 function. This point can only be unequivocally addressed by generating knock-in mice with mutations in the p53 REs of specific target genes, to clearly determine whether p53 activation of a given gene is important for p53 tumor suppressor activity.
Collaborating p53 functions in tumor suppression
An important issue not addressed by the mouse knockout experiments described above is the possibility that multiple gene products, potentially involved in different p53 effector functions, collaborate in tumor suppression, and that as a result, the striking tumor predisposition of p53-null mice might be explained by combined loss of several key p53 effector functions. The p53R172P mutant, which can activate p21 – the major cell cycle regulator transcriptionally activated by p53 – but not apoptosis target genes (Text Box 1) has helped to address this question [75]. Mice homozygous for this mutant allele and also null for p21 were generated and demonstrated that p53 uses p21 cooperatively with the apoptotic pathway in tumor suppression. p53R172P/R172P/p21−/− mice develop tumors with shorter latency than either p53R172P/R172P or p21−/− mice [76]. In fact, the tumor initiation rates and the tumor spectra are similar in p53R172P/R172P/p21−/− and p53−/− mice, although the p53R172P/R172P/p21−/− mice display longer overall survival, possibly due to residual activation of certain p53 target genes by p53R172P [76]. A similar rationale led to generation of Puma−/−p21−/− mice. Remarkably, these compound mutant mice are not abnormally cancer prone, despite complete deficiencies in DNA damage-induced p53-mediated apoptosis and cell cycle arrest [A. Strasser, personal communication]. These data suggest that functions of p53 other than the responses to acute DNA damage may be important in tumor suppression. Importantly, these findings are not incompatible with the notion that p53-triggered cell cycle arrest and apoptosis are critical for tumor suppression, although potentially through transcriptional programs distinct from those delineated under conditions of acute DNA damage.
Analysis of p53 TAD mutant knock-in mice
p53 target gene knockout mice have failed to fully resolve the role of p53 transcriptional activation in tumor suppression. While analysis of these mice has revealed tumor phenotypes in a very context-dependent manner, it is unclear whether these tumor phenotypes relate to loss of function as direct p53 target genes. Furthermore, the possibility that transactivation-independent functions of p53 are required for tumor suppression cannot be excluded. As mentioned above, p53 has two discrete TADs, and recently, a new approach utilizing p53 TAD mutant knock-in mouse strains has helped to address the contribution of transcriptional activation to p53 tumor suppression function and has identified novel p53 target genes with likely roles in p53-mediated tumor suppression.
Knock-in mice expressing p53 mutants carrying specific amino acid substitutions within the two p53 TADs were generated to dissect the roles of these TADs in various contexts of p53 function. Microarray analysis of HRasV12-MEFs expressing the different p53 mutants allowed characterization of p53 mutant transcriptional activity in a model of oncogene-induced senescence. Mutation of the first TAD (termed p5325,26, with L25Q;W26S mutations) results in severely impaired transactivation of nearly all known p53-dependent genes, including p21, Puma, and Noxa, although induction of a small subset of p53-dependent genes is similar to that seen in wild-type cells (Figure 3a; [14, 15]). The majority of genes induced by both p5325,26 and wild-type p53 represent novel p53 targets, with a few exceptions, including Bax. This limited transactivation capability allows the p5325,26 mutant to mediate only certain p53 effector functions [14, 15]. For example, p5325,26 is unable to mount responses to acute DNA damage, either in cell cycle arrest or apoptosis, but is capable of tumor suppression in diverse cell lineages in mouse models [15, 77]. Surprisingly, these data indicate that robust transcriptional activation of many well-characterized p53 target genes is dispensable for tumor suppression, and that instead, potent transactivation of novel p53 target genes, minimal transactivation of canonical p53 target genes, or both, are important for tumor suppression. While substitutions at F53Q;F54S within the second TAD (p5353,54) alone do not compromise p53 transactivation capability or biological activity, mutation of both TADs (p5325,26,53,54) results in a transcriptional profile resembling that of p53-null cells. The p5325,26,53,54 mutant is ineffective in p53 effector function in vitro and in tumor suppression in vivo [15, 77], demonstrating that the ability of p53 to activate transcription is indeed required for tumor suppression. Notably, our studies are not incompatible with an additional role for p53 at the mitochondria in tumor suppression.
Figure 3. Functional analysis of p53 transactivation domain (TAD) mutants identifies p53 target genes involved in tumor suppression.
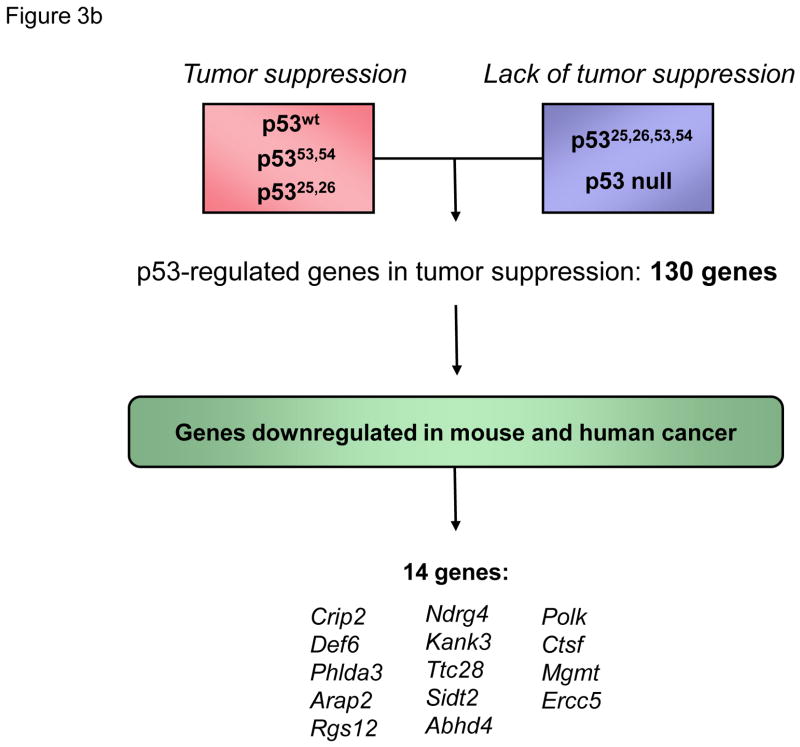
(a) Transactivation domain 1 (TAD1) and robust transactivation of classical p53 target genes are required for responses to acute DNA damage, including apoptosis and cell cycle arrest. A p53 TAD1 mutant, p5325,26, is unable to efficiently activate expression of canonical p53 target genes, including p21, Noxa, Perp, and Puma. p5325,26 is also unable to induce apoptosis or cell cycle arrest in response to acute DNA damage. Transactivation by either TAD1 or TAD2 allows p53 responses to oncogene activation. p5325,26 can activate expression of only a limited number of mostly novel target genes, but can promote tumor suppression in a number of mouse models. The tumor suppressor capability of p5325,26 can be explained by its ability to robustly activate a limited set of novel direct p53 target genes (Sidt2, Phlda3, Abhd4, etc.). The capacity of p5325,26 to promote very low level activation of various classical p53 target genes may also contribute to tumor suppression. The “?” denotes additional, still-unknown genes critical for responses to acute DNA damage and oncogene activation. (b) Comparison of gene expression profiles of p53 wild-type and p53-null HrasV12-MEFs results in more than 1000 differentially expressed genes. To enrich for genes with specific roles in tumor suppression, we leveraged gene expression profiling data generated with the p5325,26 mutant, which activates only a small subset of p53 target genes, yet is a potent tumor suppressor. Using transcriptomics analysis of HrasV12-MEFs, we identified genes induced at least 2 fold and within 1.5 standard deviations by p53wt, p5325,26, and p5353,54, which all have tumor suppressor activity, relative to p5325,26,53,54 or p53-null samples, which lack tumor suppressor activity. This list of 130 genes was then filtered for those commonly downregulated in human and mouse tumors, according to the EBI’s Gene Atlas database. A group of 14 candidate genes with likely roles in p53-mediated tumor suppression was defined by this analysis.
Because the p5325,26 mutant is deficient for efficient transactivation of most but not all p53 targets, yet is a functional tumor suppressor, it provides a powerful and unique tool to pinpoint those p53 targets most critical for p53 tumor suppressor activity. This point is underscored by our observation that comparing expression profiles of wild-type and p53-null cells identifies >1000 differentially expressed genes, whereas identifying those genes efficiently regulated by both wild-type p53 and p5325,26 defines a limited set of ~130 tumor suppression- associated genes regulated by p53. Thus, to identify such tumor suppression target genes, gene expression profiles of cells expressing p53 variants functional in tumor suppression (p5325,26, p5353,54, p53wt) were compared with those in cells with p53 mutants defective for tumor suppression (p5325,26,53,54, p53-null) [15], thereby generating a list of mostly novel p53- dependent genes likely to be important for tumor suppression (Figure 3b). For further refinement, the list was filtered for genes known to be downregulated in human and mouse cancers, to arrive at a group of 14 candidate tumor suppressors. Included in this list are genes involved in several major functional categories: cell signaling (Phlda3, Rgs12), cytoskeletal function (Crip2, Kank3, Arap2), and DNA repair (Polk, Mgmt, Ercc5). One of these, Phlda3, was previously described as a direct p53 target involved in apoptosis and frequently lost in human lung cancers [78]. These genes represent bona fide p53 target genes as nearly all contain p53 consensus sites, are directly bound by wild-type p53 by ChIP, and are regulated in a p53- dependent manner in response to DNA damage or oncogene expression in both mouse and human cells. Importantly, this gene list could be used to accurately predict the p53 status of human breast cancer samples, suggesting that the signatures derived from analysis of mouse cells were effective in identifying genes with relevance to human cancer. Moreover, overexpression and knockdown approaches revealed that several of these genes behave like tumor suppressors. These analyses therefore define a new network of p53 targets important for tumor suppression (Figure 3). Given the function of these genes in different cellular processes, such as cell cycle regulation, migration, and DNA repair, p53 tumor suppression is likely to rely on the coordinate engagement of multiple diverse pathways (Figure 1). Future investigation of these new p53 target genes will more clearly unravel their precise roles in tumor suppression.
The tapestry of p53 tumor suppression will be further illuminated by deciphering the molecular basis for different TAD requirements at different target genes. p53 activates transcription by recruiting cofactors involved in transcriptional initiation and chromatin modification to the transcriptional start sites of target genes. Biochemical analysis of human p53 suggests that TAD1 and TAD2 differentially interact with some cofactors, while collaborating in recruitment of others. For example, TAD 1 interacts with TATA binding protein (TBP) [79], TAD2 specifically binds transcription factor IIH (TFIIH) [80], and both TADs contribute to interaction with the histone acetyltransferases (HATs) p300 and CBP [81, 82] (Figure 2). Defining the cofactor binding specificities of each TAD at different promoters will provide a framework for understanding context-specific p53 responses, including tumor suppression.
Summary and Future Perspectives
Defining the molecular mechanisms underlying p53 function in tumor suppression is critical for broadly understanding cancer development. While p53 is unequivocally involved in eliciting senescence or apoptosis in response to cellular stress signals, the molecular details of p53 action in tumor suppression have remained surprisingly elusive. It has been challenging to map the transcriptional effectors of p53 tumor suppressor function because of the subtle tumor phenotypes of single or double p53 target gene knockout mutant mice, an observation leading to the notion that the combined actions of proteins encoded by a host of p53 target genes mediate p53’s tumor suppressor function. Studies of p53 TAD mutant knock-in mice have helped to address this point specifically by employing p5325,26, which is severely impaired for activation of the majority of p53 target genes, allowing an effective phenocopy of the knockdown of numerous p53 targets in one mouse. Intriguingly, this mutant is completely competent to suppress the development of a wide range of tumor types, in association with activation of a small subset of novel direct p53 target genes, some with demonstrated tumor suppressor activity. Thus, these studies have shed light on how p53 suppresses tumorigenesis by elaborating a new network of potential p53 tumor suppressor mediators, whose future analysis will contribute to a better understanding of which of the p53 tumor suppression-associated target genes and which cellular processes are most key for suppressing cancer development in diverse contexts. Beyond unraveling the mechanisms of p53-mediated tumor suppression, analysis of the TAD knock-in mice has elucidated a clear distinction in the transcriptional programs for p53 responses to acute DNA damage and oncogenic signaling. These findings have important therapeutic applications, by suggesting strategies to mitigate the detrimental p53-dependent side effects resulting from DNA-damaging radiation and chemotherapies while preserving p53 tumor suppressor function. Collectively, these studies lay the groundwork for fully elaborating the intricate circuitry fundamental for p53 cellular responses and tumor suppression.
Acknowledgments
We thank Colleen Brady, Daniela Kenzelmann Broz, Jeanine Frey, and Dadi Jiang for critical reading of the manuscript. We apologize to authors whose work could not be cited due to space constraints.
References
- 1.Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316– 323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 4.Halazonetis TD, et al. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 5.Christophorou MA, et al. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 6.Efeyan A, et al. Tumour biology: Policing of oncogene activity by p53. Nature. 2006;443:159. doi: 10.1038/443159a. [DOI] [PubMed] [Google Scholar]
- 7.Hinkal G, et al. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS One. 2009;4:e6654. doi: 10.1371/journal.pone.0006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 9.Venot C, et al. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene. 1999;18:2405–2410. doi: 10.1038/sj.onc.1202539. [DOI] [PubMed] [Google Scholar]
- 10.Candau R, et al. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, et al. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem. 1998;273:13030–13036. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]
- 12.Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson TM, et al. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37:145–152. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- 15.Brady CA, et al. Distinct p53 Transcriptional Programs Dictate Acute DNA-Damage Responses and Tumor Suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riley T, et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 17.Smeenk L, et al. Role of p53 serine 46 in p53 target gene regulation. PLoS One. 2011;6:e17574. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 19.Xiang H, et al. Bax involvement in p53-mediated neuronal cell death. J Neurosci. 1998;18:1363–1373. doi: 10.1523/JNEUROSCI.18-04-01363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong MJ, et al. Atm and Bax cooperate in ionizing radiation-induced apoptosis in the central nervous system. Proc Natl Acad Sci U S A. 2000;97:889–894. doi: 10.1073/pnas.97.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson MD, et al. Evidence for involvement of Bax and p53, but not caspases, in radiation-induced cell death of cultured postnatal hippocampal neurons. J Neurosci Res. 1998;54:721–733. doi: 10.1002/(SICI)1097-4547(19981215)54:6<721::AID-JNR1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Mccurrach ME, et al. bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad Sci U S A. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard DM, et al. Damage-induced apoptosis in intestinal epithelia from bcl-2-null and bax-null mice: investigations of the mechanistic determinants of epithelial apoptosis in vivo. Oncogene. 1999;18:7287–7293. doi: 10.1038/sj.onc.1203150. [DOI] [PubMed] [Google Scholar]
- 24.Knudson CM, et al. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 25.Knudson CM, et al. Bax accelerates tumorigenesis in p53-deficient mice. Cancer Res. 2001;61:659–665. [PubMed] [Google Scholar]
- 26.Shibata MA, et al. Haploid loss of bax leads to accelerated mammary tumor development in C3(1)/SV40-TAg transgenic mice: reduction in protective apoptotic response at the preneoplastic stage. Embo J. 1999;18:2692–2701. doi: 10.1093/emboj/18.10.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin C, et al. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 28.Eischen CM, et al. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol. 2001;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dansen TB, et al. Specific requirement for Bax, not Bak, in Myc-induced apoptosis and tumor suppression in vivo. J Biol Chem. 2006;281:10890–10895. doi: 10.1074/jbc.M513655200. [DOI] [PubMed] [Google Scholar]
- 30.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda E, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 32.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 33.Jeffers JR, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 34.Villunger A, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 35.Qiu W, et al. PUMA regulates intestinal progenitor cell radiosensitivity and gastrointestinal syndrome. Cell Stem Cell. 2008;2:576–583. doi: 10.1016/j.stem.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michalak EM, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–696. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrison SP, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemann MT, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci U S A. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ploner C, et al. Noxa: at the tip of the balance between life and death. Oncogene. 2008;27(Suppl 1):S84–92. doi: 10.1038/onc.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibue T, et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 2003;17:2233–2238. doi: 10.1101/gad.1103603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naik E, et al. Ultraviolet radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J Cell Biol. 2007;176:415–424. doi: 10.1083/jcb.200608070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akhtar RS, et al. BH3-only proapoptotic Bcl-2 family members Noxa and Puma mediate neural precursor cell death. J Neurosci. 2006;26:7257–7264. doi: 10.1523/JNEUROSCI.0196-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michalak EM, et al. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Attardi LD, et al. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. Embo J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, et al. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 46.Attardi LD, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- 47.Ihrie RA, et al. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Beaudry VG, et al. Loss of the p53/p63 regulated desmosomal protein Perp promotes tumorigenesis. PLoS Genet. 2010;6:e1001168. doi: 10.1371/journal.pgen.1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finnberg N, et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol Cell Biol. 2005;25:2000–2013. doi: 10.1128/MCB.25.5.2000-2013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finnberg N, et al. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J Clin Invest. 2008;118:111–123. doi: 10.1172/JCI29900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim IR, et al. DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis. 2009;14:1039–1049. doi: 10.1007/s10495-009-0375-1. [DOI] [PubMed] [Google Scholar]
- 52.Adachi M, et al. Enhanced and accelerated lymphoproliferation in Fas-null mice. Proc Natl Acad Sci U S A. 1996;93:2131–2136. doi: 10.1073/pnas.93.5.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillen-Ahlers H, et al. Fas/CD95 deficiency in ApcMin/+ mice increases intestinal tumor burden. PLoS One. 2010;5:e9070. doi: 10.1371/journal.pone.0009070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, et al. CD95 promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harper JW, et al. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 56.El-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 57.Deng C, et al. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 58.Brugarolas J, et al. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 59.Pantoja C, Serrano M. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene. 1999;18:4974–4982. doi: 10.1038/sj.onc.1202880. [DOI] [PubMed] [Google Scholar]
- 60.Martin-Caballero J, et al. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61:6234–6238. [PubMed] [Google Scholar]
- 61.Jones JM, et al. Heterozygosity of p21WAF1/CIP1 enhances tumor cell proliferation and cyclin D1-associated kinase activity in a murine mammary cancer model. Cell Growth Differ. 1999;10:213–222. [PubMed] [Google Scholar]
- 62.Jackson RJ, et al. p21Cip1 nullizygosity increases tumor metastasis in irradiated mice. Cancer Res. 2003;63:3021–3025. [PubMed] [Google Scholar]
- 63.Brugarolas J, et al. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cole AM, et al. p21 loss blocks senescence following Apc loss and provokes tumourigenesis in the renal but not the intestinal epithelium. EMBO Mol Med. 2010;2:472–486. doi: 10.1002/emmm.201000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hollander MC, et al. Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 66.Hildesheim J, et al. Gadd45a protects against UV irradiation-induced skin tumors, and promotes apoptosis and stress signaling via MAPK and p53. Cancer Res. 2002;62:7305–7315. [PubMed] [Google Scholar]
- 67.Tront JS, et al. Gadd45a suppresses Ras-driven mammary tumorigenesis by activation of c-Jun NH2-terminal kinase and p38 stress signaling resulting in apoptosis and senescence. Cancer Res. 2006;66:8448–8454. doi: 10.1158/0008-5472.CAN-06-2013. [DOI] [PubMed] [Google Scholar]
- 68.Tront JS, et al. Gadd45a functions as a promoter or suppressor of breast cancer dependent on the oncogenic stress. Cancer Res. 2010;70:9671–9681. doi: 10.1158/0008-5472.CAN-10-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Stanchina E, et al. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 70.Ferbeyre G, et al. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 71.Pearson M, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 72.Wang ZG, et al. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 73.Rego EM, et al. Role of promyelocytic leukemia (PML) protein in tumor suppression. J Exp Med. 2001;193:521–529. doi: 10.1084/jem.193.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doumont G, et al. G1 checkpoint failure and increased tumor susceptibility in mice lacking the novel p53 target Ptprv. Embo J. 2005;24:3093–3103. doi: 10.1038/sj.emboj.7600769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu G, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- 76.Barboza JA, et al. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci U S A. 2006;103:19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang D, et al. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc Natl Acad Sci U S A. 2011;108:17123–17128. doi: 10.1073/pnas.1111245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawase T, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–550. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 79.Thut CJ, et al. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 80.Di Lello P, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 81.Ferreon JC, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teufel DP, et al. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci U S A. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Attardi LD, Donehower LA. Probing p53 biological functions through the use of genetically engineered mouse models. Mutat Res. 2005;576:4–21. doi: 10.1016/j.mrfmmm.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 84.Symonds H, et al. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 85.Cosme-Blanco W, et al. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007;8:497–503. doi: 10.1038/sj.embor.7400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dankort D, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 89.Ihrie RA, et al. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr Biol. 2003;13:1985–1990. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 90.O’connor L, et al. CD95 (Fas/APO-1) and p53 signal apoptosis independently in diverse cell types. Cancer Res. 2000;60:1217–1220. [PubMed] [Google Scholar]
- 91.Adnane J, et al. Loss of p21WAF1/CIP1 accelerates Ras oncogenesis in a transgenic/knockout mammary cancer model. Oncogene. 2000;19:5338–5347. doi: 10.1038/sj.onc.1203956. [DOI] [PubMed] [Google Scholar]
- 92.Dulic V, et al. Uncoupling between phenotypic senescence and cell cycle arrest in aging p21-deficient fibroblasts. Mol Cell Biol. 2000;20:6741–6754. doi: 10.1128/mcb.20.18.6741-6754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Philipp J, et al. Tumor suppression by p27Kip1 and p21Cip1 during chemically induced skin carcinogenesis. Oncogene. 1999;18:4689–4698. doi: 10.1038/sj.onc.1202840. [DOI] [PubMed] [Google Scholar]
- 94.Elyada E, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 95.Bulavin DV, et al. Loss of oncogenic H-ras-induced cell cycle arrest and p38 mitogen-activated protein kinase activation by disruption of Gadd45a. Mol Cell Biol. 2003;23:3859–3871. doi: 10.1128/MCB.23.11.3859-3871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]