Abstract
The Ser/Thr kinase Raf-1 is a protooncogene product that is a central component in many signaling pathways involved in normal cell growth and oncogenic transformation. Upon activation, Raf-1 phosphorylates mitogen-activated protein kinase kinase (MEK), which in turn activates mitogen-activated protein kinase/extracellular signal-regulated kinases (MAPK/ERKs), leading to the propagation of signals. Depending on specific stimuli and cellular environment, the Raf-1–MEK–ERK cascade regulates diverse cellular processes such as proliferation, differentiation, and apoptosis. Here, we describe a MEK–ERK-independent prosurvival function of Raf-1. We found that Raf-1 interacts with the proapoptotic, stress-activated protein kinase ASK1 (apoptosis signal-regulating kinase 1) in vitro and in vivo. Deletion analysis localized the Raf-1 binding site to the N-terminal regulatory fragment of ASK1. This interaction allows Raf-1 to act independently of the MEK–ERK pathway to inhibit apoptosis. Furthermore, catalytically inactive forms of Raf-1 can mimic the wild-type effect, raising the possibility of a kinase-independent function of Raf-1. Thus, Raf-1 may promote cell survival through its protein–protein interactions in addition to its established MEK kinase function.
Diverse signaling pathways, such as those mediated by tyrosine kinase receptors and heterotrimeric G protein-coupled receptors, converge on Raf-1 through Ras and other mechanisms (1, 2). Raf-1 activation initiates a mitogen-activated protein kinase (MAPK) cascade that comprises a sequential phosphorylation of the dual-specific MAPK kinases (MEKs) and the extracellular signal-regulated kinases (ERKs). In turn, the Raf-1–MEK–ERK cascade regulates diverse cellular processes such as proliferation and differentiation. Recently, Raf-1 activation of the MEK–ERK pathway has been associated with inhibition of apoptosis, leading to cell survival (3–6). Consistent with a critical role of the MEK–ERK pathway in survival signaling, treatment of cells with either MEK inhibitors or dominant inhibitory MEKs blocks the antiapoptotic function of Raf (5, 6). The prosurvival function of the MEK–ERK pathway appears to be mediated by dual mechanisms. A transcription-dependent mechanism involves the activation of cAMP response element-binding protein by ribosomal S6 kinases, leading to increased transcription of prosurvival genes, whereas a transcription-independent mechanism allows phosphorylation of proapoptotic proteins such as Bad, leading to its inactivation (7–9). In support of this model, genetic analysis in Drosophila demonstrated that activated ERK pathway inhibits the expression and activity of the proapoptotic protein Hid (10, 11). Thus, the Raf-activated MEK–ERK pathway may promote cell survival by targeting proteins critical for mediating apoptosis.
ASK1 (apoptosis signal-regulating kinase 1) is an important mediator of apoptotic signaling initiated by a variety of death stimuli, including tumor necrosis factor α, Fas activation, oxidative stress, and DNA damage (12–16). Consistent with its role in apoptotic signaling, dominant negative mutants of ASK1 can inhibit tumor necrosis factor α and Fas ligation-induced cell death (12, 13), and overexpression of ASK1 is sufficient to cause apoptosis in a number of cell lines through a mitochondria-dependent caspase activation pathway (17). Thus, suppression of ASK1 may provide a general mechanism for cell survival. Indeed, multiple mechanisms have been described that directly control ASK1 function. For example, the binding of reduced thioredoxin has been shown to inhibit ASK1-induced apoptosis, which may couple intracellular redox state to the regulation of ASK1 activity (14, 15). The phosphoserine-binding protein 14-3-3 can inhibit the proapoptotic function of ASK1 through binding to Ser-967 of ASK1, which is phosphorylated by an unknown survival signaling kinase (18). Here, we describe a physical and functional interaction of Raf-1 with ASK1, suggesting a novel prosurvival mechanism for Raf-1 independent of the MEK–ERK pathway.
Materials and Methods
Plasmids.
Expression vectors for ASK1 and its mutants have been described (12, 18). Wild-type (WT) MEK1WT, constitutively active mutant MEK1C, and dominant negative mutant MEK1dn were gifts from K. Guan, Univ. of Michigan (19). pcDNA3–FLAG–Raf-1, Raf-N, and Raf-C have been described (20). Mutations RafK375W and RafS259A/S621A were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) with pcDNA3–FLAG–Raf-1 as a template. Hemagglutinin (HA)–ASK1 NT (6) and ΔC (6) were generated by PCRs and subcloned into pcDNA3. HA–ASK1 ΔN (678–1375) was constructed in the Gateway cloning expression vector pDEST26 (Invitrogen). pcDNA3–HA–ASK1 ΔK (1–819/1057–1375) was generated by digestion of pcDNA3–HA–ASK1 with BamHI and BglII and religation. HA–ASK1–CT (1071–1375) was constructed by EcoRI digestion and religation of pcDNA3–HA–ASK1.
Cell Culture and DNA Transfection.
HeLa, COS7, and L929 cells were cultured in DMEM (Mediatech, Washington, DC) with 10% FBS (Atlanta Biologicals, Norcross, GA). Transfection was performed with FuGENE 6 (Roche Molecular Biochemicals) according to the manufacturer's instructions.
Assays for Cell Death.
For the nuclear morphology assays (18), 2 × 105 HeLa cells were cultured in 35-mm dishes containing glass coverslips. Cells were cotransfected with pTJM9 (0.4 μg) encoding enhanced green fluorescent protein (eGFP) and test plasmids (1.6 μg total) or supplemented with pcDNA3. Eighteen hours after transfection, the medium was changed to serum-free DMEM. Twenty-four hours later, cells on the glass coverslips were washed, fixed (0.5% glutaraldehyde/2% formaldehyde in PBS), stained with 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield mounting medium (Vector Laboratories), and visualized by using a fluorescence microscope as described (18). Transfected cells with fragmented nuclei were scored for apoptosis in a blind fashion. Cells remaining in the dishes were lysed and immunoblotted with various antibodies by using the ECL system (Amersham Pharmacia). For the β-galactosidase (β-gal)-based cell morphology assay, 2 × 105 HeLa cells were cultured in 35-mm plates and cotransfected with a lacZ expression vector (0.4 μg) and test plasmids (1.6 μg total). Twenty-four hours after transfection, cells were fixed, stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and scored for apoptosis as described (13). Parallel samples were collected for Western blotting.
The DNA content-based apoptosis assay was performed with a fluorescence-activated cell sorter as described (21). Briefly, COS7 cells (2 × 105) were cotransfected with pEGFP-F (0.4 μg; CLONTECH) encoding a farnesylated eGFP and test plasmids (1.6 μg total). Twenty-four hours after transfection, total cells were trypsinized, resuspended in PBS, fixed in ice-cold ethanol followed by overnight incubation, treated with RNase A (Sigma–Aldrich), and stained with propidium iodide (Sigma–Aldrich). Samples were analyzed with a FACSort flow cytometer (Becton Dickinson). The DNA content of transfected cells was determined by using WINMDI 2.8 software (J. Trotter, Scripps Research Institute, La Jolla, CA).
Immunoprecipitation and Western Blotting.
Forty-eight hours after transfection, 4 × 105 COS7 cells were lysed in 300 μl of lysis buffer (0.2% Nonidet P-40/10 mM Hepes, pH 7.4/150 mM NaCl/5 mM NaF/2 mM Na3VO4/5 mM Na4P2O7/10 μg/ml aprotonin/10 μg/ml leupeptin/1 mM phenylmethylsulfonyl fluoride). Cell extracts were clarified by centrifugation and used for immunoprecipitation with various antibodies and protein G-Sepharose (Amersham Pharmacia). Immunocomplexes were washed four times with lysis buffer containing 1% Nonidet P-40 or RIPA buffer (1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/137 mM NaCl/20 mM Tris⋅HCl, pH 7.5) and resolved on SDS/PAGE (12.5%) for Western blotting. The enzyme-linked immunoblotting procedures were performed essentially as described (18). Corresponding secondary antibodies were used against each primary antibody: horseradish peroxidase-conjugated goat anti-mouse IgG for monoclonal antibodies and horseradish peroxidase-conjugated goat anti-rabbit IgG for polyclonal antibodies (Santa Cruz Biotechnology). Cross-reacting materials were visualized by using the ECL detection reagents (Amersham Pharmacia).
Results
Overexpression of Raf-1 Inhibits ASK1-Induced Apoptosis.
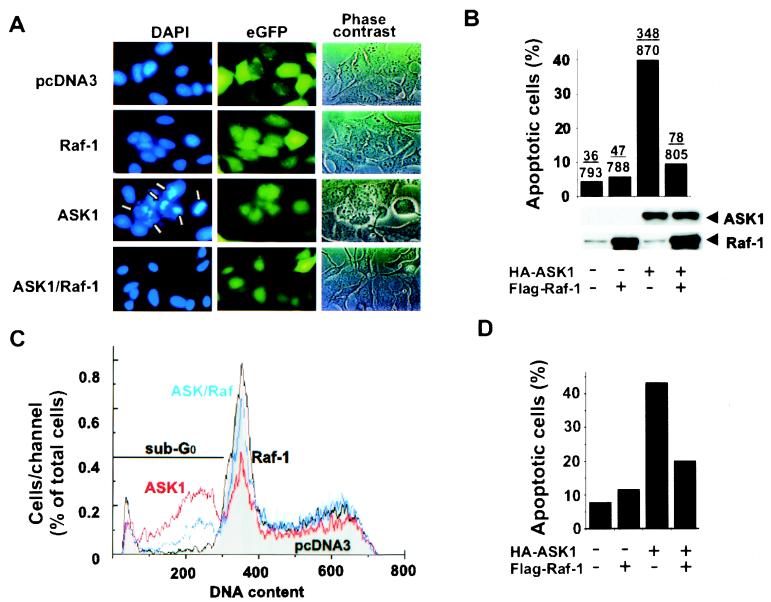
To investigate the role of the Raf-1 pathway in suppressing apoptotic signaling, we tested the effect of Raf-1 on ASK1-induced apoptosis. HeLa cells were transiently transfected with HA-tagged human ASK1 alone or together with FLAG-tagged Raf-1. Apoptotic cell death was monitored by nuclear morphology. Cells transfected with the control vector or a vector encoding Raf-1 showed homogenous DAPI staining and normal nuclear morphology (Fig. 1A). In contrast, expression of ASK1 induced the appearance of condensed chromatin and fragmented nuclei characteristic of apoptosis consistent with previously published data (18). When cells were transfected with Raf-1 together with ASK1, however, the fraction of cells with apoptotic nuclear morphology was decreased, which was quantified as shown in Fig. 1B. These results suggest that Raf-1 can inhibit the apoptotic activity of ASK1. This decrease did not appear to result from decreased levels of ASK1 protein (Fig. 1B Lower). Similar results were obtained with COS7 cells by using a DNA content-based flow cytometry assay (Fig. 1 C and D). When adherent cells undergo apoptosis, they often exhibit cell shrinkage and rounded-up morphology. These features form the basis of an alternative cell morphology-based death assay (13), which uses β-gal as a marker (Fig. 2A). Consistent with nuclear morphology and DNA content assays, coexpression of Raf-1 diminished ASK1-induced cell death (Fig. 2B). These data demonstrate that a Raf-1-mediated signaling pathway may play a negative role in controlling ASK1-dependent apoptosis.
Figure 1.
Expression of Raf-1 inhibits ASK1-induced apoptosis. HeLa cells were transfected with the plasmids pcDNA3–HA–ASK1 (1.2 μg) or pcDNA3–FLAG–Raf-1 (0.4 μg) along with an eGFP expression vector (0.4 μg) as indicated. Eighteen hours posttransfection, cells were placed in serum-free medium for an additional 24 h before staining with DAPI. Nuclear morphology of transfected cells was examined by fluorescence microscopy as described (18), and apoptotic nuclei are indicated by arrows (A). The fraction of transfected cells with fragmented nuclei was quantified in a blind manner (B Upper). Cell lysates from each sample were subjected to SDS/PAGE and Western blotting with anti-Raf-1 (SC133; Santa Cruz Biotechnology) or anti-HA (12CA5) antibodies (B Lower). (C) COS7 cells were transfected with plasmids as in A along with an eGFP-F expression vector. Twenty-four hours after transfection, total cells were harvested, their DNA was stained with propidium iodide, and eGFP and propidium iodide signals were measured on a FACSort flow cytometer. Transfected cells (eGFP-positive) were placed in various phases of the cell cycle based on their DNA content. Apoptotic cells with fragmented DNA (subG0) are indicated. (D) Data from C were compiled to show the amount of apoptosis caused by expression of transfected plasmids.
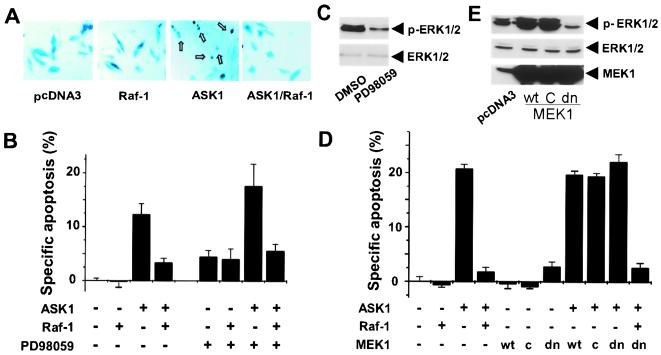
Figure 2.
MEK activity is not required for Raf-1 to inhibit ASK1. (A) Morphology of ASK1 transfected cells. HeLa cells were cotransfected with a β-gal expression vector and test plasmids as indicated. Twenty-four hours posttransfection, cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside. Shrunken apoptotic cells with rounded-up shape were scored as apoptotic (arrows). (B) Effect of MEK inhibition by PD98059 on Raf-1 function. Six hours after transfection as in A, HeLa cells were treated with PD98059 (60 μM) or vehicle (DMSO) for 18 h before being stained with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside and scored for apoptosis in a blind fashion. Specific apoptosis is derived by subtracting the level of apoptosis seen in pcDNA3-transfected cells. At least 500 cells were scored for each sample. Results shown are representative of three independent experiments. (C) Inhibition of ERK1/2 activation by PD98059. Cell lysates from HeLa cells treated with 60 μM PD98059 or vehicle were probed by Western blotting with either an ERK1/2 activation-specific antibody (Upper; Cell Signaling Technology) or pan-ERK1/2 antibody (Lower; Cell Signaling Technology). (D) Effect of MEK1 overexpression on ASK1-induced apoptosis. HeLa cells were transfected with a β-gal reporter together with indicated expression vectors for 12 h, serum-starved for 24 h, and scored for apoptosis as in A. Data are summary of three independent experiments. (E) Effect of MEK1 expression on ERK1/2 activation. Lysates from HeLa cells treated as in D were subjected to SDS/PAGE and Western blotting with ERK1/2 activation-specific or pan antibodies or anti-MEK antibody (Santa Cruz Biotechnology).
The MEK–ERK Pathway Is Not Required for Raf-1 to Block ASK1 Function.
Raf-1-dependent activation of the MEK–ERK pathway has been shown to promote cell survival by targeting various death pathways (6–11). To test the hypothesis that Raf-1 regulates ASK1-induced apoptosis through the MEK–ERK pathway, we used two widely used MEK antagonists, PD98059 and U0126 (22, 23). Surprisingly, treatment of cells with PD98059 (60 μM) did not decrease the ability of Raf-1 to inhibit ASK1-induced cell death, although this agent diminished the activation of ERK1/2 (Fig. 2 B and C). Similar results were obtained with U0126 (25 μM; data not shown). To confirm the observations with PD98059 and U0126, we used a dominant negative mutant of MEK1, MEK1dn, to interfere with MEK signaling (19). Expression of MEK1dn decreased the basal activation level of ERK1/2 but showed no effect on the inhibition of ASK1-induced apoptosis by Raf-1 (Fig. 2 D and E). However, the above MEK inhibitors are unable to completely block MEK–ERK signaling, and the remaining activity may be sufficient to transmit the Raf-1 survival signal. To test this possibility, we examined whether activation of the MEK–ERK pathway by overexpression of MEK1, an immediate effector of Raf-1, would be sufficient to mimic the Raf-1 effect and inhibit ASK1. As shown in Fig. 2E, expression of MEK1WT or the constitutively active mutant MEK1C activated ERK1/2. However, neither of these MEK1 constructs was capable of attenuating the proapoptotic activity of ASK1 (Fig. 2D). Thus, MEK1 cannot substitute for Raf-1 to inhibit ASK1 function. Because neither inhibition nor activation of MEK impacted the proapoptotic activity of ASK1, Raf-1 likely antagonizes ASK1 through a mechanism independent of the MEK–ERK pathway.
Raf-1 Interacts with ASK1 in Cells.
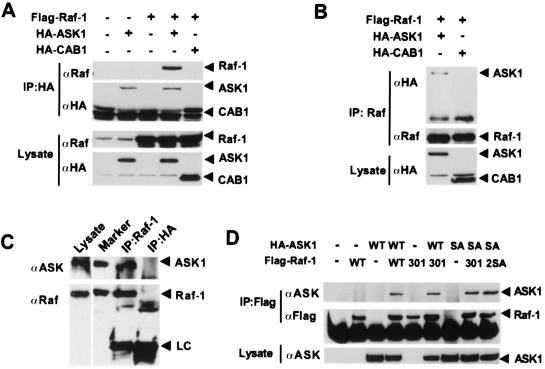
One possible mechanism for Raf-1 inhibition of ASK1 apoptotic activity is through direct interaction between the two proteins. To test this hypothesis, we performed coimmunoprecipitation experiments in COS7 cells (18). FLAG–Raf-1 was transiently expressed with either HA–ASK1 or the negative control HA–CAB1. HA immunocomplexes were isolated and examined. Raf-1 was found in the HA–ASK1 immunocomplex but was absent from the HA–CAB1 complex, suggesting that Raf-1 may specifically interact with ASK1 (Fig. 3A). To confirm the Raf-1/ASK1 association, we carried out reciprocal experiments by immunoprecipitating Raf-1 from COS7 cell lysates (Fig. 3B). HA–ASK1, but not HA–CAB1, was detected in the Raf-1 immunocomplex. These data together suggest that Raf-1 is associated with ASK1 in mammalian cells.
Figure 3.
Raf-1 specifically interacts with ASK1. (A) HA–ASK1 immunocomplex contains Raf-1. COS7 cells were transfected with the expression vector for HA–ASK1 or HA–CAB1 together with FLAG–Raf-1 (18). After 48 h, cell lysates were prepared, and ASK1 or CAB1 was immunoprecipitated with anti-HA antibody. Immunoprecipitates were washed extensively with Nonidet P-40 (1%) lysis buffer before Western blotting with antibodies to Raf-1 and HA (Upper). (Lower) Expression levels of Raf-1 and HA-ASK1 or HA-CAB1 in the lysates. (B) Raf-1 immunocomplexes contain ASK1. Polyclonal anti-Raf-1 antibody (Santa Cruz Biotechnology) was used to immunoprecipitate Raf-1 as in A. HA-ASK1 was detected in the Raf-1 immunoprecipitates with anti-HA antibody. (C) Endogenous Raf-1 and ASK1 form complexes in L929 cells. Immunoprecipitates were prepared from L929 cell lysates (left lane) using either anti-Raf-1 monoclonal antibody (Transduction Laboratories) or anti-HA antibody as a negative control and probed for ASK1 using the antibody DAV (ref. 14; Upper). Coimmunoprecipitated ASK1 and Raf-1 comigrate, respectively, with overexpressed HA–ASK1 and FLAG–Raf-1 (Marker lane). Antibody light chains (LC) in the immunoprecipitates are indicated. (D) 14-3-3 binding and Raf-1 kinase activity are not required for the Raf-1–ASK1 interaction. COS7 cells were cotransfected with plasmids encoding FLAG–Raf-1WT, catalytically inactive FLAG–RafK375M (301), or 14-3-3 binding defective FLAG–RafS259/621A (2SA) and HA-ASK1WT or 14-3-3 binding defective HA–ASK1S967A (SA). FLAG–Raf-1 complex was precipitated by using anti-FLAG antibody (Sigma) and probed with anti-ASK1 antibody (Santa Cruz Biotechnology). Expression levels of HA–ASK1 were verified by Western blotting (Lower). As a control, FLAG–RafS259/621A was found to bind HA–ASK1WT (data not shown).
To test whether Raf-1 interacts with ASK1 in the absence of experimental manipulation, we isolated the endogenous Raf-1 protein complex from L929 cells by using an anti-Raf-1 monoclonal antibody and probed for the presence of native ASK1. Indeed, the Raf-1 antibody coimmunoprecipitated endogenous ASK1 (Fig. 3C). As a control, an anti-HA monoclonal antibody failed to pull down ASK1 under the same conditions, suggesting a specific interaction of Raf-1 with ASK1. A reciprocal experiment showed the presence of Raf-1 in immunocomplexes isolated with either anti-ASK1 H300 (Santa Cruz Biotechnology) or anti-ASK1 DAV antibodies (ref. 14 and data not shown). Endogenous ASK1 was also found in complex with Raf-1 in Jurkat T cells (data not shown). Thus, Raf-1 and ASK1 associate in vivo, which supports the model that Raf-1 promotes cell survival in part by antagonizing the ASK1-mediated apoptotic signaling.
Both Raf-1 and ASK1 are capable of binding to 14-3-3 proteins (1, 18). The 14-3-3 proteins exist as dimers and could potentially bridge two distinct target proteins (24). It has been demonstrated that 14-3-3 interacts with Raf-1 through phosphorylated Ser-259 and Ser-621 (1) and with ASK1 through phosphorylated Ser-967 (18). Thus, it is possible that the Raf-1/ASK1 association is mediated by 14-3-3 dimers. To test this notion, we used Raf-1 and ASK1 mutants that are defective in 14-3-3 binding, RafS259/621A and ASK1S967A (1, 18). As shown in Fig. 3D, mutant Raf-1 and ASK1 proteins interacted as efficiently as the WT proteins did, suggesting that the Raf-1–ASK1 interaction does not require 14-3-3 proteins.
The N-Terminal Regulatory Domain of ASK1 Mediates Raf-1 Binding.
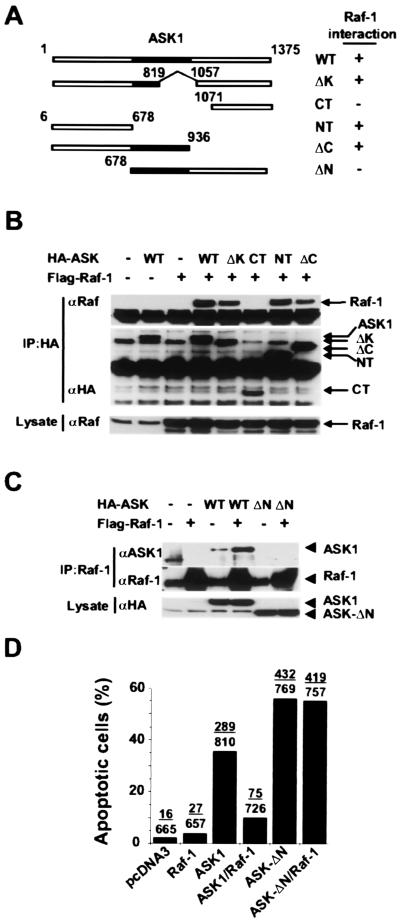
If the Raf-1–ASK1 interaction mediates the inhibitory effect of Raf-1 on ASK1-induced death, we reasoned that a mutant form of ASK1 incapable of Raf-1 binding would be refractory to Raf-1 inhibition. To test this hypothesis, we mapped the Raf-1 binding site on ASK1. ASK1 has its catalytic domain flanked by N-terminal and C-terminal regulatory domains (Fig. 4A). Various truncation mutants of ASK1 were expressed as HA-tagged fusions together with FLAG–Raf-1 in COS7 cells, and their associations were probed in a coimmunoprecipitation assay. Although all of the ASK1 proteins containing the N-terminal domain showed binding to Raf-1, no Raf-1 binding was detectable for the kinase or C-terminal domains of ASK1 (Fig. 4 B and C). Importantly, the N-terminal domain alone was sufficient to bind Raf-1. Raf-1 may inhibit ASK1 by targeting its N-terminal regulatory domain.
Figure 4.
The N-terminal domain of ASK1 mediates the Raf-1 interaction. (A) Schematic diagram of ASK1 proteins. The shaded portion of the boxes represents the ASK1 kinase domain. Association of ASK1 mutants with Raf-1 is summarized. (B) The N-terminal domain of ASK1 is required for Raf-1 binding. FLAG–Raf-1 was transiently transfected into COS7 cells with HA–ASK1WT or truncated mutants. HA–ASK1 protein complexes were immunoprecipitated and subjected to SDS/PAGE and Western blotting with anti-HA (Middle) and anti-Raf-1 antibodies (Top). Lysates from each sample were probed with anti-Raf-1 antibodies (Bottom). (C) Raf-1 does not interact with ASK1-ΔN. Raf-1 protein complexes were immunoprecipitated from each sample with anti-Raf-1 antibody and subjected to SDS/PAGE and Western blotting with anti-ASK1 antibody. Overexposure shows the interaction of endogenous Raf-1 with overexpressed HA–ASK1, but even overexpressed Raf-1 was incapable of binding to ASK1-ΔN. (D) Raf-1 cannot block ASK1-ΔN induced apoptosis. HeLa cells were transfected with plasmids as indicated together with an eGFP marker vector. The nuclear morphology-based assay described in Fig. 1 was used to score for apoptotic cells.
To test whether binding to ASK1 is necessary for the antiapoptotic activity of Raf-1, we investigated the effect of Raf-1 expression on apoptosis induced by ASK-ΔN. In a nuclear morphology-based apoptosis assay, the fraction of apoptotic cells induced by ASK1WT was drastically reduced by coexpression of Raf-1, whereas cell death induced by ASK-ΔN was nonresponsive to the coexpressed Raf-1 (Fig. 4D). These data strongly support a requirement for the Raf-1–ASK1 interaction in the inhibition of ASK1 proapoptotic function.
Raf-1 Catalytic Activity Is Not Required for Inhibition of ASK1-Induced Apoptosis.
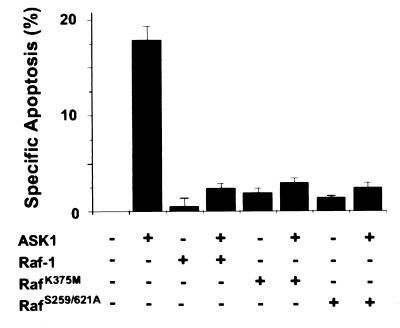
Mutations at Lys-375 and Ser-621 inactivate the kinase activity of Raf-1 (1, 2). The kinase-defective Raf-1 mutants Raf-1K375M and Raf-1S259/621A were capable of binding ASK1 (Fig. 3D). We examined whether the catalytic activity of Raf-1 was required for inhibiting ASK1-induced cell death in HeLa cells. Strikingly, these inactive Raf-1 mutants were as effective as WT in blocking the proapoptotic function of ASK1 under the conditions tested (Fig. 5). Similar results were obtained with COS7 cells in an alternative DNA content-based apoptosis assay using flow cytometry (data not shown). Together, these data suggest a kinase-independent function of Raf-1, strengthening the notion that the MEK–ERK pathway is not involved.
Figure 5.
Raf-1 inhibits ASK1-induced apoptosis independently of its catalytic function. A HeLa cell morphology-based assay as in Fig. 2 was used to score for specific apoptosis. Plasmids used were the same as described in Fig. 3C.
Discussion
Our findings suggest a mechanism by which Raf-1 promotes cell survival independently of the MEK–ERK pathway. Through protein–protein interactions, Raf-1 may directly act on a critical component of the cellular proapoptotic signaling machinery. The fact that catalytically inactive Raf-1 can replace the WT kinase to inhibit ASK1 raises the intriguing possibility that Raf-1 may have a kinase-independent function. It is conceivable that the interaction of Raf-1 with many reported targets may represent kinase-independent pathways of Raf-1 (2). Thus, Raf-1 may have dual functions: activating the MEK–ERK cascade through its enzymatic activity while inhibiting ASK1 through protein–protein interactions.
Evidence is accumulating that Raf-1 may use multiple effectors, in addition to its well established target MEK, to mediate its cellular functions. It was found that activated Raf-1, but not MEK, can drive the differentiation of hippocampal neuronal cells (25). Mutant Raf-1 that is defective in MEK activation is still capable of activating NF-κB-dependent gene expression and other selected pathways (26). These effects of Raf-1 appear to be kinase dependent, and the immediate targets of Raf-1 other than MEK remain to be identified. However, these observations together strongly support the notion that Raf-1 can transmit signals to multiple downstream pathways. Consistent with this idea, Raf-1 has been shown to interact with other critical regulatory proteins such as the cell-cycle modulators Cdc25 and Rb (27, 28) and the proapoptotic protein Bad (29). ASK1 was initially described as a MAPK kinase kinase that activates the stress-activated protein kinases SAPK/JNK and p38 (12, 30). Interaction of Raf-1 with ASK1 may allow a functional cross-talk between two antagonistic signaling pathways, which is likely to be critical for signal integration. Recent demonstration of the interaction between Raf-1 and MEKK1 supports an extensive interplay at the MAPK kinase kinase level of the signaling network (31). The concerted action of Raf-1 on several aspects of cell growth control may prevent conflicting signaling activities and lead to a meaningful biological output.
It has been postulated that apoptotic cell death is the default program of metazoan cells, which must be suppressed continuously by survival mechanisms (32). Inhibition of the proapoptotic function of ASK1 by Raf-1 may be part of the cellular machinery that maintains survival. It is conceivable that activation of ASK1-mediated apoptosis by death stimuli such as H2O2 and tumor necrosis factor α may involve the dissociation of Raf-1 from ASK1. Because Raf-1 is a vital component of a variety of growth factor-induced signaling pathways, simultaneous stimulation of the MEK–ERK pathway and inhibition of death signaling through its kinase-dependent and independent mechanisms may both be necessary to ensure cell survival and proliferation. How Raf-1 inhibits ASK1 remains to be established. It is possible that Raf-1 promotes an inactive conformation of ASK1 through the N-terminal domain of ASK1, removal of which has been shown to increase both the kinase activity of ASK1 and its lethality (13). It is also possible that Raf-1 binding interferes with the interaction of ASK1 with its effectors such as MKK3 or regulators such as Daxx (12, 13). Alternatively, it is tempting to speculate that Raf-1 may function as an adaptor protein to recruit a survival factor to inhibit ASK1 function. For example, Raf-1 interacts with Akt (33, 34), a phosphoinositide 3-kinase regulated prosurvival kinase, and thus Raf-1 may recruit Akt to phosphorylate ASK1, allowing a general survival mechanism to intercept a death-signaling pathway (35).
Together, our data show that Raf-1 interacts with ASK1, and this interaction allows Raf-1 to inhibit a critical mediator of cell death independently of the MEK–ERK pathway, possibly through a kinase-independent mechanism. Investigations into the physiological roles of Raf-1 must now consider not only its MEK kinase activity but also Raf-1-mediated protein–protein interactions.
Acknowledgments
We are grateful to Drs. David Baltimore, Kangliang Guan, Hidenori Ichijo, and Richard Kahn for valuable reagents and Dr. Shane Masters for critical reading of the manuscript. We also thank members of the Fu lab for assistance and suggestions. This work was supported in part by National Institutes of Health Grants GM53165 and GM60033 and American Heart Association Grant 9950226N.
Abbreviations
- ASK1
apoptosis signal-regulating kinase 1
- β-gal
β-galactosidase
- DAPI
4′,6-diamidino-2-phenylindole
- eGFP
enhanced green fluorescent protein
- ERK
extracellular signal-regulated kinase
- HA
hemagglutinin
- MAPK
mitogen-activated protein kinase
- MEK
MAPK kinase/ERK kinase
- WT
wild type
References
- 1.Morrison D K, Cutler R E. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 2.Kolch W. Biochem J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland J L, Troppmair J, Packham G, Askew D S, Lloyd P, Gonzalez-Garcia M, Nunez G, Ihle J N, Rapp U R. Oncogene. 1994;9:2217–2226. [PubMed] [Google Scholar]
- 4.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 5.Erhardt P, Schremser E J, Cooper G M. Mol Cell Biol. 1999;19:5308–5315. doi: 10.1128/mcb.19.8.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Gall M, Chambard J C, Breittmayer J P, Grall D, Pouyssegur J, Van Obberghen-Schilling E. Mol Biol Cell. 2000;11:1103–1112. doi: 10.1091/mbc.11.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonni A, Brunet A, West A E, Datta S R, Takasu M A, Greenberg M E. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 8.Scheid M P, Schubert K M, Duronio V. J Biol Chem. 1999;274:31108–31113. doi: 10.1074/jbc.274.43.31108. [DOI] [PubMed] [Google Scholar]
- 9.Shimamura A, Ballif B A, Richards S A, Blenis J. Curr Biol. 2000;10:127–135. doi: 10.1016/s0960-9822(00)00310-9. [DOI] [PubMed] [Google Scholar]
- 10.Kurada P, White K. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- 11.Bergmann A, Agapite J, McCall K, Steller H. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- 12.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 13.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 14.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh Y, Cooper J A. J Biol Chem. 1998;273:17477–17482. doi: 10.1074/jbc.273.28.17477. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, Seimiya H, Naito M, Mashima T, Kizaki A, Dan S, Imaizumi M, Ichijo H, Miyazono K, Tsuruo T. Oncogene. 1999;18:173–180. doi: 10.1038/sj.onc.1202276. [DOI] [PubMed] [Google Scholar]
- 17.Hatai T, Matsuzawa A, Inoshita S, Mochida Y, Kuroda T, Sakamaki K, Kuida K, Yonehara S, Ichijo H, Takeda K. J Biol Chem. 2000;275:26576–26581. doi: 10.1074/jbc.M003412200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Chen J, Fu H. Proc Natl Acad Sci USA. 1999;96:8511–8515. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto T, Stewart S, Han M, Guan K L. EMBO J. 1998;17:1717–1727. doi: 10.1093/emboj/17.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Wang H, Liu D, Liddington R, Fu H. J Biol Chem. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]
- 21.Spector D L, Goldman R D, Leinwand L A. Cells: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 22.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favata M F, Horiuchi K Y, Manos E J, Daulerio A J, Stradley D A, Feeser W S, Van Dyk D E, Pitts W J, Earl R A, Hobbs F, et al. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 24.Fu H, Subramanian R R, Masters S C. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 25.Kuo W L, Abe M, Rhee J, Eves E M, McCarthy S A, Yan M, Templeton D J, McMahon M, Rosner M R. Mol Cell Biol. 1996;16:1458–1470. doi: 10.1128/mcb.16.4.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson G, Bumeister R, Henry D O, Cobb M H, White M A. J Biol Chem. 2000;275:37303–37306. doi: 10.1074/jbc.C000570200. [DOI] [PubMed] [Google Scholar]
- 27.Galaktionov K, Jessus C, Beach D. Genes Dev. 1995;9:1046–1058. doi: 10.1101/gad.9.9.1046. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Ghosh R N, Chellappan S P. Mol Cell Biol. 1998;18:7487–7498. doi: 10.1128/mcb.18.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H G, Rapp U R, Reed J C. Cell. 1996;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang X S, Diener K, Jannuzzi D, Trollinger D, Tan T H, Lichenstein H, Zukowski M, Yao Z. J Biol Chem. 1996;271:31607–31611. doi: 10.1074/jbc.271.49.31607. [DOI] [PubMed] [Google Scholar]
- 31.Karandikar M, Xu S, Cobb M H. J Biol Chem. 2000;275:40120–40127. doi: 10.1074/jbc.M005926200. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann S, Moelling K. Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 34.Rommel C, Clarke B A, Zimmermann S, Nunez L, Rossman R, Reid K, Moelling K, Yancopoulos G D, Glass D J. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- 35.Kim A H, Khursigara G, Sun X, Franke T F, Chao M V. Mol Cell Biol. 2001;21:893–901. doi: 10.1128/MCB.21.3.893-901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]