Abstract
Abiotic stresses such as drought cause a reduction of plant growth and loss of crop yield. Stomatal aperture controls CO2 uptake and water loss to the atmosphere, thus playing important roles in both the yield gain and drought tolerance of crops. Here, a rice homologue of SRO (similar to RCD one), termed OsSRO1c, was identified as a direct target gene of SNAC1 (stress-responsive NAC 1) involved in the regulation of stomatal aperture and oxidative response. SNAC1 could bind to the promoter of OsSRO1c and activate the expression of OsSRO1c. OsSRO1c was induced in guard cells by drought stress. The loss-of-function mutant of OsSRO1c showed increased stomatal aperture and sensitivity to drought, and faster water loss compared with the wild-type plant, whereas OsSRO1c overexpression led to decreased stomatal aperture and reduced water loss. Interestingly, OsSRO1c-overexpressing rice showed increased sensitivity to oxidative stress. Expression of DST, a reported zinc finger gene negatively regulating H2O2-induced stomatal closure, and the activity of H2O2-scavenging related enzymes were significantly suppressed, and H2O2 in guard cells was accumulated in the overexpression lines. OsSRO1c interacted with various stress-related regulatory and functional proteins, and some of the OsSRO1c-interacting proteins are predicted to be involved in the control of stomatal aperture and oxidative stress tolerance. The results suggest that OsSRO1c has dual roles in drought and oxidative stress tolerance of rice by promoting stomatal closure and H2O2 accumulation through a novel pathway involving regulators SNAC1 and DST.
Key words: Drought, Oryza sativa, oxidation, ROS, stomata, SRO.
Introduction
Abiotic stresses such as drought, high salinity, and heat cause extensive losses to agricultural production worldwide. These losses frequently result from the simultaneous occurrence of different abiotic stresses as well as from increased frequency of extreme weather conditions (Mittler and Blumwald, 2010). Plants have evolved a range of physiological, biochemical, and molecular responses to confer tolerance to environmental stresses. Guard cells are located in the leaf epidermis in pairs to form stomatal pores, which allow influx of CO2 as a raw material for photosynthesis and water loss via transpiration to the atmosphere (Hetherington and Woodward, 2003). In response to drought stress, plants synthesize abscisic acid (ABA), which triggers closing of stomatal pores, thus reducing water loss (Schroeder et al., 2001b ). In the process of ABA-mediated stomatal closure, H2O2 plays a vital role as a secondary messenger by elevating calcium levels in guard cells through the activation of plasma membrane calcium channels (Pei et al., 2000; Wang and Song, 2008). The ABA-activated SnRK2 protein kinase OST1 (open stomata 1) interacts with and phosphorylates the NADPH oxidase (AtrbohF), which functions in ABA-mediated reactive oxygen species (ROS) generation in guard cells (Kwak et al., 2003; Sirichandra et al., 2009). Double mutation of two NADPH oxidase genes (AtrbohD and AtrbohF) impairs ABA-promoted H2O2 production and stomatal closure (Kwak et al., 2003). Recently, an ABA-independent stomatal closure mechanism was reported in rice, in which a zinc finger transcription factor, DST, negatively regulates H2O2-induced stomatal closure by directly regulating the expression of genes related to H2O2 scavenging (Huang et al., 2009).
Recently, the SRO (Similar to RCD One) protein family was identified as a group of plant-specific proteins involved in stress and developmental responses in Arabidopsis (Jaspers et al., 2010). RCD1 (radical-induced cell death 1) was the first identified SRO protein in Arabidopsis. The loss of function of RCD1 resulted in pleiotropic phenotypes including increased sensitivity to apoplastic ROS, resistance to chloroplastic ROS formation by methyl viologen (MV), sensitivity to salt and glucose, tolerance to freezing, altered nitric oxide and hormone (ABA, jasmonic acid, and ethylene) responses, as well as developmental phenotypes such as aberrant leaf and rosette morphology, early flowering, and defects in root architecture and reproductive development (Overmyer et al., 2000; Ahlfors et al., 2004, 2009; Fujibe et al., 2004; Katiyar-Agarwal et al., 2006; Jaspers et al., 2009; Teotia and Lamb, 2009). RCD1 interacts with SOS1 and a large number of transcription factors that are involved in both development and stress-related processes, reflecting the phenotypes of the rcd1 mutant (Katiyar-Agarwal et al., 2006; Jaspers et al., 2009; Teotia and Lamb, 2009). SRO1, the paralogue gene of RCD1, has part of the function of RCD1. These two genes play redundant roles in several aspects of development including germination, vegetative growth, root architecture, and embryogenesis (Jaspers et al., 2009; Teotia and Lamb, 2009). SRO1 is also involved in abiotic stress responses. Both rcd1-1 and sro1-1 plants are resistant to osmotic stress (Teotia and Lamb, 2009), whereas, the sro1-1 mutant showed resistance to apoplastic ROS and salt stress, in contrast to the rcd1-3 mutant (Katiyar-Agarwal et al., 2006; Teotia and Lamb, 2009). This suggests that the two homologous genes have both redundant and independent functions under different stress conditions. Another gene of the Arabidopsis SRO family, SRO5, produces a natural small interfering (siRNA) by pairing with its neighbouring gene P5CDH and participates in a regulatory network during ROS-mediated salt responses (Borsani et al., 2005). However, the function of SRO protein in drought stress response is unknown and none of the SRO homologues has been identified in rice.
Transcription factors are one of the most important regulatory proteins involved in abiotic stress responses. Members of the DREB, MYB, bZIP, zinc finger, and NAC families have been characterized as having roles in plant tolerance to abiotic stresses (Hu et al., 2006; Wang et al., 2008; Xiang et al., 2008; Huang et al., 2009; Su et al., 2010). A previous study suggested that SNAC1, an NAC transcription factor, is predominantly induced in guard cells by drought. Overexpression of SNAC1 resulted in increased stomatal closure and improved drought and salt tolerance (Hu et al., 2006). In this study, a rice homologue of SRO, OsSRO1c, was identified and was characterized as an SNAC1-regulated downstream gene. It was found that OsSRO1c is predominantly expressed in guard cells under drought stress. The ossro1c mutant showed enhanced sensitivity to drought. Overexpression of OsSRO1c increased stomatal closure and reduced water loss by regulation of H2O2 homeostasis. The data indicated that OsSRO1c is involved in oxidative stress and modulates the stress response through interaction with various stress-related proteins.
Materials and methods
Plant materials
The japonica rice cultivars Dongjing (DJ) and Zhonghua11 (ZH11) were used in this study. Mutant ossro1c-1 (DJ background) and ossro1c-2 (ZH11 background) seeds were obtained from the POSTECH RISD (http://www.postech.ac.kr/life/pfg/risd/) and Shanghai T-DNA Insertion Population (SHIP; http://ship.plantsignal.cn/), respectively. Homozygous mutant and the wild-type (WT) genotypes segregated from the heterozygous mutant were identified by PCR analysis using a pair of genomic primers flanking the insertion site and a primer on the T-DNA.
Stress treatments and physiological measurements
To measure the transcript level of OsSRO1c under stress and phytohormone treatments and to uncover the function of OsSRO1c in the stress response, various treatments were performed. Details of the various treatments are provided in Supplementary Methods S1 available at JXB online.
Water loss rates of detached leaves from the WT and the mutant were measured by monitoring the fresh weight loss at the indicated time points. Thermal images of the plant were taken with an uncooled infrared thermal camera (ThermaCAM A320, FLIR, USA). Quantification of ABA was performed by the Applied Biosystems 4000 Q TRAR LC-MS system (Applied Biosystems, USA) according to previously described methods (Du et al., 2010). Details of the activity assay for ROS-scavenging enzymes and H2O2 quantitative measurement are provided in Supplementary Methods S1 at JXB online.
Physiological measurements of guard cells
Leaves of 50-day-old plants with the same period of dehydration stress or normal growth were fixed by 2.5% glutaraldehyde, and stomatal pictures were obtained by scanning electron microscopy (JSM-6390LV, JEOL, Japan). For the stomatal conductance measurement, flag leaves of the plant at the booting stage were used for stomatal conductance measurement with an SC-1 porometer (Decagon, USA). The measurement was performed in the field under a constant water concentration of 66±1% and a constant temperature of 31.6±0.5 °C. H2O2 production in guard cells was detected using 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA; Molecular Probes) as described previously (Huang et al., 2009). The fluorescence was observed by confocal laser-scanning microscopy (TCS SP2, Leica, Germany). All confocal images were taken under identical conditions and the guard cell region was selected to quantify the mean grey value of guard cells.
Other methods
Details of the methods for plasmid construction, rice transformation, chromatin immunoprecipitation (ChIP) assay, RNA isolation and reverse transcription–PCR (RT–PCR0, subcellular localization and bimolecular fluorescence complementation (BiFC) assays in rice protoplasts, and yeast one-hybrid and two-hybrid assays are provided in Supplementary Methods S1 at JXB online.
Results
OsSRO1c is directly regulated by SNAC1
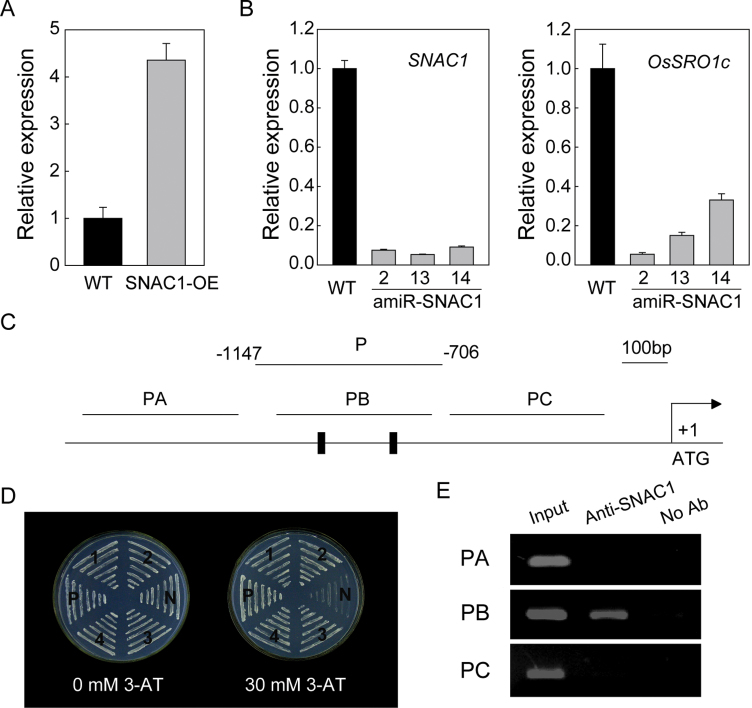
Previous microarray analysis indicated that >80 genes were up-regulated in SNAC1-overexpressing plants (Hu et al., 2006) and, among these up-regulated genes, the gene with the locus name LOC_Os03g12820, designated as OsSRO1c based on its homologues in Arabidopsis, was further characterized. Real-time quantitative RT–PCR (qPCR) analysis confirmed the up-regulation of OsSRO1c in SNAC1-overexpressing plants (Fig. 1A). In SNAC1-amiRNA (artificial microRNA) transgenic rice plants, OsSRO1c was significantly repressed (Fig. 1B). These results suggest that the expression of OsSRO1c is mainly regulated by SNAC1.
Fig. 1.
OsSRO1c is directly regulated by SNAC1. (A and B) Expression of OsSRO1c in SNAC1-overexpressing and amiRNA plants. The expression levels are normalized to an Actin endogenous control. Error bars indicate the SE based on three replicates. (C) Diagram of the OsSRO1c promoter showing the DNA fragments used for the yeast one-hybrid assay (P) and ChIP-PCR (PA–PC). A black rectangle indicates the core DNA-binding sequence (CDBS) of the NAC protein identified by Tran et al. (2004). (D) SNAC1 binds to the OsSRO1c promoter in yeast. N, negative control (p53HIS2 plus pGAD-SNAC1); P, positive control (p53HIS2 plus pGAD-Rec2-53); 1–4, four different colonies containing pGAD-SNAC1 and pHIS-POsSRO1c. (E) ChIP assay showing that SNAC1 can bind the OsSRO1c promoter in vivo. Nuclei from wild-type ZH11 were immunoprecipitated by anti-SNAC1 or without antibody. The precipitated chromatin fragments were analysed by PCR using three primer sets (PA–PC) amplifying three OsSRO1c promoter regions as indicated in C. One-tenth of the input chromatin was analysed as a control.
The NAC recognition sequence (NACRS) and core DNA-binding sequence (CDBS) for NAC protein have been identified in Arabidopsis (Tran et al., 2004). The high resolution crystal structure of the SNAC1 NAC domain has been reported recently, and shares a structural similarity with the reported structure of the Arabidopsis ANAC NAC domain (Chen et al., 2011). To test whether SNAC1 can bind to the promoter of OsSRO1c, the OsSRO1c promoter containing the CDBS was fused upstream to the HIS3 minimal promoter and co-transformed with the pGAD-SNAC1 plasmid (Hu et al., 2006) into the yeast strain Y187. The co-transformants could grow on SD/Leu−/Trp−/His− medium with 30mM 3-aminotriazole (3-AT), whereas the negative control could not (Fig. 1D), indicating that SNAC1 could bind to the OsSRO1c promoter in yeast. Most importantly, ChIP of protein extracts from rice ZH11 by the anti-SNAC1 antibody specifically precipitated the promoter sequence containing the CDBS (Fig. 1E). These results strongly support that OsSRO1c is a direct target gene of SNAC1.
OsSRO1c belongs to the plant-specific SRO family
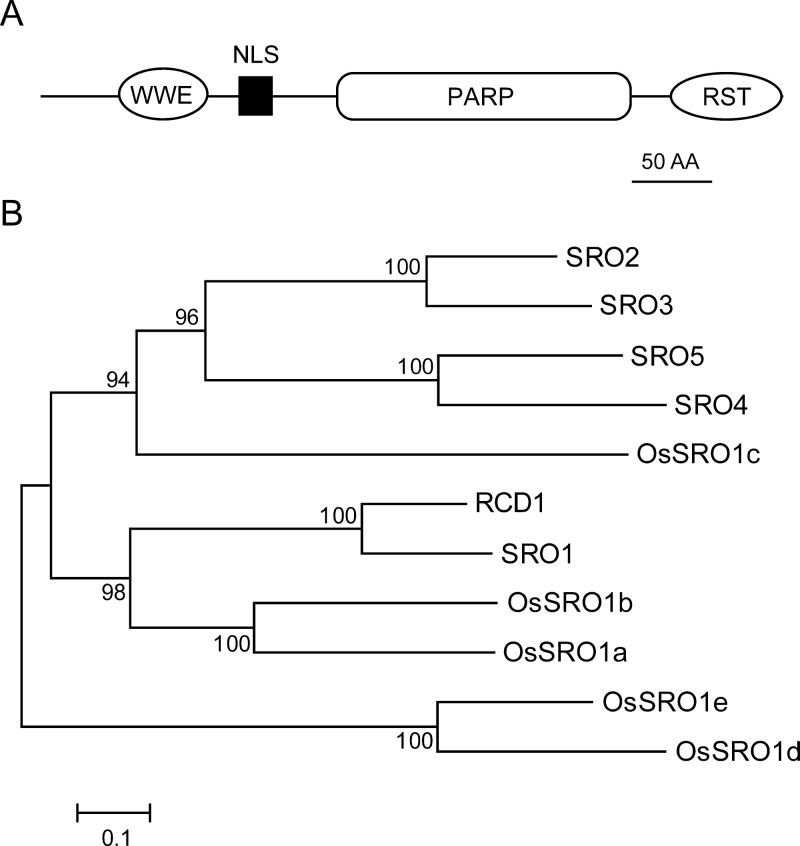
OsSRO1c encodes a protein of 463 amino acids with a mol. wt of 50.65kDa. Based on a sequence search against the Pfam database (http://pfam.sanger.ac.uk/), OsSRO1c consists of a putative N-terminal WWE domain (PF02825), a poly (ADP-ribose) polymerase catalytic domain (PARP; PF00644), and a C-terminal RCD1-SRO-TAF4 domain (RST domain; PF12174) (Fig. 2A). Therefore, OsSRO1c was classified as a homologue of the recently identified plant-specific SRO proteins that contain the PARP domain and the RST domain (Jaspers et al., 2010).
Fig. 2.
Protein sequence analysis of OsSRO1c. (A) Schematic diagram of functional domains and the predicted nuclear localization signal (NLS) of OsSRO1c. WWE, presumed protein–protein interaction domain characterized by tryptophan and glutamic acid residues (PF02825); PARP, PARP catalytic domain (PF00644); RST, RCD-SRO-TAF4 domain (PF12174). (B) Phylogenetic tree of SRO proteins in Arabidopsis and rice. The phylogenetic tree was created in MEGA4 software with the Neighbor–Joining method. Numbers indicate percentage values after 1000 replications. The scale bar indicates amino acid substitutions per position.
The SRO protein family has six members in Arabidopsis. In the japonica rice genome, five members including OsSRO1c were annotated based on sequence analysis (Jaspers et al., 2010). In addition to the PARP domain and RST domain, all the rice SRO proteins have the N-terminal WWE domain and belong to group I (Jaspers et al., 2010). Phylogenetic tree analysis showed that OsSRO1c and SRO2–SRO5 were in the same group (Fig. 2B). OsSRO1c has high sequence similarity to both the PARP and RST domains of Arabidopsis SRO proteins (Supplementary Fig. S1A at JXB online). However, a large number of amino acid variations exist in the RST domain. Although the SRO proteins contain the PARP domain, bioinformatic analysis of the PARP domain fold structure and biochemical assay of RCD1 suggested that SROs may not have ADP-ribosyl transferase activity (Jaspers et al., 2010). Compared with HsPARP1 and AtPARP1 that contain conserved histidine (H), tyrosine (Y), and glutamic acid (E) required for forming the ADP-ribosyl transferase catalytic triad (Doucet-Chabeaud et al., 2001), OsSRO1c and RCD1 do not contain the three conserved amino acids (Supplementary Fig. S1B), suggesting that OsSRO1c may not have ADP-ribosyl transferase activity either.
OsSRO1c, responsive to multiple stresses, is induced by drought in guard cells
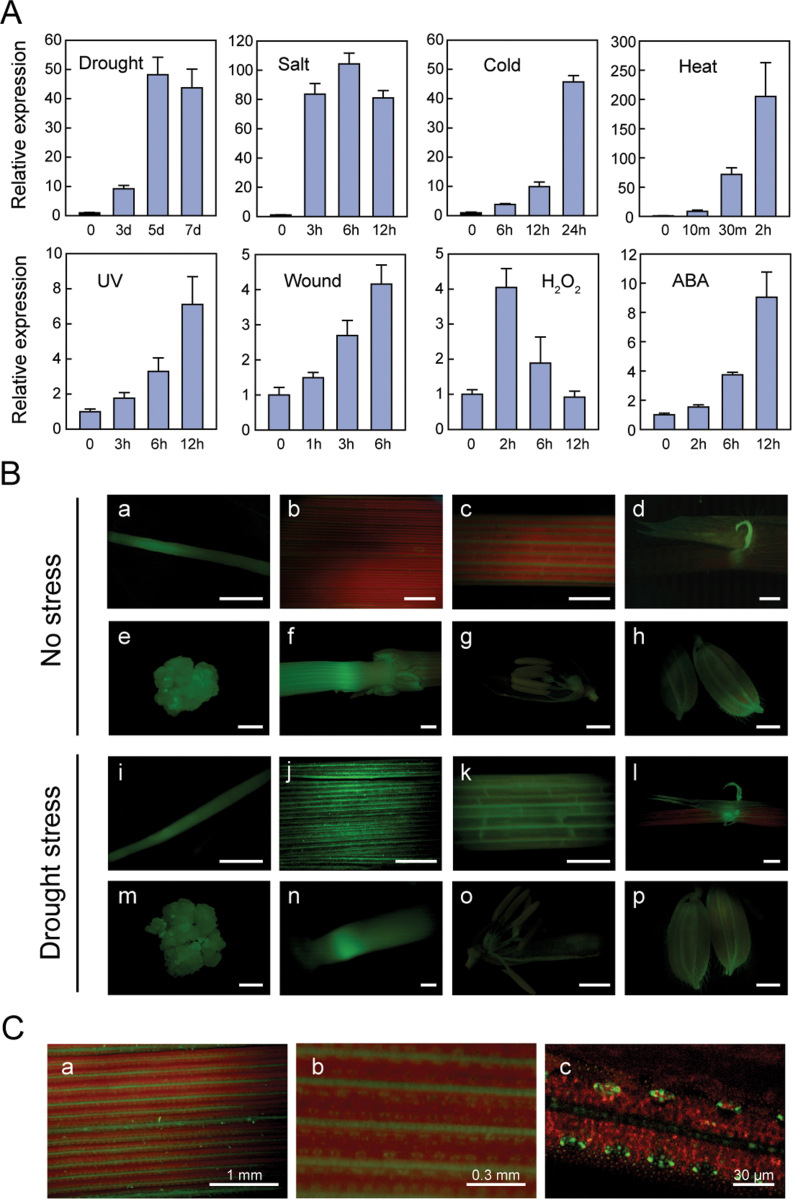
As a target gene of the stress-responsive transcription factor SNAC1, the expression of OsSRO1c in response to stress was investigated. As shown in Fig. 3A, OsSRO1c was strongly (>40-fold) induced by drought, salt, cold, and heat treatments and slightly induced (4- to 9-fold) by UV, wounding, H2O2, and ABA treatment. These results indicated that expression of OsSRO1c is responsive to multiple stresses. Expression of OsSRO1c was also analysed by qPCR in 16 organs/tissues (Supplementary Fig. S2 at JXB online). The results indicated that OsSRO1c had relatively high expression in calli, collar, stem, internodes, sheath, flag leaf, secondary branches of the panicle, and the spikelet hull.
Fig. 3.
Stress-inducible expression of OsSRO1c. (A) Expression of OsSRO1c under various abiotic stresses and ABA treatments. Four-leaf stage seedlings were subjected to drought stress (grow without water supply), salt (200mM NaCl), cold (4 ºC), heat (42 ºC), UV, wounding, H2O2 (1%), and ABA (100 µM). Relative expression levels of OsSRO1c were examined by real-time quantitative RT–PCR at the indicated time points. Error bars indicate the SE based on three replicates. (B) Expression pattern of GFP driven by the OsSRO1c promoter in transgenic rice plants under normal conditions (Ba–Bh) or dehydration stress (Bi–Bp). Shown are root (Ba and Bi), leaves (Bb and Bj), leaf sheath (Bc and Bk), ligule and auricle (Bd and Bl), calli (Be and Bm), stem and nodes (Bf and Bn), stamen and pistil (Bg and Bo), and hull (Bh and Bp). The scale bar indicates 2mm. (C) Optical (Ca and Cb) and confocal (Cc) microscopic analyses of GFP induction in guard cells by dehydration. The leaves of POsSRO1c: GFP transgenic rice were observed after dehydration stress for 5h.
To confirm the expression profiles of OsSRO1c, transgenic rice containing the POsSRO1c:GFP (green fluorescent protein gene under control of the OsSRO1c promoter) construct were generated and the expression pattern of GFP was monitored under normal and stress conditions (Fig. 3B). The GFP signal was observed in root, auricle, stem, internodes, and vascular bundle of the leaf sheath, and the spikelet hull under normal growth conditions, which is consistent with the qPCR analysis of OsSRO1c expression in 16 organs/tissues (Supplementary Fig. S2 at JXB online). When transgenic plants were drought stressed to leaf-rolling, strong induction of GFP expression was observed in the leaf blade (Fig. 3B). More focused observation revealed that the signal was localized specifically in the vascular bundles and guard cells (Fig. 3C). Such a drought-induced expression pattern of OsSRO1c in guard cells is very similar to that of SNAC1 (Hu et al., 2006), further supporting that OsSRO1c is a target gene of SNAC1.
Sequence analysis of OsSRO1c using the program NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/; Nguyen Ba et al., 2009) revealed a potential nuclear localization signal (NLS) GGRKRERDEAGSEVKGEDRRRR (Fig. 2A). To determine the subcellular localization of OsSRO1c, GFP-tagged OsSRO1c protein and cyan fluorescent protein (CFP)-tagged GHD7 protein were co-expressed in rice protoplasts. GHD7 is a known nuclear protein in rice (Xue et al., 2008) and CFP–GHD7 was used a positive control. As shown in Supplementary Fig. S3A at JXB online, the green fluorescence produced by sGFP–OsSRO1c overlapped with the cyan fluorescence produced by CFP–GHD7, indicating that OsSRO1c protein is targeted to the nucleus. To confirm this result, transgenic rice consistently expressing the OsSRO1c–EGFP fusion gene were generated. As shown in Supplementary Fig. S3B, fluorescence was observed only in the nucleus. These results suggest that OsSRO1c is a nuclear protein.
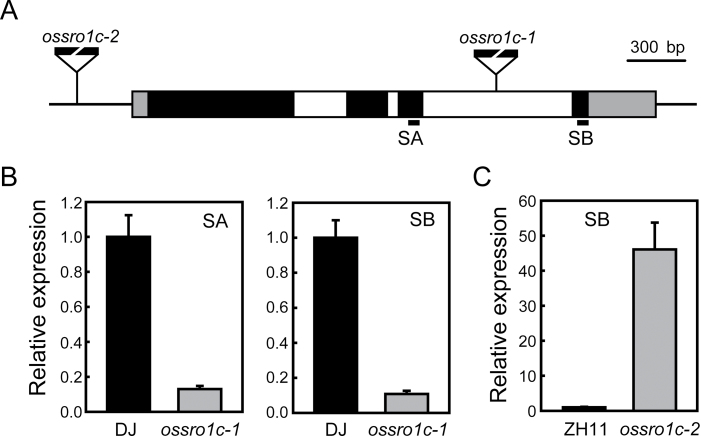
The ossro1c-1 mutant is sensitive to drought stress
Because OsSRO1c is a direct target gene of SNAC1 that confers stress tolerance, it is necessary to discover the function of OsSRO1c in stress response. Two allelic T-DNA insertion mutants ossro1c-1 (DJ background) and ossro1c-2 (ZH11 background) were collected (see the Materials and methods). The T-DNA was inserted in the third intron (2238bp downstream of ATG) in ossro1c-1 and in the promoter (308bp upstream of ATG) in ossro1c-2 (Fig. 4A). The expression of OsSRO1c was significantly repressed in the ossro1c-1 mutant in both upstream and downstream regions of the insertion site (Fig. 4B). However, ossro1c-2 showed dramatically increased (~50-fold) expression of OsSRO1c compared with the WT (Fig. 4C) and thus was used as a gain-of-function mutant for further studies.
Fig. 4.
Identification of ossro1c T-DNA insertion mutants. (A) Schematic diagram of the OsSRO1c gene and two alleles of T-DNA insertion mutants, ossro1c-1 and ossro1c-2. Exons, introns, and untranslated regions are indicated in black, white, and grey, respectively. (B and C) Expression of OsSRO1c in ossro1c-1 and ossro1c-2. SA and SB indicate the regions upstream and downstream of the insertion site, respectively. Error bars indicate the SE based on three replicates.
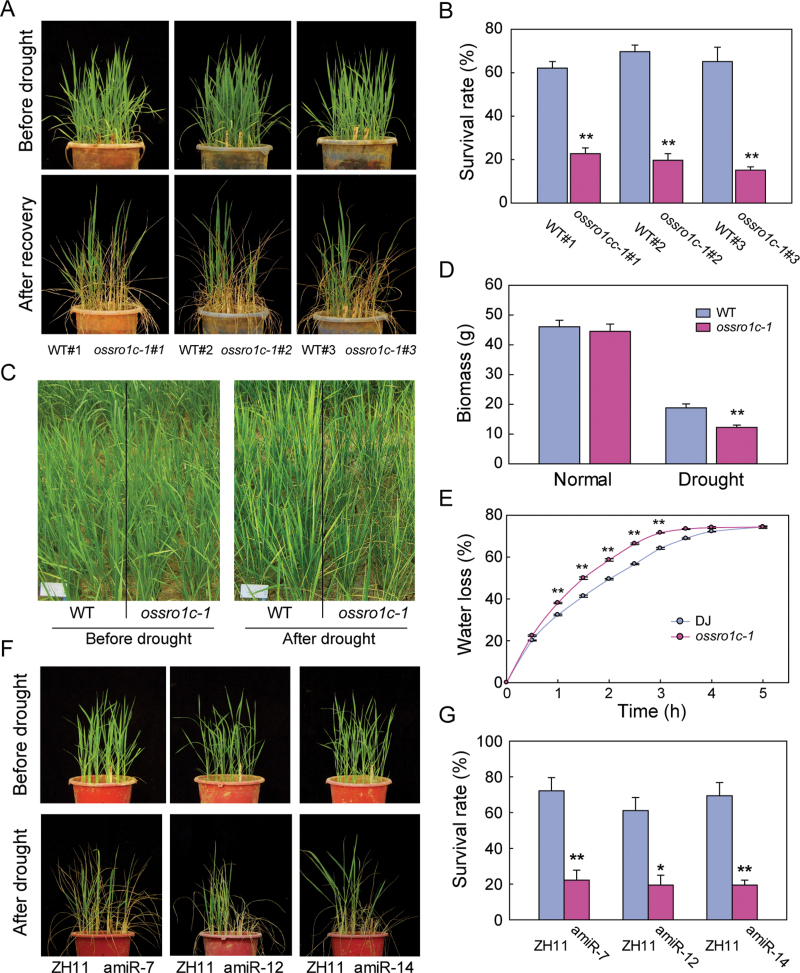
The ossro1c-1 mutant was tested for drought resistance. Three homozygous mutant lines (ossro1c/ossro1c) and three WT lines (OsSRO1c/OsSRO1c) derived from the segregants of the heterozygous (OsSRO1c/ossro1c) mutant were chosen for drought testing. Plants at the four-leaf stage were subjected to drought treatment by stopping irrigation for 10 d. All the ossro1c-1 mutant lines showed more severe wilting after rewatering compared with WT plants (Fig. 5A). After recovery for 7 d, 60–70% of WT plants were recovered, whereas only 15–20% of the mutant plants were recovered (Fig. 5B), significantly (t-test, P < 0.01) less than the WT. When grown in a paddy field under a removable rain-off shelter and drought stressed at the later tillering stage, ossro1c-1 mutant plants showed earlier leaf-rolling and wilting than WT plants (Fig. 5C). After severe drought stress in the field, the biomass of the ossro1c-1 mutant was significantly lower than that in WT plants (t-test, P < 0.01; Fig. 5D). These results suggest that the ossro1c-1 mutant is impaired in drought resistance. Water loss rates of detached leaves from WT plants and ossro1c-1 mutants were measured. The results showed that the detached leaves of mutants lost water faster than WT leaves (Fig. 5E). This result was also confirmed by infrared thermal image analysis, in which the detached leaves and drought-stressed seedlings of ossro1c-1 showed a lower surface temperature than WT leaves, indicating more transpirational water loss in ossro1c-1 mutants (Supplementary Fig. S4 at JXB online).
Fig. 5.
Increased drought sensitivity of ossro1c-1 and amiR-OsSRO1c plants. (A) Increased drought sensitivity of ossro1c-1 at the seedling stage. Three homozygous mutant lines (ossro1c-1#1, #2, #3) and three WT lines (WT#1, #2, #3) derived from the segregants of heterozygous mutant were used in drought stress testing. Four-leaf stage plants were not watered for 10 d, followed by rewatering for 7 d. (B) Survival rate of WT siblings and ossro1c-1 mutants after drought stress (n=3). (C) Increased drought sensitivity of ossro1c-1 at a later tillering stage. (D) Biomass of the WT and ossro1c-1 mutants under normal and drought stress (n=12). (E) Water loss from detached leaves of the WT and ossro1c-1 mutants at the indicated time points (n=3). (F) Increased drought sensitivity of amiR-OsSRO1c plants. Four-leaf stage plants were not watered for 10 d, followed by rewatering for 7 d. (G) Survival rate of ZH11 and amiR-OsSRO1c plants after drought stress (n=3). Data represent the mean± SE. *P < 0.05; **P < 0.01, t-test.
To confirm further the functions of OsSRO1c in rice, an amiRNA strategy (Warthmann et al., 2008) was used to knock down the expression of OsSRO1c in transgenic rice. The expression of OsSRO1c in 15 T0 OsSRO1c-amiRNA (amiR-OsSRO1c) plants was checked by qPCR, and OsSRO1c was found to be significantly repressed in half of the T0 amiR-OsSRO1c lines (Supplementary Fig. S5A at JXB online). Three independent lines (amiR-OsSRO1c-7, amiR-OsSRO1c-12, and amiR-OsSRO1c-14) were selected for drought testing. The results showed that these amiR-OsSRO1c lines were also hypersensitive to drought stress at the seedling stage (Fig. 5F). The survival rate of WT controls was 60–70%, whereas only ~20% of the amiR-OsSRO1c lines were recovered (Fig. 5G).
Drought stress often causes physiological stresses such as osmotic stresses. To investigate if OsSRO1c is involved in osmotic stress tolerance, the ossro1c-1 mutant, amiR-OsSRO1c lines, and WT seedlings were exposed to osmotic stress using 150mM mannitol. The osmotic stress caused significantly more (t-test, P < 0.01) reduced growth of ossro1c-1 mutant than of WT seedlings (Supplementary Fig. S6A, B at JXB online). A similar result was obtained for amiR-OsSRO1c lines (Supplementary Fig. S6C, D). These results suggest that OsSRO1c is also involved in osmotic stress tolerance in rice.
Overexpression of OsSRO1c decreased transpirational water loss
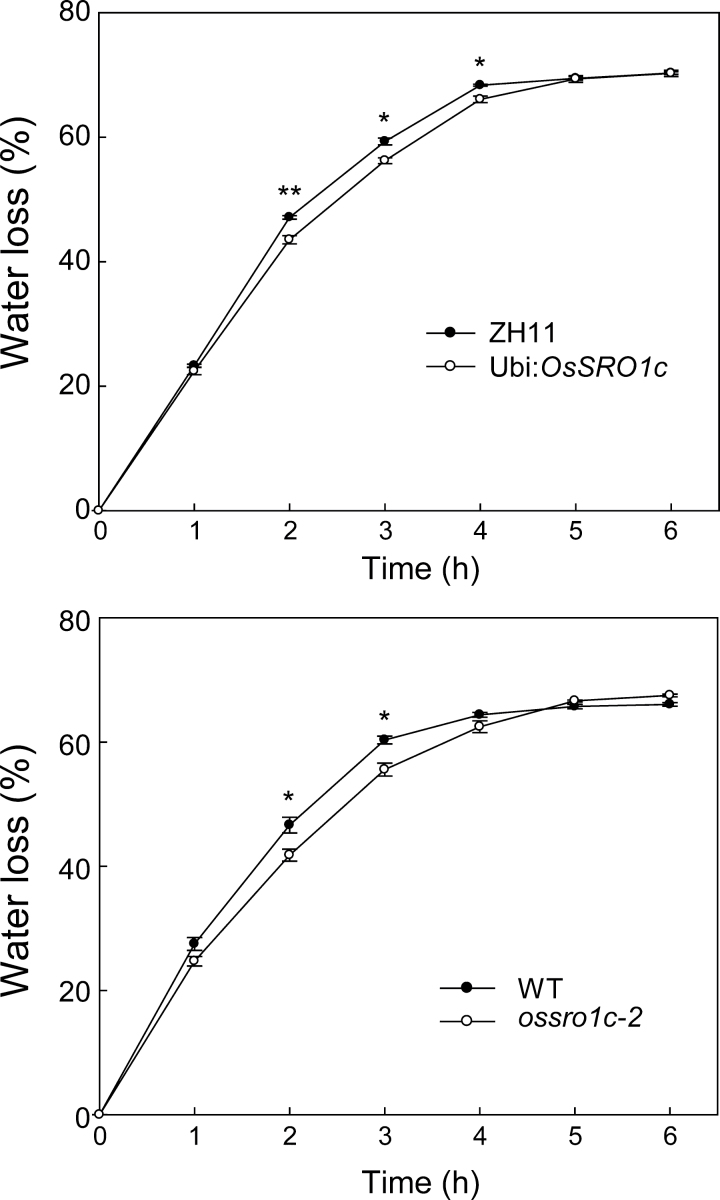
Because OsSRO1c was up-regulated by drought predominantly in guard cells and ossro1c-1 mutants showed an increased water loss rate under drought stress, experiments were carried out to determine whether overexpression of OsSRO1c could result in decreased water loss. The full-length OsSRO1c under the control of the maize ubiquitin promoter was introduced into rice ZH11; 22 independent transgenic plants were obtained and the overexpression of OsSRO1c was confirmed by qPCR. Three independent transgenic lines (OsSRO1c-OE-3, OsSRO1c-OE-11, and OsSRO1c-OE-14) with overexpression of the gene (Supplementary Fig. S5B at JXB online) were selected for testing. First, water loss rates of detached leaves from OsSRO1c-overexpressing lines, the ossro1c-2 (gain-of-function) mutant, and WT plants were measured. The results showed that the detached leaves of both OsSRO1c-overexpressing lines and the ossro1c-2 mutant lost water more slowly than WT plants (Fig. 6). Infrared thermal images showed that both the OsSRO1c-overexpressing and ossro1c-2 mutant plants had higher surface temperatures than WT plants (Supplementary Fig. S7), indicating reduced transpirational water loss in the plants with elevated OsSRO1c expression.
Fig. 6.
Overexpression of OsSRO1c reduced water loss. Water loss from detached leaves of wild-type and OsSRO1c-overexpressing plants or ossro1c-2 mutants at the indicated time points (n=3). Data represent the mean ±SE. *P < 0.05; **P < 0.01, t-test.
OsSRO1c modulates stomatal closure and H2O2 homeostasis
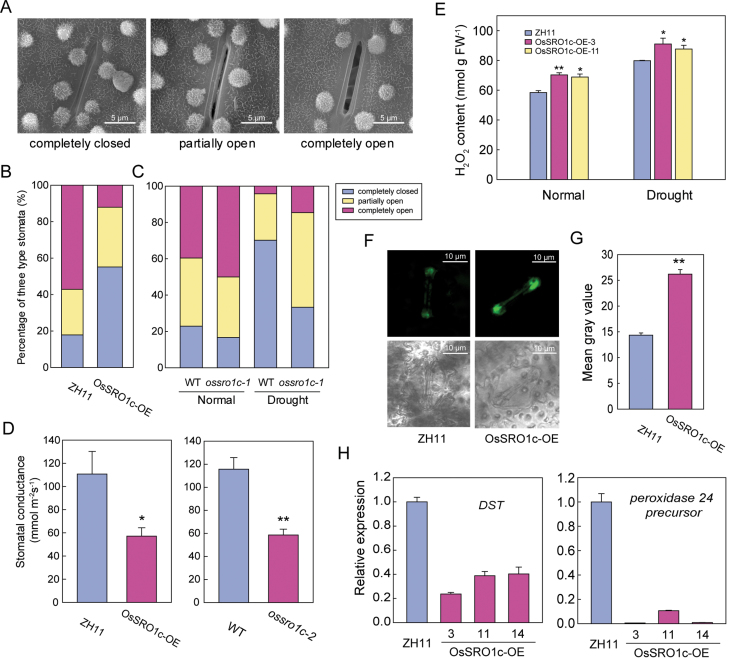
The function of OsSRO1c in the control of water loss prompted an investigation into stomatal aperture, the major factor affecting the water-holding capacity in rice leaves. The stomatal apertures of OsSRO1c-overexpressing plants, ossro1c-1 mutant plants, and WT plants were checked by using scanning electron microscopy. The results showed that 55.2% of stomata were completely closed in the OsSRO1c-overexpressing plants, whereas only 17.9% were completely closed in the WT plants. On the other hand, only 12.2% of stomata were completely open in the OsSRO1c-overexpressing plants, but 57.1% were completely open in WT plants (Fig. 7B). The percentage of partially open stomata was not significantly different between OsSRO1c-overexpressing and WT plants. These results suggest that overexpression of OsSRO1c can promote stomatal closure. Furthermore, stomatal conductance was lower in OsSRO1c-overexpressing plants or ossro1c-2 mutants than in WT plants (Fig. 7D). For the ossro1c-1 mutant, the percentages of the three types of stomata were not obviously different compared with the WT plants under normal conditions. When drought stressed to leaf-rolling, 70% of stomata rapidly closed in WT plants, but only 33.3% of stomata were closed in the ossro1c-1 mutant (Fig. 7C). This result is in agreement with the faster water loss rate of detached leaves from the ossro1c-1 mutant.
Fig. 7.
Down-regulation of DST and accumulation of H2O2 induces stomatal closure in OsSRO1c-overexpressing plants. (A) Scanning electron microscopy images of three levels of stomatal opening. (B) The percentage of three levels of stomatal opening in WT (ZH11) and OsSRO1c-overexpressing plants (n=56 stomata for ZH11; n=58 stomata for OsSRO1c-overexpressing plants). (C) The percentage of three levels of stomatal opening in WT and ossro1c-1 mutants under normal and drought stress (n=48 stomata for the WT under normal conditions; n=47 stomata for the WT under drought stress; n=42 stomata for ossro1c-1 under normal conditions; n=48 stomata for ossro1c-1 under drought stress). (D) Stomatal conductance of WT and OsSRO1c-overexpressing plants or ossro1c-2 mutants (n=12). (E) H2O2 content of ZH11 and OsSRO1c-overexpressing plants under normal conditions and drought stress (n=3). (F) Detection of H2O2 production in guard cells of ZH11 and OsSRO1c-overexpressing plants with H2DCFDA. (G) Quantitative analysis of H2O2 production in guard cells of ZH11 and OsSRO1c-overexpressing plants (n=47 stomata for ZH11; n=48 stomata for OsSRO1c-overexpressing plants). (H) Expression of DST and peroxidase 24 precursor in OsSRO1c-overexpressing plants. Data represent the mean ±SE. *P < 0.05; **P < 0.01, t-test.
The phytohormone ABA triggers stomatal closing in response to drought stress (Schroeder et al., 2001b ). However, the endogenous ABA content showed no significant difference between WT and OsSRO1c-overexpressing plants (Supplementary Fig. S8A at JXB online). Moreover, no significant difference was found in stomatal closure between WT and ossro1c-1 mutant or OsSRO1c-overexpressing plants under ABA treatment (Supplementary Fig. S8B). Therefore, the role of OsSRO1c in promoting stomatal closure may not due to an increased ABA level or sensitivity.
H2O2 is another signal molecule that induces stomatal closure (McAinsh et al., 1996; Apel and Hirt, 2004). A higher level of H2O2 accumulation was detected in the OsSRO1c-overexpressing plants under both drought and unstressed conditions (Fig. 7E). In addition, more H2O2 accumulation was detected in the guard cells of OsSRO1c-overexpressing plants, as indicated by the ROS indicator, H2DCFDA (Fig. 7F, G). These results indicated that the increased stomatal closure in OsSRO1c-overexpressing plants was probably due to accumulation of H2O2 in guard cells. The accumulation of H2O2 in guard cells leading to stomatal closure was also found in the rice dst mutant, and DST acts as negative regulator of H2O2 accumulation (Huang et al., 2009). To determine whether the function of OsSRO1c is related to DST, the expression of DST and its downstream gene was checked in the OsSRO1c-overexpressing plants. The result showed that DST was significantly repressed in OsSRO1c-overexpressing plants (Fig. 7H). The functional downstream gene of DST, peroxidase 24 precursor, that encodes a peroxidase to scavenge H2O2 in guard cells (Huang et al., 2009), was also down-regulated in the OsSRO1c-overexpressing plants (Fig. 7H). These results indicate that OsSRO1c may positively regulate H2O2-induced stomatal closure by suppressing DST.
OsSRO1c is involved in oxidative stress response
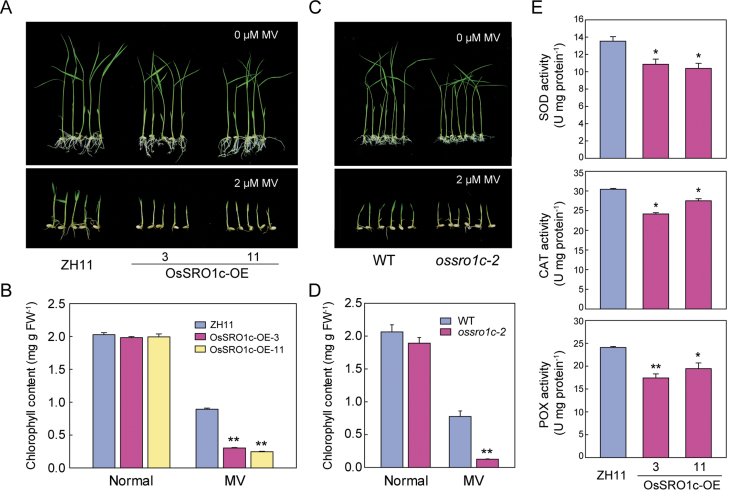
Because overexpression of OsSRO1c resulted in H2O2 elevation, the function of OsSRO1c in oxidative stress was investigated further. Germinated WT and OsSRO1c-overexpressing plants and ossro1c-2 mutants were sown on Murashige and Skoog (MS) medium containing 2 µM MV (an oxidative stress inducer). After 7 d, OsSRO1c-overexpressing plants and ossro1c-2 mutants exhibited an etiolating phenotype with more severe decreases of chlorophyll compared with WT plants (Fig. 8A–D). In contrast to the overexpression plants, the ossro1c-1 mutants and amiR-OsSRO1c plants showed increased resistance to oxidative stress (Supplementary Fig. S9 at JXB online). Ascorbate peroxidase (APX), superoxide dismutase (SOD), and catalase (CAT) are major ROS-scavenging enzymes, which have important roles in oxidative stress tolerance (Apel and Hirt, 2004). Among these ROS-scavenging enzyme-encoding genes, two SOD genes, Fe-SOD and SodCc2, were significantly repressed in OsSRO1c-overexpressing plants (Supplementary Fig. S10). The activities of ROS-scavenging enzymes, namely SOD, CAT, and peroxidase (POX), were also measured in OsSRO1c-overexpressing plants. The results showed that OsSRO1c-overexpressing plants had a significantly lower activity of SOD, CAT, and POX than did WT plants (Fig. 8E). The above results suggest that OsSRO1c overexpression has a negative role in resistance to oxidative stress, which may be associated with the suppression of ROS-scavenging enzyme genes and the activity of ROS-scavenging enzymes.
Fig. 8.
Overexpression of OsSRO1c increased the sensitivity to oxidative stress. (A and C) OsSRO1c-overexpressing plants (A) and ossro1c-2 mutants (C) were sensitive to MV stress. Seeds germinated for 3 d were transplanted in either MS medium or MS medium supplemented with 2 µM MV and measured 1 week later. (B and D) Total chlorophyll contents of WT and OsSRO1c-overexpressing plants (B) or ossro1c-2 mutants (D) under normal conditions and MV stress (n=3). FW, fresh weight. (E) Activity of SOD, CAT, and POX in ZH11 and OsSRO1c-overexpressing plants (n=3). Data represent the mean ±SE. *P < 0.05; **P < 0.01, t-test.
OsSRO1c interacts with diverse proteins
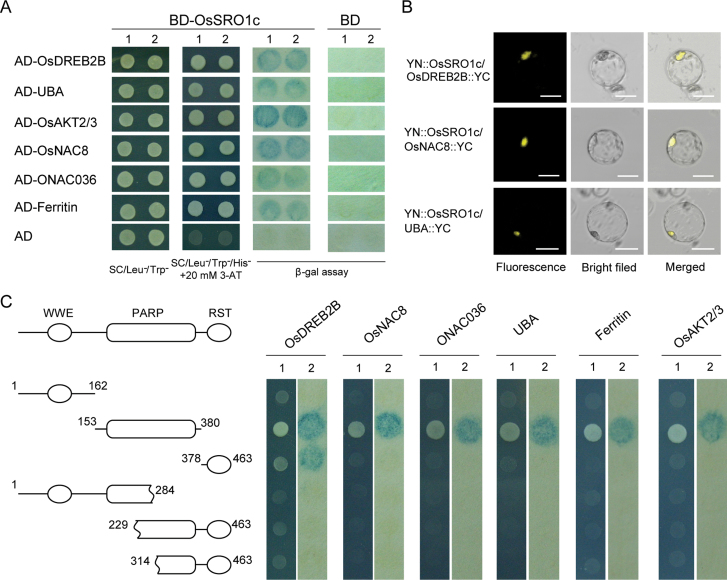
To further understand the function of OsSRO1c, OsSRO1c-interacting proteins (SRIPs) were isolated by yeast two-hybrid screening. A cDNA library of mixed rice leaves treated with multiple stresses (drought, high salt, cold, and ABA) was screened using the full-length OsSRO1c protein as bait. Fourteen positive clones representing six unique SRIPs were isolated. Half of the positive clones were identified to be OsDREB2B, an AP2/ERF transcription factor. The other five SRIPs were two NAC transcription factors (OsNAC8 and ONAC036), ubiquitin-associated (UBA) protein, ferritin, and potassium channel OsAKT2. The interaction of OsSRO1c with the SRIPs was rechecked by a yeast two-hybrid assay (Fig. 9A). The interactions of OsSRO1c with OsDREB2B, OsNAC8, and UBA were also confirmed by BiFC assay in living rice cells (Fig. 9B).
Fig. 9.
OsSRO1c-interacting proteins (SRIPs) identified by yeast two-hybrid and BiFC assays. (A) Yeast two-hybrid assay of OsSRO1c–SRIP interactions. AD-SRIPs co-transformed with BD empty vector are used as negative controls. 1 and 2 show two different colonies of each pairwise interaction test. (B) BiFC assay of three pairs of OsSRO1c–SRIP interactions. Bar=10 µm. (C) The PARP domain of OsSRO1c is required for OsSRO1c–SRIP interactions. The schematic diagram on the left indicates different truncated fragments, and numbers indicate the start and end amino acid position of the truncated fragment. The growth of yeast cells on selective medium (1) and expression of the lacZ reporter gene (blue on X-gal assay) (2) are shown.
Because both OsNAC8 and OsDREB2B are from large gene families, tests were carried out to determine whether OsSRO1c can interact with other members of the NAC family or DREB family. Four members of the DREB family (OsDREB1A, OsDREB1B, OsDREB2A, and OsDREB2B) were tested in the yeast two-hybrid system. The result indicated that OsSRO1c predominantly interacts with OsDREB2B but does not interact with other DREB proteins (Supplementary Fig. S11 at JXB online). Nine stress-responsive NAC transcription factors, namely SNAC1, SNAC2, OsNAC4, OsNAC5, OsNAC8, ONAC036, ONAC073, ONAC088, and ONAC131, were also tested; the results showed that OsSRO1c interacts only with OsNAC8 and ONAC036 (Supplementary Fig. S11). In addition, the fragments of OsDREB2B, OsNAC8, and ONAC036 isolated from yeast two-hybrid screening only had the C-terminus of the proteins, indicating that the conserved N-terminal AP2 or NAC domain may not be required for protein interaction and the variable C-terminal domains may be important to determine the specific interaction of OsSRO1c with certain members of the two families.
Because OsSRO1c has three domains, which domain of OsSRO1c is critical for the interaction was also checked. Three constructs containing one of the domains were created for interaction assay. The results suggested that the PARP domain of OsSRO1c can interact with all the SRIPs, whereas the RST domain could interact only with OsDREB2B, and none of the proteins interacted with the WWE domain (Fig. 9C). To verify the interaction role of the PARP domain, truncation constructs containing a partial PARP domain plus RST domain (or WWE domain) were tested in the yeast two-hybrid system, and none of the constructs showed a positive interaction (Fig. 9C). These results indicate that the PARP domain of OsSRO1c is required for the interaction with the protein identified above, whereas the RST domain may be required specifically in the interaction with OsDREB2B.
Discussion
Direct regulation of OsSRO1c by SNAC1
Plants have developed complicated regulatory networks to respond to various abiotic stresses. Expression of various genes is activated by transcription factors and enhances plant resistance to environmental stress. SNAC1 is an important transcription factor involved in drought response, and SNAC1-overexpressing plants significantly enhance drought resistance by increasing stomatal closure to prevent water loss (Hu et al., 2006). Here, the identification of an SRO protein (OsSRO1c) in rice, which was characterized as a direct target of SNAC1, is reported. The expression of OsSRO1c was up-regulated in SNAC1-overexpressing plants and repressed in SNAC1-amiRNA plants, and SNAC1 protein could bind to the promoter of OsSRO1c in both yeast and rice (Fig. 1). SNAC1 and OsSRO1c are positively co-expressed genes (PCC=0.6391) by co-expression analysis based on abiotic stress microarray data in the Rice Oligonucleotide Array Database (ROAD) (http://www.ricearray.org). Both SNAC1 and OsSRO1c were induced by various abiotic stresses and expressed in guard cells under drought conditions (Hu et al., 2006). The drought-sensitive phenotypes of the ossro1c-1 mutant and amiR-OsSRO1c plants were in agreement with the drought resistance phenotype of SNAC1-overexpressing plants. Moreover, the SNAC1-overexpressing plants and OsSRO1c-overexpressing plants both increased stomatal closure and reduced water loss under drought stress (Hu et al., 2006). All these results indicated that OsSRO1c was a direct target of SNAC1.
Dual functions of OsSRO1c in drought resistance
Many stresses that disrupt the cellular homeostasis of cells enhance the production of ROS. The enhanced production of ROS can pose a threat to cells, but it is also thought that ROS act as signals for the activation of stress response and defence pathways (Mittler, 2002). In rice guard cells, zinc finger protein DST targeted the promoter of peroxidase 24 precursor and regulated H2O2 homeostasis (Huang et al., 2009). In this work, overexpression of OsSRO1c resulted in down-regulation of DST and its target gene peroxidase 24 precursor, accumulation of H2O2 in guard cells, increased stomatal closure, and reduced water loss (Figs 6, 7). On the other hand, a few genes encoding ROS-scavenging enzymes, such as glutathione S-transferase GSTU6 and peroxidase 16 precursor, that were up-regulated in the dst mutant and may contribute to the removal of excess ROS (Huang et al., 2009), were down-regulated in OsSRO1c-overexpressing plants (Supplementary Fig. S10 at JXB online). Down-regulation of these genes may contribute to the excess accumulation of ROS, and thus has negative effects on the drought and oxidative stress tolerance. Actually, it was observed that the OsSRO1c-overexpressing plants and ossro1c-2 mutants showed more severe damage under drought and oxidative stress treatment (Fig. 8; Supplementary Fig. S12). Therefore, OsSRO1c plays dual roles in drought resistance of rice by boosting H2O2 acclimation under stress conditions. On one hand, H2O2 acclimation in guard cells acts as a signal molecule to promote stomatal closure and reduce water loss; on the other hand, excess H2O2 acclimation causes oxidative stress in cells and results in cellular damage and death. The drought resistance phenotype of SNAC1-overexpressing plants is the overall effect of the stress-related downstream genes including OsSRO1c. The major contribution of OsSRO1c to the drought resistance phenotype of SNAC1-overexpressing plants is its role in promoting stomatal closure because the negative role of OsSRO1c in tolerance to oxidative stress caused by severe drought was not obvious in the SNAC1-overexpressing plants (Hu et al., 2006). One possible explanation is that the negative effect of OsSRO1c on oxidative stress may be compromised by other SNAC1-regulated genes in SNAC1-overexpressing plants. An alternative explanation is that the expression level of OsSRO1c in SNAC1-overexpressing plants is not as high as in OsSRO1c-overexpressing plants and therefore no significant oxidative stress occurred by overexpression of SNAC1.
The role of OsSRO1c is largely ABA independent
Because of the important roles of the stomatal apparatus for plants, intensive studies have focused on uncovering the molecular mechanisms of stomatal movement, and many ion channels, plant hormones, and signalling components function together in controlling stomatal closure and opening (Schroeder et al., 2001a ; Acharya and Assmann, 2009; Kim et al., 2010). However, knowledge of the regulation of stomatal movement remains fragmented, and this is an important subject especially for finding solutions to the fine control of stomatal closure without significant sacrifice of photosynthesis under water-limited conditions. So far, a total of nine plant transcriptional regulators have been characterized for their roles in regulating stomatal movements, and two of them, SNAC1 and DST, were identified in rice (Cominelli et al., 2010). In this study, it was found that a direct downstream gene of SNAC1, OsSRO1c, promoted stomatal closure through repression of the expression of DST and accumulation of H2O2 in guard cells (Fig. 7). Furthermore, DST was also repressed in SNAC1-overexpressing plants (Supplementary Fig. S13 at JXB online). Therefore, these two transcription factors may be involved in the same pathway mediating H2O2-induced stomatal closure. ABA has important roles in triggering stomatal closure. OsSRO1c was induced by ABA; however, the endogenous ABA level showed no change in OsSRO1c-overexpressing plants (Supplementary Fig. S8A). Stomatal response to ABA was not changed in either ossro1c-1 mutant or OsSRO1c-overexpressing plants (Supplementary Fig. S8B). In addition, the seedling growth of WT, mutant, and overexpression plants showed no significant difference when they were treated with ABA (Supplementary Fig. S14). These results indicated that the role of OsSRO1c in modulating stomatal closure may be mainly via an ABA-independent pathway. Regulation of stomatal closure via H2O2 as a secondary messenger generated through NADPH oxidase has been reported as being induced by ABA (Kwak et al., 2003; Sirichandra et al., 2009). However, the present data imply that an ABA-independent pathway mediating H2O2-induced stomatal closure may exist in rice.
Comparison of OsSRO1c with Arabidopsis SRO proteins
SRO is a recently identified protein family with a unique domain architecture conserved in plants. Except for three members (RCD1, SRO1, and SRO5) in Arabidopsis, most of the SRO gene family members have not been characterized. OsSRO1c is significantly different from RCD1 and SRO5 not only in functional domains but also in the spectrum of functions in stress resistance. The RST domain of OsSRO1c varies greatly compared with that of RCD1 and other Arabidopsis SRO proteins (Supplementary Fig. S1 at JXB online). Previous studies suggest that SRO proteins RCD1, SRO1, and SRO5 have roles in salt, osmotic, and oxidative stress (Overmyer et al., 2000; Ahlfors et al., 2004; Borsani et al., 2005; Katiyar-Agarwal et al., 2006; Teotia and Lamb, 2009). It was found that OsSRO1c has an important role in regulating stomatal closure and drought resistance. OsSRO1c appears to be the first member of the SRO family identified with a function in drought resistance. The ossro1c-1 mutant is sensitive to drought stress but has no significant phenotype under high salinity stress (not shown). Although both RCD1 and OsSRO1c are involved in osmotic and oxidative stress, they also have differences. For example, the ossro1c-1 mutant is sensitive to osmotic stress (Supplementary Fig. S6 at JXB online), but the rcd1 mutant is resistant to osmotic stress (Teotia and Lamb, 2009). OsSRO1c-overexpressing plants and the activation mutant plants are sensitive to MV (Fig. 8), which is in contrast to the phenotype of the loss-of-function mutant, but the RCD1-overexpressing plant exhibited a weak rcd1 phenotype and was as sensitive to MV as the WT plant (Fujibe et al., 2006).
Interaction proteins of OsSRO1c and their roles in stress tolerance
The functional diversity of the SRO family may be related to the diverse interaction proteins of different SRO proteins. A large number of RCD1-, SRO1-, and SRO5-interacting proteins have been identified, and most of them are transcription factors (Jaspers et al., 2009, 2010). In this work, three transcription factors and three functional proteins were identified as SRIPs. OsDREB2B was predominant among the positive clones in yeast two-hybrid screening, suggesting that it may be a major SRIP just as DREB2A is for RCD1. Pairwise interaction tests of OsSRO1c against four OsDREBs and nine ONACs suggest that specificity exists for the interaction between SROs and certain members of the transcription factor families (Supplementary Fig. S11 at JXB online). The recently identified C-terminal conserved RST domain of RCD1 was proposed as a functional protein interaction domain (Jaspers et al., 2009). However, the present results indicate that the PARP domain of OsSRO1c is required for interaction with SRIPs, whereas the RST domains only mediate the interaction with OsDREB2B (Fig. 9C). The WWE domain is predicted to mediate protein–protein interaction and has been demonstrated by the interaction of the WWE domain of Deltex protein with the ankyrin repeat of the Notch receptor in Drosophila (Zweifel et al., 2005). Although the C-terminus of OsAKT2 contains an ankyrin repeat domain, no interaction was observed between the C-terminus of OsAKT2 and the putative WWE domain of OsSRO1c (Fig. 9C).
Among the SRIPs, only OsDREB2B was functionally characterized. Overexpression of OsDREB2B significantly improved the tolerance of transgenic rice and Arabidopsis to drought stress (Chen et al., 2008; Matsukura et al., 2010). This evidence supports the important biological significance of OsSRO1c with its interaction proteins. It was further checked whether OsDREB2B (and OsNAC8 and ONAC036 as well) can bind the promoter of DST or its target gene (peroxidase 24 precursor), but the result was negative (not shown). Because OsSRO1c itself has no transcriptional activity (Fig. 9A), OsSRO1c may regulate the expression of DST (and other genes related to ROS production as well) in an indirect manner, most probably by interacting with some unknown transcription factors that regulate DST or other ROS-related genes. Although the functions of the other five SRIPs remain unknown, some clues support their involvement in stress tolerance. Ferritin in Arabidopsis was suggested to play an important role in oxidative stress resistance by controlling iron homeostasis (Ravet et al., 2009). The Arabidopsis AKT2/3 subunit constitutes the Ca2+ sensitivity of the guard cell K+ uptake channel (Ivashikina et al., 2005). According to the whole-genome transcriptional profiles of rice cell types, OsAKT2/3 is the only Shaker-type potassium channel gene in rice with significant expression in stomata (Rice Cellular Expression Profile Project, http://bioinformatics.med.yale.edu/riceatlas), implying that it may have a role in stomatal movement. OsSRO1c may affect the functions of its interacting proteins by post-translational modification or formation of a protein complex to affect the activities of transcription factors and channel proteins or the stability of the interaction proteins.
In conclusion, OsSRO1c is a direct target gene of SNAC1 and has dual roles in the regulation of stomatal closure and oxidative stress tolerance by regulating H2O2 homeostasis in rice. An ongoing effort is to determine how the interactions of OsSRO1c with diverse proteins control H2O2 homeostasis under stresses, and thus, to understand completely the role of OsSRO1c in drought and oxidative stress resistance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Multiple sequence alignment and sequence comparison.
Figure S2. Tissue and organ expression pattern of OsSRO1c.
Figure S3. Subcellular localization of OsSRO1c.
Figure S4. Thermal images of detached leaves and drought-stressed seedlings of mutants.
Figure S5. Transcript level of OsSRO1c in transgenic plants.
Figure S6. Suppression of OsSRO1c showed increased sensitivity to osmotic stress.
Figure S7. Thermal images of the overexpression plants.
Figure S8. ABA content and stomatal responses to ABA in overexpression plants.
Figure S9. Loss of OsSRO1c function enhanced resistance to oxidative stress.
Figure S10. Quantitative PCR analysis of expression of ROS-scavenging genes.
Figure S11. Pairwise interaction test of OsSRO1c with rice DREBs and NACs.
Figure S12. Increased drought sensitivity of the overexpression lines.
Figure S13. Expression of DST in SNAC1-overexpressing plants.
Figure S14. ABA sensitivity of mutant and overexpression plants.
Table S1. Primer sequences used in this study.
Methods S1. Supplementary methods for plasmid construction, rice transformation, stress treatments, physiological measurements, yeast assay, BiFC assay, and RT–PCR.
Acknowledgements
We thank Professors Gynheung An (POSTECH, Korea) and Hongwei Xue (Shanghai Institute of Plant Physiology and Ecology) for providing the mutants ossro1c-1 and ossro1c-2, respectively, and Professor Jian Xu, Mr Lei Wang, and Rongjian Ye (Huazhong Agricultural University) for providing plasmid pM999-33, 35:CFP-GHD7, and DX2181, respectively. This work was supported by grants from the National Program for Basic Research of China (2012CB114305), the National Program on High Technology Development (2012AA10A303), the National Natural Science Foundation of China (30921091and 31271316), and the National Program of China for Transgenic Research (2011ZX08009-003-002, 2011ZX08001-003).
References
- Acharya BR, Assmann SM. 2009. Hormone interactions in stomatal function. Plant Molecular Biology 69, 451–462 [DOI] [PubMed] [Google Scholar]
- Ahlfors R, Brosche M, Kollist H, Kangasjarvi J. 2009. Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana . The Plant Journal 58, 1–12 [DOI] [PubMed] [Google Scholar]
- Ahlfors R, Lang S, Overmyer K, et al. 2004. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein–protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. The Plant Cell 16, 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55, 373–399 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell 123, 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP. 2008. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnology Letters 30, 2191–2198 [DOI] [PubMed] [Google Scholar]
- Chen Q, Wang Q, Xiong L, Lou Z. 2011. A structural view of the conserved domain of rice stress-responsive NAC1. Protein and Cell 2, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Tonelli C. 2010. Transcription factors controlling stomatal movements and drought tolerance. Transcription 1, 41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. 2001. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis . Molecular Genetics and Genomics 265, 954–963 [DOI] [PubMed] [Google Scholar]
- Du H, Wang N, Cui F, Li X, Xiao J, Xiong L. 2010. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiology 154, 1304–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT. 2004. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiology 134, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibe T, Saji H, Watahiki MK, Yamamoto KT. 2006. Overexpression of the RADICAL-INDUCED CELL DEATH1 (RCD1) gene of Arabidopsis causes weak rcd1 phenotype with compromised oxidative-stress responses. Bioscience, Biotechnology, and Biochemistry 70, 1827–1831 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI. 2003. The role of stomata in sensing and driving environmental change. Nature 424, 901–908 [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. 2006. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA 103, 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. 2009. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes and Development 23, 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashikina N, Deeken R, Fischer S, Ache P, Hedrich R. 2005. AKT2/3 subunits render guard cell K+ channels Ca2+ sensitive. Journal of General Physiology 125, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosche M, et al. 2009. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. The Plant Journal 60, 268–279 [DOI] [PubMed] [Google Scholar]
- Jaspers P, Overmyer K, Wrzaczek M, Vainonen JP, Blomster T, Salojarvi J, Reddy RA, Kangasjarvi J. 2010. The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants. BMC Genomics 11, 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu JK. 2006. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis . Proceedings of the National Academy of Sciences, USA 103, 18816–18821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. 2010. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology 61, 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis . EMBO Journal 22, 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K. 2010. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Molecular Genetics and Genomics 283, 185–196 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Clayton H, Mansfield TA, Hetherington AM. 1996. Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiology 111, 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7, 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E. 2010. Genetic engineering for modern agriculture: challenges and perspectives. Annual Review of Plant Biology 61, 443–462 [DOI] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. 2009. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Jr, Kangasjarvi J. 2000. Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. The Plant Cell 12, 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI. 2000. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734 [DOI] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. 2009. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis . The Plant Journal 57, 400–412 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. 2001a. Guard cell signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology 52, 627–658 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. 2001b. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410, 327–330 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, et al. 2009. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Letters 583, 2982–2986 [DOI] [PubMed] [Google Scholar]
- Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM. 2010. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiology 153, 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotia S, Lamb RS. 2009. The paralogous genes RADICAL-INDUCED CELL DEATH 1 and SIMILAR TO RCD ONE 1 have partially redundant functions during Arabidopsis thaliana development. Plant Physiology 151, 180–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, et al. 2004. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell 16, 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Song CP. 2008. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytologist 178, 703–718 [DOI] [PubMed] [Google Scholar]
- Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C. 2008. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Molecular Biology 67, 589–602 [DOI] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Herve P. 2008. Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3, e1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. 2008. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiology 148, 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767 [DOI] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Barrick D. 2005. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure 13, 1599–1611 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.