Abstract
TFII-I is an unusual transcription factor possessing both basal and signal-induced transcriptional functions. Here we report the characterization of a TFII-I-related factor (MusTRD1/BEN) that regulates transcriptional functions of TFII-I by controlling its nuclear residency. MusTRD1/BEN has five or six direct repeats, each containing helix–loop–helix motifs, and, thus, belongs to the TFII-I family of transcription factors. TFII-I and MusTRD1/BEN, when expressed individually, show predominant nuclear localization. However, when the two proteins are coexpressed ectopically, MusTRD1/BEN locates almost exclusively to the nucleus, whereas TFII-I is largely excluded from the nucleus, resulting in a loss of TFII-I-dependent transcriptional activation of the c-fos promoter. Mutation of a consensus nuclear localization signal in MusTRD1/BEN results in a reversal of nuclear residency of the two proteins and a concomitant gain of c-fos promoter activity. These data suggest a means of transcriptional repression by competition at the level of nuclear occupancy.
Keywords: transcription factor, nuclear translocation, transcriptional repression
In recent years it has become increasingly clear that the regulation of nuclear entry or exit of transcription factors provides an important control point for the expression of target genes, because these factors must be transported from the cytoplasm, where they are synthesized, to the nucleus, where they function. For example, sequestration of transcription factors in the cytoplasm in a latent but inducible form has been shown to be a significant point of control of gene expression in a variety of systems ranging from yeast to flies to mammals (1). Different mechanisms have been proposed to bring about such a regulation (1–5). One common theme among all of these mechanisms is that a reversible protein modification allows alteration of the nucleocytoplasmic shuttling pathway in response to an extracellular signal (6, 7). Here we show the regulation of the nuclear entry of a transcription factor, TFII-I, by a mechanism that appears to be independent of reversible protein modifications.
TFII-I is a ubiquitously expressed transcription factor that appears to have broad biological functions and may be involved in Williams–Beuren syndrome and X-linked agammaglobulinemia (8, 9). Based on its unique interactions at both the Inr element (10–12) and upstream regulatory sites (11, 13–15), TFII-I is postulated to be a transcriptional cofactor that integrates signals from the regulatory components to the basal machinery (11). In B cells, a significant fraction of cytoplasmic TFII-I is associated constitutively with Bruton's tyrosine kinase (16, 9). Upon Ig receptor crosslinking, TFII-I is released from Bruton's tyrosine kinase to enter the nucleus (9), suggesting that its transcriptional activity could be regulated through the alteration of its subcellular localization. Here we describe a repressor (MusTRD1/BEN) that appears to specifically regulate the nucleocytoplasmic shuttling of TFII-I. Nuclear exclusion of TFII-I results in a repression of its target reporter gene, and such repression can be alleviated when MusTRD1/BEN is retained in the cytoplasm through targeted mutation.
The existence of a TFII-I-related protein was reported by several groups as a gene located within a 1.4- to 1.6-Mb heterozygous deletion of chromosomal subband 7q11.23 that results in Williams–Beuren syndrome and was called WBSCR11 (17), GTF2IRD1 (18), or GTF3 (19). BEN (binding factor for early enhancer) was isolated as a transcription factor that binds to early enhancer of the Hoxc8 gene (20). The ORF of BEN encodes a protein of 1,072 aa and contains six helix–loop–helix (HLH) domains, a hydrophobic leucine zipper-like motif, and a serine stretch. The murine BEN gene is structurally similar to the human gene TFII-I and appears to be an alternative splice isoform of the human WBSCR11/GTF2RD1 (20). The gene product also has been reported as a retinoblastoma-associated nuclear protein (CREAM-1) with potential transcriptional activator functions (21). O'Mahoney and colleagues (22) reported it as a troponin I enhancer binding protein, muscle TFII-I repeat domain-containing protein 1 (MusTRD1). However, the predicted MusTRD1 protein was 458 aa long, whereas WBSCR11, GTF2IRD1, GTF3, and CREAM-1 were either 944 or 959 aa long, and their corresponding protein products all were ubiquitously expressed. The difference in size is due to alternative splicing (18). Subsequent sequence analysis of MusTRD1 cDNA (22) and ectopic expression of its protein product by us suggested a 120-kDa protein with 944 aa containing five repeats. Thus, MusTRD1/GTF3 is the human ortholog of the corresponding mouse BEN/GTF2IRD1. Henceforth we call this protein MusTRD1/BEN.
Materials and Methods
Cell Culture.
COS7 and HeLa cell lines were grown in DMEM (GIBCO/BRL) containing 10% FBS (GIBCO/BRL), 50 units/ml of penicillin, and 50 μg/ml of streptomycin (GIBCO/BRL).
Constructs.
To make pEBG-MusTRD1/BEN containing a C-terminal polyhis tag, a full-length MusTRD1 cDNA was PCR-amplified from pGEM-7Zf(+)-EH67 (kindly provided by Edna Hardeman, Children's Medical Research Institute, Wentworthville, Australia), with the use of oligonucleotides 1 and 2 shown in Table 1. The product was inserted into a pEBG vector as a ClaI–NotI fragment. For pEBB-GFP-MusTRD1/BEN, the full-length MusTRD1 cDNA was PCR-amplified with the use of oligonucleotides 1 and 3 and inserted as a ClaI–NotI fragment into a pEBB vector. The green fluorescent protein (GFP) was PCR-amplified from pEGFP-N1 (CLONTECH) with the use of the primers 4 and 5 (Table 1) and inserted in-frame into pEBB-MusTRD1/BEN as a SpeI–ClaI fragment at the N terminus. In MusTRD1/BEN ΔNLS3, the region between amino acids 883 to 889 was deleted by double-round PCR mutagenesis with the use of primers 6–9 (Table 1). In MusTRD1/BEN Δss, the region between amino acids 897 and 908 was deleted with the use of primers 6 and 9–11 (Table 1). Both PCR products were inserted into pEBG-MusTRD1/BEN or pEBB-GFP-MusTRD1/BEN as XbaI–BsiWI fragments.
Table 1.
Oligonucleotides used to generate plasmids
| Expression vector | Restriction site | Nucleotide sequence |
|---|---|---|
| pEBG-MusTRD1 | ClaI | (1) 5′-CCATCGATGGAATGGCCTTGCTGGGTAAG-3′ |
| NotI | (2) 5′-ATAGTTTAGCGGCCGCCTAGTGGTGGTGGTGGTGGTGGTAATTAAGAGGTCCCGGGAG-3′ | |
| pEBB-MusTRD1 | ClaI | (1) 5′-CCATCGATGGAATGGCCTTGCTGGGTAAG-3′ |
| NotI | (3) 5′-ATAAGAATGCGGCCGCTGTAATTAAGAGGTCCCGGGAG-3′ | |
| pEBB-GFP-MusTRD1 | SpeI | (4) 5′-GGACTAGTCCATGGTGAGCAAGGGCGAG-3′ |
| ClaI | (5) 5′-CCATCGATCTTGTACAGCTCGTCCATGC-3′ | |
| pEBG-MusTRD1-ΔNLS3 and | BsiWI | (6) 5′-CCAACACGTACGACATCCAC-3′ |
| pEBB-GFP-MusTRD1-ΔNLS3 | ΔNLS3 UP | (7) 5′-TCCTTCCGAGACAATGCTGCTGTC-3′ |
| ΔNLS3 LP | (8) 5′-GACAGCAGCATTGTCTCGGAAGGA-3′ | |
| XmaI | (9) 5′-AGAGGTCCCGGGAGCTGC-3′ | |
| pEBG-MusTRD1-Δss and | BsiWI | (6) 5′-CCAACACGTACGACATCCAC-3′ |
| pEBB-GFP-MusTRD1-Δss | Δss UP | (10) 5′-CTGAATCTGGGTTGACGGAATTTCC-3′ |
| Δss LP | (11) 5′-GGAAATTCCGTCAACCCAGATTCAGTG-3′ | |
| XmaI | (9) 5′-AGAGGTCCCGGGAGCTGC-3′ |
Transient Transfection and Luciferase Assays.
Transfections were performed with Lipofectamine (GIBCO/BRL). Ten micrograms of pEBG-II-I and 12 μg of any of the pEBG-MusTRD1/BEN constructs were used per 100-mm-diameter plate. For the c-fos luciferase assays, pSVOAΔ5′ containing a murine c-fos promoter (15) (600 ng) and pRL-TK (35 ng) were transfected either alone or with pEBG-MusTRD1/BEN or ΔNLS3 (1,000 ng) and pEBG-TFII-I (800–3,200 ng) (12). Before harvesting, cells were serum-starved for 12 h and stimulated with 25 ng/ml of recombinant human epidermal growth factor (Sigma) for 4 h. To test for GAL4 transcriptional activity, a GAL4 reporter, pFR-Luc (Stratagene) (200 ng), and pRL-TK (35 ng) were transfected alone or with the GAL4 expression vector pMA242 (200 ng) (24) and/or pEBG-MusTRD1/BEN (500 ng). To test for USF1 activity, a vector containing four tandem E-boxes from the secretin promoter pT81-[Ebox]4-Luc (25) (400 ng) and pRL-TK (35 ng) were transfected either alone or with the USF1 expression vector pCX-USF1 (26) (400–800 ng) and/or pEBG-MusTRD1/BEN (1,000 ng). To assay for Sp1 activity, pGL3-promoter (Promega) (40 ng) and pRL-TK (35 ng) were transfected alone or with the Sp1 expression vector pPac-Sp1 (kind gift from Robert Tjian, Univ. of California, Berkeley) (400–800 ng) and pEBG-MusTRD1/BEN (1,000 ng). Total transfected DNA was kept constant by empty vectors pEBB or pEBG. Luciferase activity was assessed with a Dual Luciferase Kit (Promega). Experiments were done at least three times, with triplicate sets included each time.
Protein Analysis.
Nuclear and cytoplasmic extracts were prepared as described (27). Identical amounts of protein from each sample were subjected to SDS/PAGE and immunoblotting. The mouse monoclonal anti-glutathione S-transferase (GST-2; Sigma) antibody was used at 1:4,000 dilution. Both rabbit anti-TFII-I and sheep anti-MusTRD1/BEN (a kind gift from Edna Hardeman) antibodies were used at 1:2,500 dilution. Detection was done by enhanced chemiluminescence (NEN) with standard methods.
Immunostaining.
COS7 cells were transfected with 200 ng of tagless TFII-I (V. Cheriyath and A.L.R., unpublished observations) in the absence or in the presence of 200 ng of GFP-tagged MusTRD1/BEN constructs. Forty hours after transfection, cells were fixed in 4% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with 10% FBS in PBS for 1 h. Nontransfected HeLa cells were fixed in 4% paraformaldehyde for 60 min. Cells were incubated with primary antibody [rabbit anti-TFII-I, 1:6,000; affinity-purified rabbit anti-BEN, 1:200; mouse monoclonal anti-GAL4-TA (C10; Santa Cruz Biotechnology) and rabbit anti-USF1 (a kind gift from Robert Roeder, Rockefeller Univ., New York), 1:1,000; rabbit anti-Sp1 (sc-59; Santa Cruz Biotechnology), 1:10,000] for 2 h, washed three times with PBS-T (PBS/0.1% Triton X-100) followed by Alexa 594 goat anti-rabbit or goat anti-mouse IgG (H + L) (Molecular Probes) (1:20,000), washed three times with PBS-T, and mounted in 4′,6-diamidino-2-phenylindole-mounting buffer (50% glycerol/5 μg 4′,6-diamidino-2-phenylindole/ml in PBS). Preparations were visualized by fluorescence and confocal microscopy as described (9).
Results
Structural Features and Localization of MusTRD1/BEN.
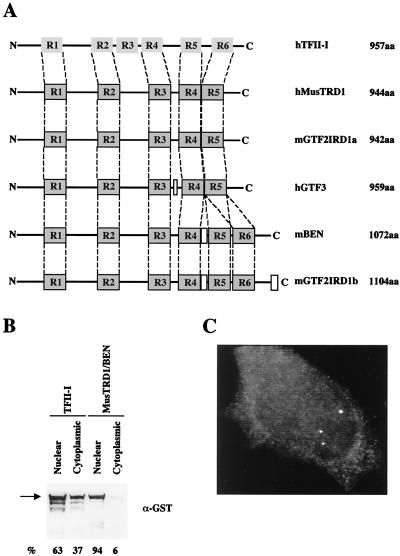
Fig. 1A shows the relationship of the six repeats of TFII-I to the repeats of MusTRD1/BEN and its family members. Note that each repeat in TFII-I (and, by inference, in MusTRD1/BEN) contains a putative HLH domain (11). The prosite search (28) revealed an additional Myc-type HLH motif between amino acids 458 and 466 and a stretch of 12 serines between positions 897 and 908 in MusTRD1/BEN. Finally, a psort search (29) predicted three conserved nuclear localization signals (NLS) (30) at positions 407–413 (NLS1), 715–721 (NLS2), and 883–889 (NLS3). The serine stretch adjacent to NLS3 is perhaps the most interesting feature in MusTRD1/BEN. Similar serine stretches are also present in transcriptional activators such as ICP4, IE180, IE62, Nopp140, PC4, Sox-4, and Sp4 (31–37); nuclear shuttling proteins such as Nopp 140 (38); and transcriptional repressors belonging to the polycomb group of proteins, such as Pc1 and cramped (39, 40). Ectopic expression and subsequent Western blot analysis of TFII-I and MusTRD1/BEN revealed that whereas TFII-I is distributed between cytoplasm (37%) and nucleus (63%), MusTRD1/BEN is almost exclusively nuclear (94%) (Fig. 1B). Accordingly, fluorescent microscopy of endogenous MusTRD1/BEN in HeLa cells shows specific staining only in the nucleus (Fig. 1C).
Figure 1.
Structural features of MusTRD1/BEN. (A) The relative localization of the six repeat domains in human TFII-I (hTFII-I, 957 aa) and the five or six repeat domains in MusTRD1/BEN (hMusTRD1, 944aa) and its isoforms (mGTF2IRD1a, 942 aa; hGTF3, 959 aa; mBEN, 1,072 aa; mGTF2IRD1b, 1,104 aa) is indicated. Although there is only 30–32% identity with TFII-I throughout the entire protein, the total homology among the repeats is 39%. (B) Ectopically expressed GST-TFII-I or GST-MusTRD1/BEN in COS7 cells was visualized by anti-GST antibody. The percentage subcellular distribution of each protein is indicated at the bottom. (C) Endogenous nuclear expression of MusTRD1/BEN in HeLa cells was visualized with the use of an affinity-purified rabbit polyclonal anti-BEN antibody.
Transcriptional Function of MUSTRD1/BEN: Changes in Subcellular Localization of TFII-I by MUSTRD1/BEN.
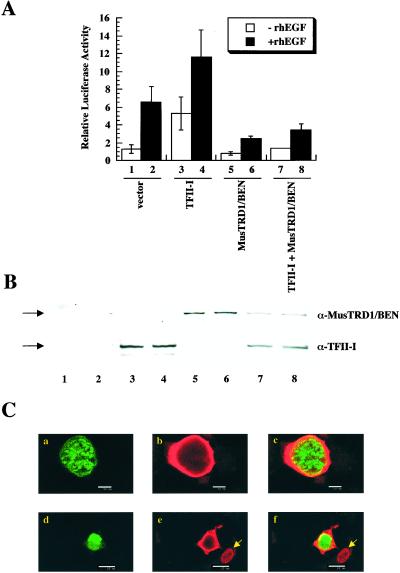
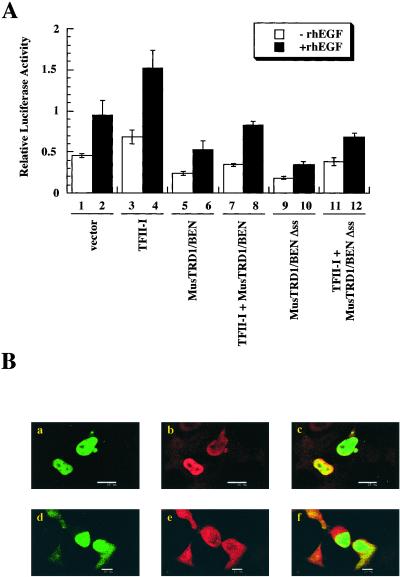
We next tested the transcriptional function(s) of MusTRD1/BEN on the c-fos promoter, the activity of which is up-regulated by TFII-I both in the absence and in the presence of epidermal growth factor (15). In contrast to TFII-I, which stimulates the c-fos promoter both in the absence (Fig. 2A, compare lanes 1 and 3) and in the presence (Fig. 2A, compare lanes 2 and 4) of epidermal growth factor, MusTRD1/BEN represses the c-fos promoter both in the absence (Fig. 2A, compare lanes 1 and 5) and in the presence (Fig. 2A, compare lanes 2 and 6) of epidermal growth factor. Most notably, MusTRD1/BEN represses even in the presence of TFII-I (Fig. 2A, compare lanes 3 and 4 with 7 and 8). The ectopic expression of these proteins was tested by Western blot to confirm that the differences in activity were not due to significant differences in their expression patterns (Fig. 2B). Although there appears to be a slight drop in the expression of both proteins when coexpressed, this drop is not seen consistently (see Fig. 3B).
Figure 2.
MusTRD1/BEN represses TFII-I transcriptional activity and promotes TFII-I nuclear exclusion. (A) c-fos-luciferase activity of TFII-I and/or MusTRD1/BEN in the presence (filled columns) or in the absence (empty columns) of 25 ng/ml of recombinant human epidermal growth factor. Lanes 1 and 2, empty vector; 3 and 4, TFII-I; 5 and 6, MusTRD1/BEN; 7 and 8, TFII-I + MusTRD1/BEN. (B) Cell extracts from A were Western blotted with an anti-MusTRD1/BEN (α-MusTRD1/BEN) antibody and then stripped and reprobed with anti-TFII-I antibody (α-TFII-I). (C) Confocal microscopic analysis of cotransfected GFP-MusTRD1/BEN and GST-TFII-I COS7 cells. Cells were stained with anti-TFII-I antibody and a secondary antibody coupled to Alexa 594. Duplicate experiments (either a–c or d–f) are shown. a and d show GFP-MusTRD1/BEN (green); b and e show TFII-I (red); and c and f are the superimposition of a and b (c) or d and e (f), and colocalization is in yellow (a–c, scale bar = 10 μm; d–f, scale bar = 25 μm). Nuclear localization of an ectopic TFII-I (but not ectopic MusTRD1/BEN)-expressing cell is shown by an arrow.
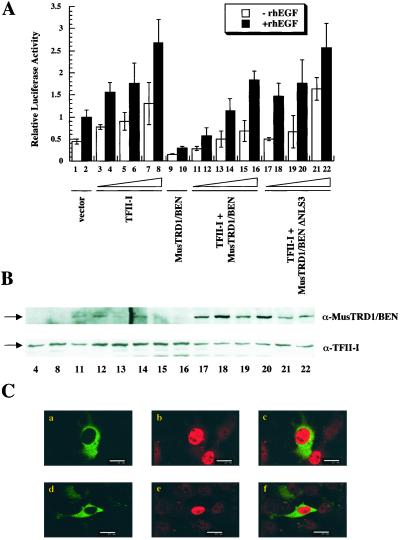
Figure 3.
MusTRD1/BEN ΔNLS3 remains at the cytoplasm and induces neither nuclear exclusion nor transcriptional repression of TFII-I. (A) c-fos-luciferase activity of wild type or ΔNLS3 MusTRD1/BEN in the presence or in the absence of TFII-I and either in the presence (filled columns) or in the absence (empty columns) of 25 ng/ml of recombinant human epidermal growth factor. Lanes 1 and 2, empty vector; 3–8, increasing concentrations of TFII-I; 9 and 10, MusTRD1/BEN, 1,000 ng; 11–16, MusTRD1/BEN, 1,000 ng + increasing concentrations of TFII-I; 17–22, MusTRD1/BEN ΔNLS3, 1,000 ng + increasing concentrations of TFII-I: (3, 4, 11, 12, 17, and 18, 800 ng; 5, 6, 13, 14, 19, and 20, 1,600 ng; 7, 8, 15, 16, 21, and 22, 3,200 ng). (B) Cell extracts from indicated lanes in A were Western blotted with an anti-MusTRD1/BEN (α-MusTRD1/BEN) antibody and then stripped and reprobed with anti-TFII-I antibody (α-TFII-I). (C) Confocal images of coexpressed GFP-MusTRD1/BEN ΔNLS3 and GST-TFII-I. Cells were stained as in Fig. 2B. Duplicate images (either a–c or d–f) are shown. a and d show the distribution of GFP-MusTRD1/BEN ΔNLS3 (green); b and e show TFII-I (red); c and f show a superimposition of images a and b (c) or d and e (f). (Scale bar = 25 μm.)
The lack of any significant physical association between the two proteins by GST pull-down and its lack of specific DNA binding to TFII-I recognition elements (data not shown) led us to postulate that MusTRD1/BEN indirectly causes changes in subcellular distribution of TFII-I that might contribute to its repressive function. Immunohistochemical analysis of GFP-MusTRD1/BEN coexpressed with GST-TFII-I in COS7 cells and monitored by confocal microscopy revealed a dramatic redistribution of TFII-I (Fig. 2C, shown in red, b and e) in the presence of MusTRD1/BEN (Fig. 2C, green, a and d). Whereas MusTRD1/BEN exhibited a predominant nuclear staining, TFII-I was largely excluded from the nucleus in the presence of MusTRD1/BEN. A very small proportion of the two proteins showed colocalization (Fig. 2C, seen as yellow, c and f). TFII-I showed a predominant nuclear staining in the absence of MusTRD1/BEN (e.g., see Fig. 2C e and f, arrowhead). Upon careful analysis from several experiments, we found two major patterns of TFII-I redistribution in the presence of MusTRD1/BEN: a uniform nuclear pattern and a partial nuclear plus perinuclear ring-like pattern (Table 2). Interestingly, although there is a 50% reduction in nuclear TFII-I in the presence of ectopic MusTRD1/BEN, the ring-like distribution of TFII-I is also seen in some cells, even in the absence of MusTRD1/BEN, albeit with much less frequency. Whether this distribution pattern is due to the endogenous expression of MusTRD1/BEN in COS cells remains to be determined.
Table 2.
Subcellular distribution of TFII-I in cells expressing nuclear MusTRD1/BEN
| Condition | Uniform nuclear
|
Nuclear + perinuclear ring
|
||
|---|---|---|---|---|
| Average, % | SD, % | Average, % | SD, % | |
| TFII-I (tagless) | ||||
| + pEBB vector | 62.8 | 5.9 | 37.1 | 5.9 |
| TFII-I (tagless) | ||||
| + GFP-MusTRD1/BEN | 30.2 | 2.9 | 59.3 | 5.5 |
| TFII-I (tagless) | ||||
| + GFP-MusTRD1/BEN Δss | 52.8 | 6 | 38.9 | 6.4 |
To test the hypothesis that the presence of MusTRD1/BEN in the nucleus is required for the impediment of nuclear residency of TFII-I, we mutated the NLS3 in MusTRD1/BEN. The transcriptional function was tested by coexpression of TFII-I with either this mutant or wild-type MusTRD1/BEN (Fig. 3A). Whereas TFII-I activated the c-fos promoter in a dose-dependent manner (Fig. 3A, lanes 3–8), the wild-type MusTRD1/BEN repressed even at the highest amount of TFII-I (Fig. 3A, lanes 11–16). Consistent with our hypothesis, MusTRD1/BEN ΔNLS3 did not significantly repress the c-fos activity at any concentrations of TFII-I (Fig. 3A, lanes 17–22) and is primarily localized in the cytoplasm (Fig. 3C, a and d). More importantly, cotransfected TFII-I (Fig. 2D, b and e) is primarily localized in the nucleus, and the levels are similar to the control levels (i.e., in the absence of MusTRD1/BEN; Fig. 3B).
Repression by MUSTRD1/BEN Is Specific for TFII-I.
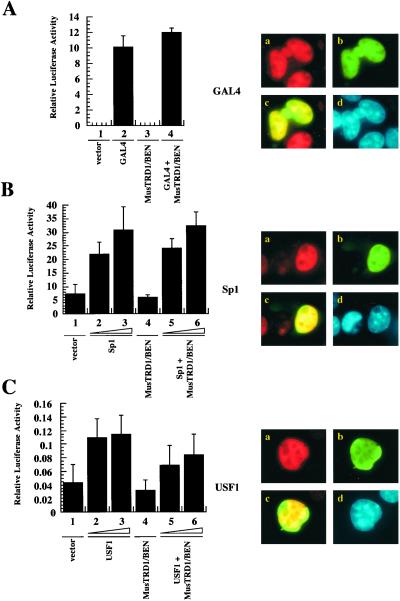
Next we tested the specificity of repression by MusTRD1/BEN by fluorescence microscopy. In transient transfection assays, MusTRD1/BEN failed to repress a GAL4-dependent promoter (compare lane 2 with lane 4, Fig. 4A Left) (12). Moreover, MusTRD1/BEN (green) does not inhibit nuclear localization of GAL4 (red), and, thus, the two proteins completely colocalize (yellow) in the nucleus (blue) (Fig. 4A Right). Likewise, neither the transcriptional activation (Fig. 4B Left) nor the subcellular distribution of Sp1 (Fig. 4B Right) was affected by MusTRD1/BEN. Given that both TFII-I and MusTRD1/BEN exhibit multiple HLH motifs, we tested another HLH activator, USF1 (26), to ensure that the repression is specific for TFII-I and not for HLH proteins in general. This reporter was slightly repressed (20–30%) (Fig. 4C Left), although in localization studies USF1 and MusTRD1/BEN showed complete nuclear colocalization (Fig. 4C Right). Because TFII-I can be a coactivator of USF1 and functions via an E-box in vivo (10, 11), the repression may be due partly to the nuclear exclusion of endogenous TFII-I by MusTRD1/BEN.
Figure 4.
MusTRD1/BEN repressive effects are specific for TFII-I. (A) Transcriptional activity of a GAL4-responsive reporter in the absence (lanes 1 and 3) or in the presence (lanes 2 and 4) of 200 ng of GAL-4 and 500 ng of MusTRD1/BEN (lanes 3 and 4). (Right) Fluorescent microscope images of cotransfected GAL-4 (red, a) and GFP-MusTRD1/BEN (green, b). Nuclear overlap of GAL4 and MusTRD1/BEN is seen in yellow (c), and the nucleus is seen by 4′,6-diamidino-2-phenylindole staining in blue (d). (B) Transcriptional activity of the pGL3 promoter either in the absence (lanes 1 and 4) or in the presence of 400 ng (lanes 2 and 5) or 800 ng (lanes 3 and 6) of Sp1 and Sp1 + 1,000 ng of MusTRD1/BEN (lanes 5 and 6). (Right) Images of cotransfected Sp1 (red, a) and GFP-MusTRD1/BEN (green, b). Nuclear overlap of Sp1 and MusTRD1/BEN is seen in yellow (c), and the nucleus is in blue (d). (C) As in B, but with the pT81-[E-box]4-luciferase reporter construct and USF1 as the transcriptional activator.
MUSTRD1/BEN: A Bimodal Repressor.
To test whether the serine stretch in MusTRD1/BEN provides its preferential nuclear residency over TFII-I, we deleted the serine stretch (Δss), coexpressed it with TFII-I, and analyzed both transcriptional function, and colocalization potentials of the mutant protein. However, contrary to our expectations, we found that the Δss mutant still behaved as a repressor of TFII-I-dependent transcription (Fig. 5A). Although the Δss still behaved as a repressor, it could not prevent the nuclear localization of TFII-I (Fig. 5B, compare a and d with b and e), and the two proteins colocalized in the nucleus (Fig. 5B c and f). Taken together, these data suggest a bimodal repressive action of MusTRD1/BEN in which it prevents the proper nuclear residency of TFII-I and inhibits the transcriptional activation of nuclear TFII-I (Fig. 5).
Figure 5.
Inhibition of TFII-I basal and epidermal growth factor-stimulated c-fos promoter activation by MusTRD1/BEN wild type and MusTRD1/BEN Δss. (A) c-fos-luciferase activity with either wild type or Δss MusTRD1/BEN (1,000 ng) in the absence (empty columns) or in the presence (filled columns) of 25 ng/ml recombinant human epidermal growth factor and TFII-I (800 ng). Lanes 1 and 2, empty vector; 3 and 4, TFII-I; 5 and 6, MusTRD1/BEN; 7 and 8, TFII-I + MusTRD1/BEN; 9 and 10, MusTRD1/BEN Δss; 11 and 12, TFII-I + MusTRD1/BEN Δss. (B) Cells were stained as in Fig. 2B and were visualized by confocal microscopy. Duplicate images are shown. (a and d) GFP-MusTRD1/BEN Δss (green). (b and e) TFII-I localization (red). (c and f) Superimposition of either a and b (c) or d and e (f). Colocalization is seen in yellow. (These images were taken with the same ×60 objective, but the scale bar = 25 μm in a–c and 10 μm in d–f.)
To get a quantitative idea of the nuclear residency of TFII-I in the presence of MusTRD1/BEN and its mutants, we analyzed immunostained cotransfected COS7 cells by fluorescence microscopy (Table 2). Whereas in the absence of MusTRD1/BEN, uniform nuclear TFII-I distribution was observed in 62.8% of the cells, in the presence of MusTRD1/BEN it was observed in 30.2% (in each case a total of 100 cells were counted). Moreover, there was an increase from 37.1% to 59.3% in the number of cells that showed a distribution pattern of TFII-I with both nuclear and cytoplasmic staining. In contrast, 52.8% of TFII-I plus MusTRD1/BEN Δss cotransfected cells showed uniform nuclear staining that was comparable to 62.8% of control cells transfected with only TFII-I (see Table 2). Moreover, TFII-I and MusTRD1/BEN Δss cotransfected cells no longer showed increased nuclear plus perinuclear distribution of TFII-I (38.9%) when compared with the TFII-I only transfected cells (37.1%).
Discussion
The precise regulation of the differentiation and developmental program requires not only that genes be turned on, but also that they be turned off. In fact, the founding principles of transcriptional regulation by Jacob and Monod came from studies of repression in Escherichia coli (41). Given the complexity of the eukaryotic organism and its precise coordination of gene expression programs, it is not surprising that there are several general pathways of gene repression. The repressor can exert its effects via (i) competition with an activator for DNA binding; (ii) competition with an activator for a cofactor required for activation; (iii) direct interaction with an activator, thereby masking its activation domain; or (iv) remodeling of the chromatin such that the accessibility for the activator is blocked (reviewed in ref. 42). Apart from these competitive mechanisms occurring in the nucleus, a repressor can sequester an inducible activator in the cytoplasm, as exemplified by IκB (23). Here we report a potentially novel mechanism of gene repression in which the repressor selectively occupies the nucleus at the expense of the activator, resulting in a nuclear exclusion of the activator.
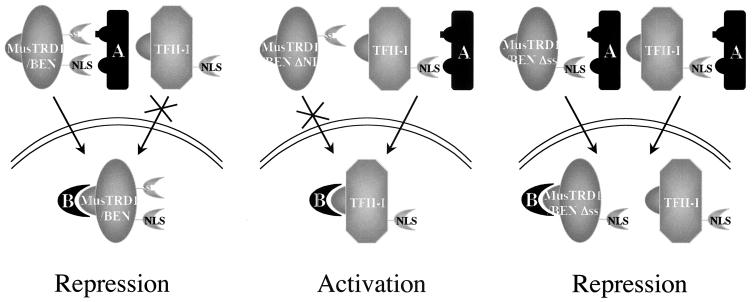
Although the precise mechanism is not yet known, our data suggest a competition with a nuclear shuttling component (Fig. 6). We propose a dual mechanism for MusTRD1/BEN-mediated inhibition. First, MusTRD1/BEN outcompetes TFII-I for a “dedicated” nucleocytoplasmic shuttling component A, resulting in nuclear exclusion of TFII-I. However, given that the endogenous TFII-I is generally higher than MusTRD1/BEN, this step may not quantitatively exclude nuclear TFII-I. Thus, in a second step, nuclear MusTRD1/BEN competes with nuclear TFII-I for a necessary and specific transcriptional cofactor, B. The dual repression mechanism might ensure a complete inhibition of TFII-I-dependent gene activation. However, we cannot formally exclude the possibility that MusTRD1/BEN accelerates the nuclear export of TFII-I. Moreover, we emphasize that although, under our assay conditions, ectopic MusTRD1/BEN behaves as a repressor for TFII-I function, it might respond differently in the context of TFII-I unresponsive genes and could even behave as an activator in other promoters and/or cell types. Thus, it is likely that MusTRD1/BEN could specifically bind to a DNA sequence that is not recognized by TFII-I. TFII-I and MusTRD1/BEN also exhibit quite distinct phosphorylation patterns, raising the possibility that they are activated by different signaling pathways (data not shown). Given that MusTRD1/BEN represses TFII-I transcriptional activity, it is not surprising that MusTRD1/BEN naturally exists in low amounts inside the cell (Fig. 1C) (19). In preliminary experiments, it also appears that BEN behaves as a repressor for Hoxc8 gene expression (D.B. and F.H.R., unpublished observations). Thus, we believe that there may be circumstances where MusTRD1/BEN would need to be up-regulated (even if only transiently) through a different signaling pathway or in a cell-specific fashion. Although we have not yet found any physiological signal that would cause an up-regulation of MusTRD1/BEN or a system where the constitutive levels of MusTRD1/BEN are naturally high, our ectopic expression system suggests that elevating the concentration of the repressor might be a potent means of regulating TFII-I transcriptional activity. Hence, the levels of MusTRD1/BEN expression could set the threshold of TFII-I activity. These questions have yet to be addressed.
Figure 6.
Model for MusTRD1/BEN-mediated repression. The interactions of factor A with both MusTRD1/BEN and TFII-I are through NLS. Only MusTRD1/BEN has a weak secondary interaction through the serine stretch (ss), giving it an advantage over TFII-I (Left). In ΔNLS3 MusTRD1/BEN, component A can only bind to and facilitate TFII-I nuclear entry, allowing activation of target genes (Middle). When the serine stretch in MusTRD1/BEN is deleted, factor A can bind equally well to MusTRD1/BEN and TFII-I, resulting in nuclear colocalization of both proteins. Nuclear MusTRD1/BEN now competes with nuclear TFII-I for a transcriptional cofactor B, ensuring repression of TFII-I target genes (Right).
Acknowledgments
We thank Peter Brodeur, Edouard Vannier, Michelle Benson, and Claire Moore for critical reading of the manuscript and Dean Dawson for use of the fluorescent microscope. We are indebted to Robert Roeder, Robert Tjian, Andrew Leiter, and especially Edna Hardeman for generously sharing antibodies and plasmids. We are grateful to Venu Cheriyath for his input. This work is supported in part by Grant AI45150 from the National Institutes of Health to A.L.R.
Abbreviations
- MusTRD1
muscle TFII-I repeat domain-containing protein 1
- GFP
green fluorescent protein
- HLH
helix–loop–helix
- NLS
nuclear localization signal
- GST
glutathione S-transferase
References
- 1.Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 2.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 3.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 4.Hood J K, Silver P A. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 5.Kaffman A, O'Shea E K. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 6.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 7.Ducret C, Maira S M, Deirich A, Wasylyk B. Mol Cell Biol. 1999;19:7076–7087. doi: 10.1128/mcb.19.10.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Jurado L A, Wang Y K, Peoples R, Coloma A, Cruces J, Francke U. Hum Mol Genet. 1998;7:325–334. doi: 10.1093/hmg/7.3.325. [DOI] [PubMed] [Google Scholar]
- 9.Novina C D, Kumar S, Bajpai U, Cheriyath V, Zhang K, Pillai S, Wortis H H, Roy A L. Mol Cell Biol. 1999;19:5014–5024. doi: 10.1128/mcb.19.7.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy A L, Meisterernst M, Pognonec P, Roeder R G. Nature (London) 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 11.Roy A L, Du H, Gregor P D, Novina C D, Martinez E, Roeder R G. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheriyath V, Novina C D, Roy A L. Mol Cell Biol. 1998;18:4444–4454. doi: 10.1128/mcb.18.8.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grueneberg D A, Henry R W, Brauer A, Novina C D, Cheriyath V, Roy A L, Gilman M. Genes Dev. 1997;11:2482–2493. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D W, Cheriyath V, Roy A L, Cochran B H. Mol Cell Biol. 1998;18:3310–3320. doi: 10.1128/mcb.18.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheriyath V, Roy A L. J Biol Chem. 2000;275:26300–26308. doi: 10.1074/jbc.M002980200. [DOI] [PubMed] [Google Scholar]
- 16.Yang W, Desiderio S. Proc Natl Acad Sci USA. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osborne L R, Campbell T, Daradich A, Scherer S W, Tsui L C. Genomics. 1999;57:279–284. doi: 10.1006/geno.1999.5784. [DOI] [PubMed] [Google Scholar]
- 18.Franke Y, Peoples R J, Francke U. Cytogenet Cell Genet. 1999;86:296–304. doi: 10.1159/000015322. [DOI] [PubMed] [Google Scholar]
- 19.Tassabehji M, Carette M, Wilmot C, Donnai D, Read A P, Metcalfe K. Eur J Hum Genet. 1999;7:737–747. doi: 10.1038/sj.ejhg.5200396. [DOI] [PubMed] [Google Scholar]
- 20.Bayarsaihan D, Ruddle F H. Proc Natl Acad Sci USA. 2000;97:7342–7347. doi: 10.1073/pnas.97.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan X, Zhao X, Qian M, Guo N, Gong X, Zhu X. Biochem J. 2000;345:749–757. [PMC free article] [PubMed] [Google Scholar]
- 22.O'Mahoney J V, Guven K L, Lin J, Joya J E, Robinson C S, Wade R P, Hardeman E C. Mol Cell Biol. 1998;18:6641–6652. doi: 10.1128/mcb.18.11.6641. , and erratum (2000) 20, 5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin A S. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Ptashne M. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 25.Mutoh H, Fung B P, Naya F J, Tsai M, Nishitani J, Leiter A B. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du H, Roy A L, Roeder R G. EMBO J. 1993;12:501–511. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogiwara A, Uchiyama I, Takagi T, Kanehisa M. Protein Sci. 1996;5:1991–1999. doi: 10.1002/pro.5560051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 31.Cheung A K. Nucleic Acids Res. 1989;17:4637–4646. doi: 10.1093/nar/17.12.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagen G, Muller S, Beato M, Suske G. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perera L P, Mosca J D, Ruyechan W T, Hayward G S, Straus S E, Hay J. J Virol. 1993;67:4474–4483. doi: 10.1128/jvi.67.8.4474-4483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wetering V D M, Oosterwegel M, Norren V K, Clevers H. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 36.Miau L H, Chang C J, Tsai W H, Lee S C. Mol Cell Biol. 1997;17:230–239. doi: 10.1128/mcb.17.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates P A, DeLuca N A. J Virol. 1998;72:7115–7124. doi: 10.1128/jvi.72.9.7115-7124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier U T, Blobel G. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto Y, Girard F, Bello B, Affolter M, Gehring W J. Development (Cambridge, UK) 1997;124:3385–3394. doi: 10.1242/dev.124.17.3385. [DOI] [PubMed] [Google Scholar]
- 40.Stroboulis J, Damjanovski S, Vermaak D, Meric F, Wolffe A P. Mol Cell Biol. 1999;19:3958–3968. doi: 10.1128/mcb.19.6.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacob F, Monod J. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 42.Maldonado E, Hampsey M, Reinberg D. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]