Abstract
A naturally occurring mutation was detected within the probe binding region targeting the envelope gene sequence of West Nile virus used in real-time polymerase chain reaction assays to test mosquito pools and other samples. A single C→T transition 6nt from the 5′ end of the 16mer in the envelope gene probe-binding region at genomic position 1,194 reduced assay sensitivity. The mutation first was detected in 2009 and persisted at a low prevalence into 2011. The mutation caused a 0.4% false negative error rate during 2011. These data emphasized the importance of confirmational testing and redundancy in surveillance systems relying on highly specific nucleic acid detection platforms.
Keywords: surveillance, mosquito pool, qRT-PCR, probe binding region, mutation
The high specificity of real-time reverse transcription polymerase chain reaction (qRT-PCR) imparts a risk for detection failure resulting from pathogen mutation. The reliance of contemporary arboviral surveillance systems on qRT-PCR assays for detection of viral RNA in mosquito pools highlights this risk because of the high mutability of RNA viruses, with inherently error-prone RNA-dependent RNA polymerases (mutation rates of ≈ 10−4 RNA copies) lacking a proofreading function (Domingo and Holland 1994). This factor, coupled with the exceedingly large population sizes generated by viruses such as West Nile virus (WNV) during acute infection (>109 infectious particles per milliliter of sera in some avian and >107 infectious particles in some infected mosquitoes), potentiates the emergence of stochastic, nonselected mutations. The high specificity of molecular detection platforms could result in loss of sensitivity if one or more mutations were to be introduced into either a primer or probe-binding region used by molecular assays; however, the sensitivity and high-throughput nature of these assays have outweighed these risks of detection failure because of the relatively low probability of a frequent mutation occurring within the relatively small primer and probe-binding regions of target genomes (representing <0.5% of the genome).
Since the arrival of WNV into California in 2003 (Reisen et al. 2004), the Center for Vectorborne Diseases at the University of California has participated in the California Mosquito-borne Encephalitis Virus Surveillance Program (Kramer 2009) by testing 233,197 mosquito pools for western equine encephalomyelitis, St. Louis encephalitis, and WNV RNA using a multiplex qRT-PCR assay. West Nile viral qRT-PCR primers and probe from the envelope gene (WN1) (Lanciotti and Kerst 2001) are used in the screening assay, with positives confirmed by a second qRT-PCR by using NS1 primers and probe (WN2) (Shi et al. 2001), by retesting with WN1 or by virus isolation. Repeated congruence among screening qRT-PCR and confirmatory assays, and the requirement for rapid throughput and low cost, eventually led to the current paradigm in which only pools with qRT-PCR WN1 cycle threshold (Ct) scores between 30 and 40 are confirmed using WN2, with the WN2 primers and probe pair on average 2–3 Ct less sensitive than WN1. Pools with Ct values below 30 are considered positive and not subjected to additional testing. During 2009 some mosquito pools with Ct scores >30 were found to have a reversal in sensitivity, with Ct for WN2 < WN1. The current paper examines this problem, determines its cause because of a single mutation, and discusses its impact on estimates of mosquito infection rates.
Materials and Methods
Surveillance test records during 2009–2011 were examined for pools where WN1 Ct scores >30 had confirmatory Ct values for WN2 <WN1. In 2010 virus was isolated from six representative pools exhibiting this switch in sensitivity by culture on Vero cells and then sent to the CDC in Ft. Collins, CO for sequencing across the primer and probe regions. A new probe was made to accommodate a single genetic polymorphism in the probe-binding region (see results section) and samples were retested with the new probe. During 2011 a subset of pools negative by multiplex screening assays were retested by a singleplex assay using the new probe to determine if decreased sensitivity because of the minor genetic polymorphism resulted in false negatives.
Results and Discussion
During the surveillance seasons of 2009–2011, 28 of 981; 18 of 1,012; and 14 of 1,385 WN1 RNA-positive Culex mosquito pools exhibited WN2 Ct values lower than WN1 upon confirmation. This anomaly was found in both Culex tarsalis Coquillett and Cx. quin-quefasciatus Say pools collected south of the Tehachapi Mountains and throughout the Central Valley from Kern to Shasta Counties (Fig. 1). WN1 Ct values for these 60 positive pools (mean ± SE, 35.5 ± 2.5) were significantly greater (paired t-test = 11.8, df = 59, P < 0.001) than WN2 (31.0 ± 3.2), and these Ct values were poorly correlated (R2 = 0.14), indicating a low agreement in estimates of WNV RNA copies based on standard curves. Sequencing was performed across the WN1 primer and probe regions from RNA extracted from six of the aberrant pools collected in 2010. A single C→T transition was identified in all six RNA samples 6nt from the 5′ end of the 16mer in the envelope gene probe-binding region at genomic position 1,194. A new probe was constructed to accommodate this mutation and was used for qRT-PCR retesting of 43 of the 60 aberrant samples from 2009 to 2011. The Ct score with the new WN1mut probe (mean = 28.7 ± 4.1) was significantly lower (paired t-test = 9.4, df = 42, P < 0.001) than results with the previous WN1 probe (35.3 ± 2.8), and slightly, but significantly (paired t = 4.9, df = 42, P < 0.001), less than values for the WN2 primer and probe set (29.6 ± 4.0). In addition, these WN1mut and WN2 values were highly correlated (R2 = 0.92), indicating accurate tracking of WNV RNA concentrations. During 2011, 18,907 pools were tested for WNV, of which 1,385 were positive. Of the 17,522 pools with WN1 > 40 Ct (considered to be WNV RNA negative), 527 (3%) were retested with the WN1mut probe and two were found to be positive, indicating an error rate of 0.38%. Upon retesting, these two pools again had WN1 >40 Ct, but WN1mut Ct values of 35.8 and 36.3. Extrapolating overall all negative pools and assuming a random distribution of the mutant virus throughout the state, we may have missed 66 positive pools because of the lost sensitivity associated with this single genetic polymorphism.
Fig. 1.
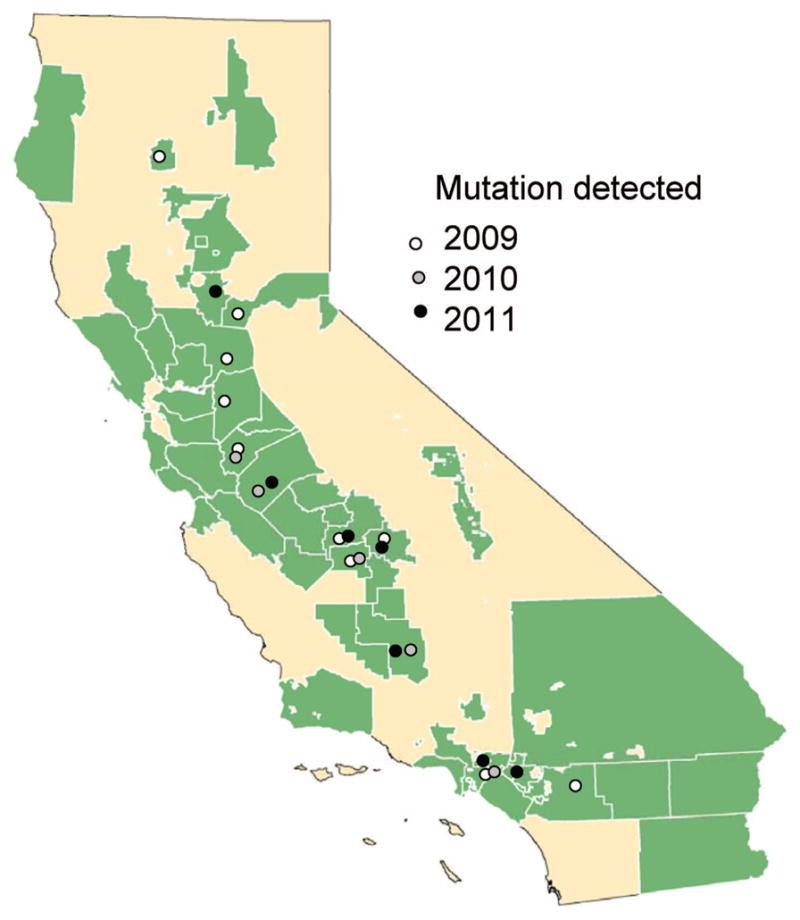
Map of California showing areas where mosquitoes are sampled by local mosquito control districts. Points show years and districts where the mutant WNV was detected in one or more pools based on the relationship between WN2 and WN1 Ct scores. (Online figure in color.)
The primer and probe targeting the envelope gene (Lanciotti and Kerst 2001) is by far the most sensitive and widely used molecular tool to detect North American lineage one WNV. Results presented herein indicated that a single mutation within the probe-binding region was sufficient to lower the sensitivity of this assay and led to at least two detection failures. Although this mutation was detected at a low rate, it was found in pools collected from the Central Valley and the Los Angeles areas during three consecutive years. In addition, 0.38% negative pools (Ct > 40) had detectable RNA when tested by the new WN1mut probe. Based on standard curves of viral titer culture versus Ct, values >35 would be estimated to have <1 plaque forming units/mL; however, it is clear that the single polymorphism within the probe would alter infection rate estimates.
Results presented herein emphasized the importance of using at least two distinctive genetic targets for screening and confirming qRT-PCR results. Arbo-viral surveillance systems that rely solely on molecular assays with similar specificities are vulnerable to similar detection failures. Future surveillance assay development should focus on the needed balance between high-throughput sensitivity and broad reactivity. Until such molecular tools are available, historic techniques, such as sentinel chicken surveillance and virus isolation, capable detection of genetic variants, will have an important role in arboviral surveillance programs.
Acknowledgments
This study was supported by the California Mosquito-borne Surveillance Program, districts comprising the Mosquito and Vector Control Association of California, and NIH Grants AI55607 and AI065359.
References Cited
- Domingo E, Holland JJ. Mutation rates and rapid evolution of RNA viruses. In: Morse SS, editor. Evolutionary biology of viruses. Raven Press; New York: 1994. pp. 161–184. [Google Scholar]
- Kramer VL. California State mosquito-borne virus surveillance and response plan. 2009 doi: 10.4269/ajtmh.2003.68.508. ( http://westnile.ca.gov/resources.php) [DOI] [PubMed]
- Lanciotti RS, Kerst AJ. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J Clin Microbiol. 2001;39:4506–4513. doi: 10.1128/JCM.39.12.4506-4513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen WK, Lothrop HD, Chiles RE, Madon MB, Cossen C, Woods L, Husted S, Kramer VL, Edman JD. West Nile virus in California. Emerg Infect Dis. 2004;10:1369–1378. doi: 10.3201/eid1008.040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Kauffman EB, Ren P, Felton A, Tai JH, DuPuis AP, Jones SA, Ngo KA, Nicholas DC, Maffei J, et al. High-throughput detection of West Nile virus RNA. J Clin Microbiol. 2001;39:1264–1271. doi: 10.1128/JCM.39.4.1264-1271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


