Abstract
Plant cell growth and morphogenesis depend on remodelling of both actin and microtubule cytoskeletons. AtFH1 (At5g25500), the main housekeeping Arabidopsis formin, is targeted to membranes and known to nucleate and bundle actin. The effect of mutations in AtFH1 on root development and cytoskeletal dynamics was examined. Consistent with primarily actin-related formin function, fh1 mutants showed increased sensitivity to the actin polymerization inhibitor latrunculin B (LatB). LatB-treated mutants had thicker, shorter roots than wild-type plants. Reduced cell elongation and morphological abnormalities were observed in both trichoblasts and atrichoblasts. Fluorescently tagged cytoskeletal markers were used to follow cytoskeletal dynamics in wild-type and mutant plants using confocal microscopy and VAEM (variable-angle epifluorescence microscopy). Mutants exhibited more abundant but less dynamic F-actin bundles and more dynamic microtubules than wild-type seedlings. Treatment of wild-type seedlings with a formin inhibitor, SMIFH2, mimicked the root growth and cell expansion phenotypes and cytoskeletal structure alterations observed in fh1 mutants. The results suggest that besides direct effects on actin organization, the in vivo role of AtFH1 also includes modulation of microtubule dynamics, possibly mediated by actin–microtubule cross-talk.
Key words: Actin, Arabidopsis, At5g25500, LatB, microtubules, SMIFH2, VAEM
Introduction
Plant growth, development, and morphogenesis are intimately associated with the dynamics of both microtubule and actin microfilament cytoskeletons (see, for example, Smith and Oppenheimer, 2005). Plant cell morphogenesis depends on mechanical properties of the cell wall, determined by organization of the cellulose microfibrils, interlinked with cortical microtubules (Emons et al., 2007). Microfilaments contribute less directly, for example via participation in membrane recycling (Bannigan and Baskin, 2005), though they are important in tip-growing cells such as root hairs (Peremyslov et al., 2010).
Root growth results from regulated cell divisions in the meristem, and anisotropic cell expansion and differentation in the elongation and differentiation zones. Mutations affecting the cytoskeleton often affect root growth or root hair development (Thitamadee et al., 2002; Gilliland et al., 2003; Abe and Hashimoto, 2005).
Formins (FH2 proteins) are key eukaryotic cytoskeletal regulators. Their hallmark FH2 domain can dimerize and nucleate actin (Blanchoin and Staiger, 2010). Seed plants have two formin clades with numerous paralogues (Deeks et al., 2002; Grunt et al., 2008); in vitro studies of several proteins demonstrated microfilament nucleation, capping, and binding (e.g. Ingouff et al., 2005; Yi et al., 2005). Metazoan formins also participate in remodelling the microtubular cytoskeleton (Bartolini and Gundersen, 2010). Similar observations were also reported for plant formins—Arabidopsis AtFH4 and AtFH14 (Deeks et al., 2010; Li et al., 2010) and rice FH5 (Yang et al., 2011; Zhang et al., 2011), which interact with microtubules using diverse mechanisms (see also Wang et al., 2012). AtFH4 is a class I formin, exhibiting the clade-specific structure with a signal peptide, a proline-rich extracellular domain, and a transmembrane domain in front of the conserved FH1 and FH2 domains (Cvrčková, 2000). It binds microtubules via a motif shared by a subgroup of class I formins, the GOE domain (Deeks et al., 2010). AtFH14 and rice FH5 are typical class II formins with a PTEN-related domain in front of FH1 and FH2 (Grunt et al., 2008); since they lack the GOE motif, they obviously bind microtubules by other means.
AtFH1 is the main housekeeping class I formin in Arabidopsis thaliana, as judged from its gene expression pattern (Zimmermann et al., 2004). It has the typical class I structure, associates with membranes (Banno and Chua, 2000; Cheung and Wu, 2004), and its extracellular domain may anchor the actin cytoskeleton across the plasmalemma into the cell wall (Martiniere et al., 2011). AtFH1 can nucleate and bundle actin (Michelot et al., 2005, 2006); it contains no known microtubule-binding motifs, and no discernible phenotype was described so far in mutants lacking AtFH1, although its transient overexpression caused loss of pollen tube polarity (Cheung and Wu, 2004).
Here the characterization of seedling root development in mutants harbouring T-DNA insertions in the AtFH1 locus is reported. While under normal conditions mutants exhibited no obvious phenotypic alterations, they were hypersensitive towards an anti-actin drug (alone or together with a microtubule inhibitor). Organization of microfilaments and microtubules in the mutant root cortex, as well as their dynamics, documented by variable-angle epifluorescence microscopy (VAEM; see Wan et al., 2011), differed from those of wild-type (wt) plants. The growth and cytoskeletal organization phenotypes were mimicked by treatment with a specific inhibitor of FH2 domain function (Rizvi et al., 2009). Thus, AtFH1 appears to participate in regulation of cytoskeletal dynamics in vivo by a mechanism involving cross-talk between actin and microtubules.
Materials and methods
Plants
Two T-DNA insertional mutants (fh1-1, SALK-032981; and fh1-2, SALK-009693) in the AtFH1 gene (At5g25500) were obtained from the SALK Institute (Alonso et al., 2003). To determine AtFH1 allelic status, PCR using primers fh1-1-LP (5’GTCTCCGTCACTGTCGTTAGC3’) with fh1-1-RP (5’TTGTTGTTTAACGACTTCGCC3’) was employed to detect the wt allele in crosses involving fh1-1, and fh1-2-LP (5’TG TTTGTGTAGGCTGCTTGTG3’) with fh1-2-RP (5’ATTCTTTCGTG GTACACACGG3’) for the wt allele in crosses of fh1-2. For mutant alleles, the RP primers were combined with the SALK primer LBb1.3: 5’ATTTTGCCGATTTCGGAAC3’ for the T-DNA insertion.
Mutants were crossed with green fluorescent protein (GFP)–MAP4 and GFP–FABD reporter lines (Marc et al., 1998; Ketelaar et al., 2004) as described (Cole et al., 2005). Media with kanamycin and BASTA® were used to select GFP–MAP4- and GFP–FABD-carrying plants, respectively, and fluorescence was evaluated microscopically. Genotyping to select fh1 homozygotes was done in the second and third generation.
RT–PCR
RNA was isolated from 7-day-old seedlings using the RNeasy Plant kit (Qiagen). First-strand cDNA synthesis and semi-quantitative reverse transcription–PCR (RT–PCR; with β-actin-specific primers for control) were performed according to Dvořáková et al. (2007) using 30 cycles, DreamTaq polymerase (Fermentas), and AtFH1-specific primers (5’GGATCCAGAAGAAAGAAGAAGATAACACAATGC3’ and 5’CTGAGCCTTCTTCGGGTCCAGG3’). The 2042bp product was visualized by agarose gel electrophoresis.
Growth conditions and inhibitor treatments
Inhibitor treatment experiments were performed according to Collings et al. (2006). Seed germination was synchronized by several days at 4 °C, followed by growth on vertical Murashige and Skoog (MS) plates for 4–5 d at 22 °C with a 16h light/8h dark cycle prior to transfer on inhibitor-containing media, which were then incubated under the same conditions for 72h, unless stated otherwise. Inhibitor stock solutions were prepared in dimethylsulphoxide (DMSO), stored at −20 °C [latrunculin B (LatB), oryzalin (Oryz), taxol, and jasplakinolide] or 4 °C (SMIFH2), and added to liquid agar to the desired concentrations; the DMSO concentration was adjusted to 0.2% (v/v). All inhibitors were purchased from Sigma. Effective doses were calculated using the R statistical software (http://www.r-project.org/index.html) according to Knezevic et al. (2007) from two or three replications of ~20 plants for each concentration.
Morphometric analyses
Root diameter and root growth (defined as increment in length in a specified interval of time) was determined from photographs taken at 24, 48, and 72h after transfer with a digital camera (Olympus C5050), measuring the distances between the root tips and marks made on the rear of the plates at tip locations at transfer time. To determine root hair density, root hairs were counted under a light microscope (BX-51, Olympus) at ×10 magnification in a 2mm region at the midpoint of the portion of root grown after transfer. Lengths of 10 root hairs from the midpoint of each measured region were measured at ×20 magnification. From the same zone, root diameter and the lengths of 10 trichoblasts and 10 atrichoblasts per root were estimated. In all experiments, 2–3 replicates of ~20 plants were used per data point. Measurements were performed using the ImageJ software (http://rsbweb.nih.gov; Abramoff et al., 2004).
Confocal microscopy and image analysis
GFP-tagged cytoskeleton was observed in roots of 5-day-old seedlings using a confocal laser scanning microscope (LCS 510; Leica) with a ×63/1.2 water immersion objective and 488nm argon laser (25 mW) excitation. Images were acquired as z-series with a 0.7–1 µm interval. Microfilament bundling and density were quantified according to van der Honing et al. (2012) and Higaki et al. (2010). Profiles of fluorescence intensity were divided into four classes of grey level (arbitrary units) to generate plots documenting microfilament bundling (low intensity represents weakly labelled bundles or single filaments; high intensity corresponds to brightly labelled bundles). Skewness of fluorescence intensity distribution (correlated with microfilament bundling because bundles exhibit brighter fluorescence) and occupancy (i.e. fraction of pixels constituting the skeletonized microfilaments relative to the total pixel number of the analysed region, proportional to the overall microfilament density) were determined using the ImageJ plugins and macros from Higaki′s laboratory (http:/hasezawa.ib.k.u-tokyo.ac.jp/zp/Kbi/HigStomata). Microtubule density was determined as the number of microtubules in an area of 500 µm2 from confocal images in five cells from several plants.
VAEM
To evaluate cytoskeletal dynamics, we used the Leica AF6000 LX fluorescence platform with integrated TIRF module, the HCX PL APO ×100/1.46 oil immersion objective, 400nm peak excitation, and 210ms exposure time. Plants were mounted in water on chambered slides; images were captured with a Leica DFC350FXR2 digital camera at 0.5 s intervals over the course of 2min and analysed with Leica Application Suite (LAS) and ImageJ software. Kymographs were generated using the Multiple Kymograph ImageJ plug-in from a time-lapse image series collected from well-focused 30 µm long ‘optical transects’ parallel to the longitudinal axis of the root (Sampathkumar et al., 2011). The distribution of microtubule growth and shrinkage rates was estimated from at least 250 microtubule ends from at least 50 atrichoblasts in each genotype or treatment.
Results
Cytoskeletal inhibitors differentially affect root growth in fh1 mutant and wild-type seedlings
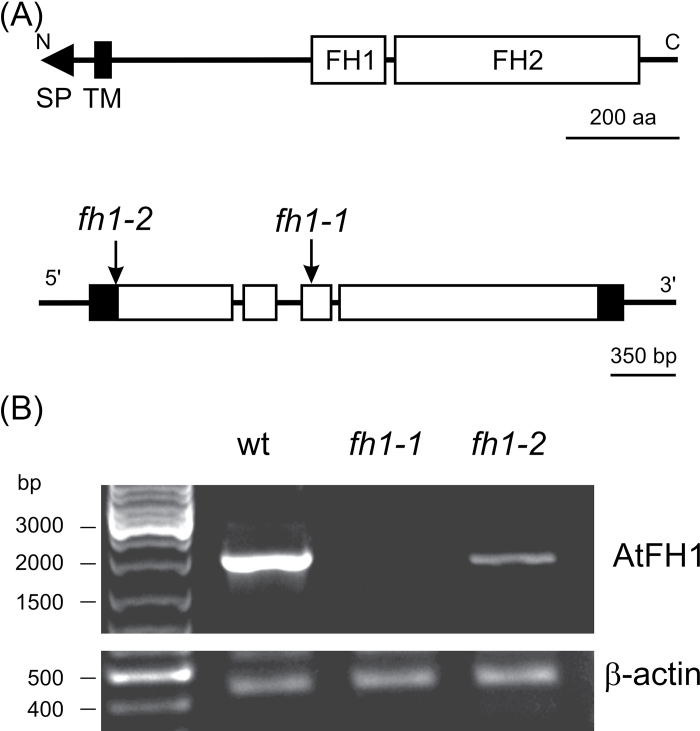
Two Arabidopsis T-DNA mutant lines, fh1-1 and fh1-2, with corresponding wt controls were characterized. The T-DNA insertion interrupts the AtFH1 gene in the third exon in fh1-1 and in the 5’ untranslated region (UTR; 27bp before start codon) in fh1-2 (Fig. 1A). In homozygous seedlings, AtFH1 mRNA was undetectable in fh1-1, while fh1-2 had a reduced transcript level (Fig. 1B).
Fig. 1.
The AtFH1 (At5g25500) locus and mutants. (A) AtFH1 protein domain structure (above); map of the AtFH1 gene and location of T-DNA insertions (below: open boxes, coding exons; filled boxes, non-coding exons; lines, introns and non-transcribed sequences). (B) AtFH1 transcripts in wt and homozygous mutant seedlings determined by semi-quantitative RT–PCR.
Under standard growth conditions in soil or in vitro, fh1-1 and fh1-2 plants do not differ noticeably from the wt. The in vitro growth media were thus supplemented with anti-cytoskeletal drugs LatB and/or Oryz to enhance expected subtle cytoskeletal defects and uncover novel mutant phenotypes.
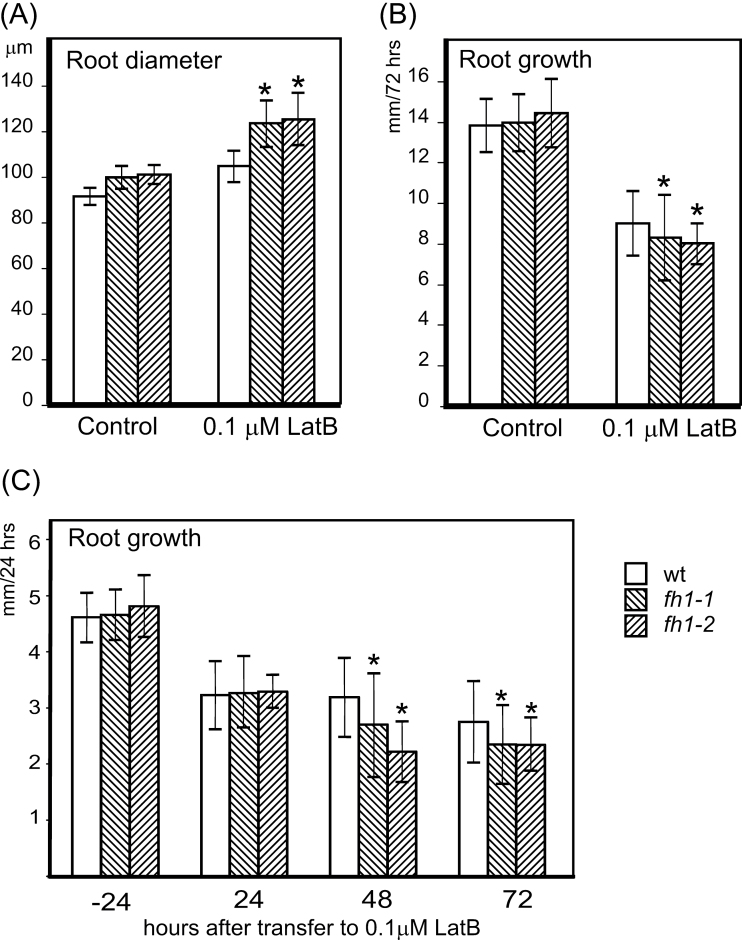
At 0.1 µM concentration, the actin polymerization inhibitor LatB caused a more severe increase in root diameter and reduction in the longitudinal root growth rate in young seedlings of both mutant lines compared with the wt; the difference developed gradually within the first 48h on LatB (Fig. 2). Higher concentrations severely affected both genotypes, and the difference between the mutant and wt was no longer significant (Supplementary Fig. S1A, B available at JXB online).
Fig. 2.
Mutants lacking AtFH1 exhibit thicker, slower growing roots than the wt when treated with 0.1 µM LatB. (A) Root diameter and (B) incremental root growth during 72h after transfer to LatB. Significant differences between any of the mutants and the wt in root diameter (t-test P < 0.0001) or root growth (t-test P < 0.05) are marked by asterisks. (C) Gradual decrease in root growth rates after 24, 48, and 72h on LatB. Significant differences between mutant and wt seedlings (t-test P < 0.0001) are marked by asterisks; data from the last 24h before transfer are shown for control.
While the microtubule-depolymerizing drug Oryz also caused root thickening and reduced root growth, its effect was similar in both fh1 and wt seedlings. (Supplementary Fig. S1C, D at JXB online). However, simultaneous addition of 0.33 µM LatB (i.e. a concentration that equally affected mutant and wt roots) increased the sensitivity of fh1 mutants to a low concentration of Oryz compared with the wt (Supplementary Fig. S1E).
Next, the inhibitor concentrations at which root diameter showed half the maximal increase (D50) and at which roots showed a 50% reduction in growth rate (L50) were estimated from dose–response curves of mutant and wt seedlings. Radial root expansion was always more sensitive to inhibitors than longitudinal growth. For LatB, both D50 and L50 were significantly lower in the fh1 mutants than in the wt (Table 1).
Table 1.
Effective doses of LatB and Oryz in mutants and the wild type.
| Treatment | D50 | L50 | ||||||
|---|---|---|---|---|---|---|---|---|
| fh1-1 | Wt | fh1-1 | Wt | |||||
| Lat B | 10.1** | 28.3 | 131.6** | 163.7 | ||||
| Oryz | 96* | 114.4 | 233.1 | 224.3 | ||||
D50, inhibitor concentration causing response half way between zero and the maximal observed diameter increase; L50, inhibitor concentration causing response half way between zero and the maximal observed growth reduction.
*Significant difference from the wt at P < 0.05; **significant difference from the wt at P < 0.001.
Treatment with cytoskeleton-stabilizing drugs (jasplakinolide for actin or taxol for microtubules) resulted in reduced root growth and increased diameter in both fh1-1 mutant and wt seedlings. Both genotypes responded similarly, although longitudinal growth of mutant roots was significantly less affected by taxol (Supplementary Fig. S2 at JXB online).
Cytoskeletal inhibitors affect cell expansion and root hair development in mutants
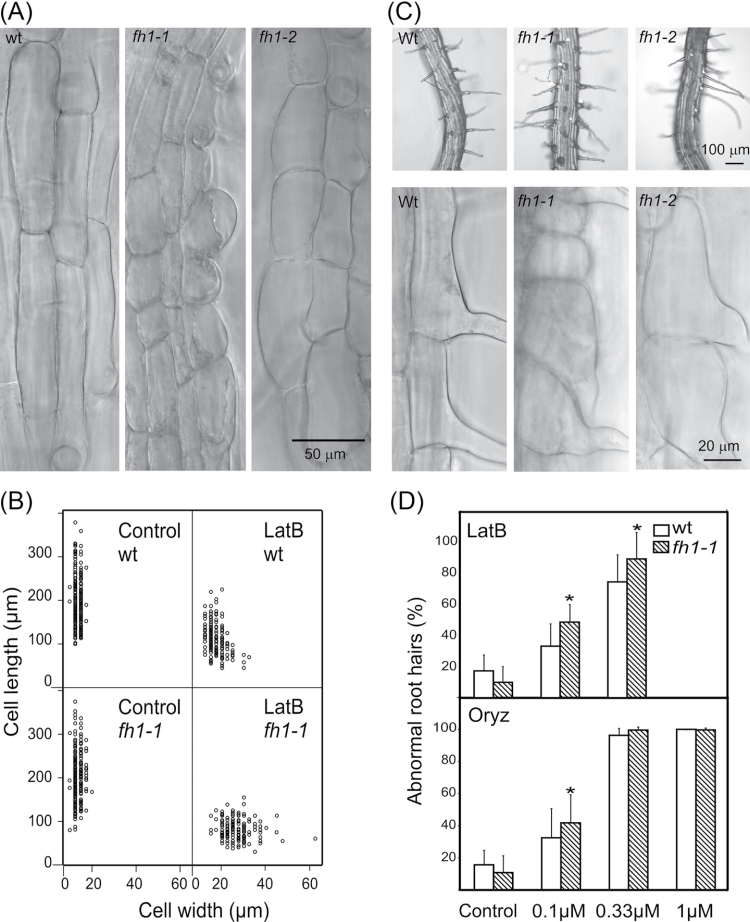
Reduced longitudinal root growth can be due to impaired cell division or elongation, or both. To evaluate the contribution of cell elongation, the length of mature trichoblasts and atrichoblasts in inhibitor-treated roots was measured. LatB-grown fh1-1 and fh1-2 roots had shorter, wider cells, suggesting that the phenotype is at least partly due to more isodiametric cell growth (Fig. 3A, B; Supplementary Table S1 at JXB online).
Fig. 3.
Effects of LatB on rhizodermis and root hair development in fh1 mutant and wt seedlings. (A) Typical appearance of elongation zone rhizodermis in wt and mutants exposed to LatB. (B) Relationship between mature rhizodermis cell length and width in fh1-1 mutant and wt seedlings in control conditions and on 0.1 µM LatB (each sample contains equal numbers of trichoblasts and atrichoblasts); compare Supplementary Table S1 at JXB online for fh1-2. (C) Abnormal root hairs found in mutant but not wt plants grown on 0.1 µM LatB. (D) Percentage of abnormal root hairs in fh1-1 and wt plants grown in LatB- and Oryz-supplemented media. Significant differences (t-test P < 0.001) are marked by an asterisk.
Mutant rhizodermis cells, especially trichoblasts, were often mis-shapen, exhibiting bulbous structures at root hair bases and/or branched root hairs (Fig. 3C). A significantly higher density of both total and abnormal root hairs was found in mutant, but not wt, seedlings grown on 0.1 µM LatB compared with drug-free control, apparently due to shorter trichoblasts. At 0.33 µM LatB, the total number of root hairs was reduced in both genotypes; mutants had more abnormal root hairs than the wt. A further increase in the LatB concentration completely inhibited root hair development. While fh1 mutants showed, on average, longer root hairs than the wt on control media or 0.1 µM LatB or Oryz, their root hairs were shorter on 0.33 µM LatB, suggesting increased sensitivity of tip growth to higher LatB doses. However, since root hair length varied substantially, the biological significance of this observation is questionable (Fig. 3D; Supplementary Table S1 at JXB online).
Actin and microtubule distribution in fh1 mutants
In the above experiments, both fh1 allelles behaved similarly, though fh1-1 had more pronounced phenotypes, in agreement with the residual gene expression in fh1-2. fh1-1 was thus chosen for introduction of in vivo fluorescent protein-tagged cytoskeletal markers (GFP–FABD for actin and GFP–MAP4 for microtubules) by crossing. Sister segregants carrying wt AtFH1 were used as controls.
The effects of the markers themselves on root growth in both fh1-1 and the wt were examined. GFP–MAP4 caused root thickening and reduction of root growth, and induced left-handed root twisting, as described previously (Granger and Cyr, 2001; Hashimoto, 2002); these effects were less pronounced in fh1-1mutants than in the wt. GFP–FABD did not show any significant effects in either fh1-1 or wt seedlings (Supplementary Fig. S3 at JXB online).
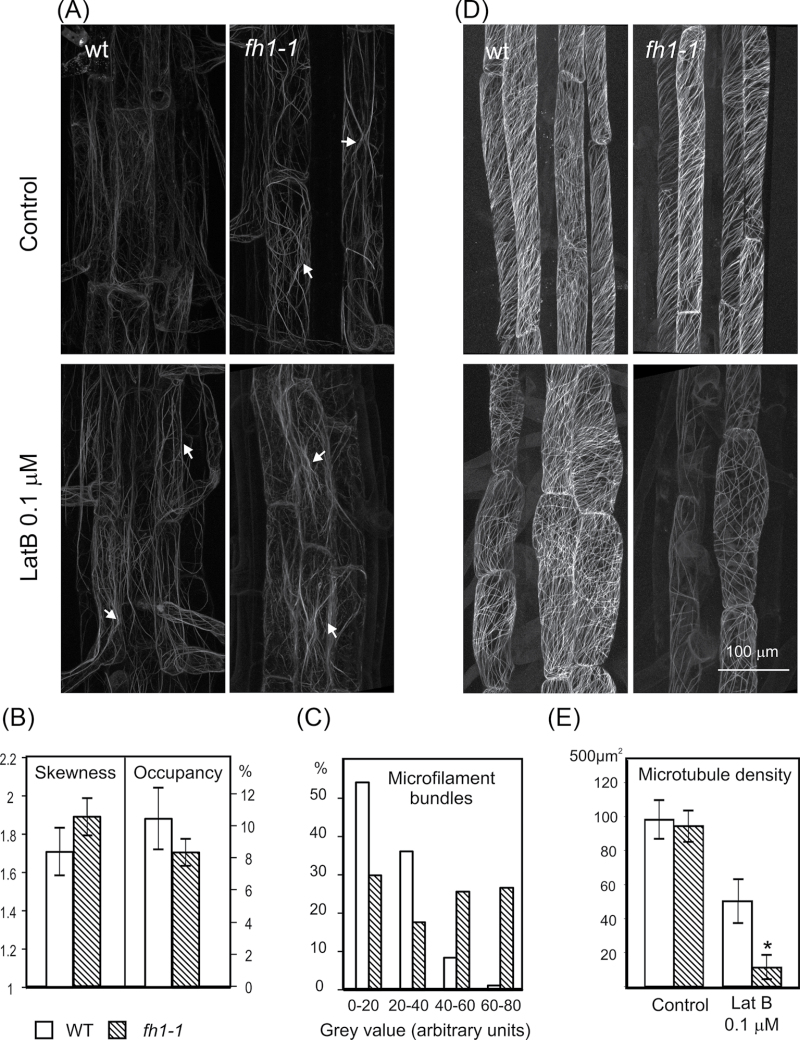
Given that this study was looking at root development, the focus here was on in vivo observations in rhizodermal cells. Thicker and more frequent actin bundles were usually observed in fh1 mutants than in wt seedlings. Low doses of LatB did not disrupt filaments but rather increased actin bundling, more obviously in mutants than in wt plants. LatB-treated wt plants thus somewhat resembled fh1 mutants grown under control conditions (Fig. 4A). Quantification of the microfilament patterns in the rhizodermis of seedlings growing on control media by estimating the skewness of fluorescence intensity distribution (correlated with the level of microfilament bundling) and pixel occupancy (giving insight into the overall density of actin cytoskeleton) showed that mutants have fewer but thicker microfilaments, consistent with increased actin bundling (Fig. 4B). The differences are even more obvious in profiles of individual bundle fluorescence intensity (Fig. 4C), confirming that fh1 mutants have fewer weakly labelled thin bundles or single filaments, and more bright thick bundles than wt plants.
Fig. 4.
Typical cytoskeleton organization in the rhizodermis of fh1-1 mutant and wt seedlings. (A) Actin labelled by GFP–FABD; arrows, actin filament bundles. (B) Actin filament bundling (skewness) and density (occupancy) under control conditions. (C) Frequency distribution of actin fluorescence peaks in four fluorescence intensity classes under control conditions. (D) Microtubules labelled by GFP–MAP4. (E) Microtubule density. Significant differences (t-test P < 0.001) are marked by an asterisk.
Surprisingly, differences in microtubule organization between the wt and mutants were more pronounced than those in microfilaments. Even on control media, and more obviously in LatB-treated plants, mutants had fewer microtubules, shorter and less organized compared with the wt (Fig. 4D). Quantitative measurements of microtubule density revealed a significant reduction in LatB-treated fh1 mutants compared with the wt (Fig. 4E).
Effect of fh1 mutation on cytoskeletal dynamics monitored by VAEM
To compare individual microfilament and microtubule dynamics in rhizodermis cells of wt and fh1 mutant plants carrying GFP–FABD and GFP–MAP4, the VAEM technique was employed. Since preliminary experiments indicated that the three developmental zones of the root tip differ in cytoskeletal dynamics, the beginning of the differentiation zone was investigated, where both cytoskeletal systems behaved consistently very dynamically.
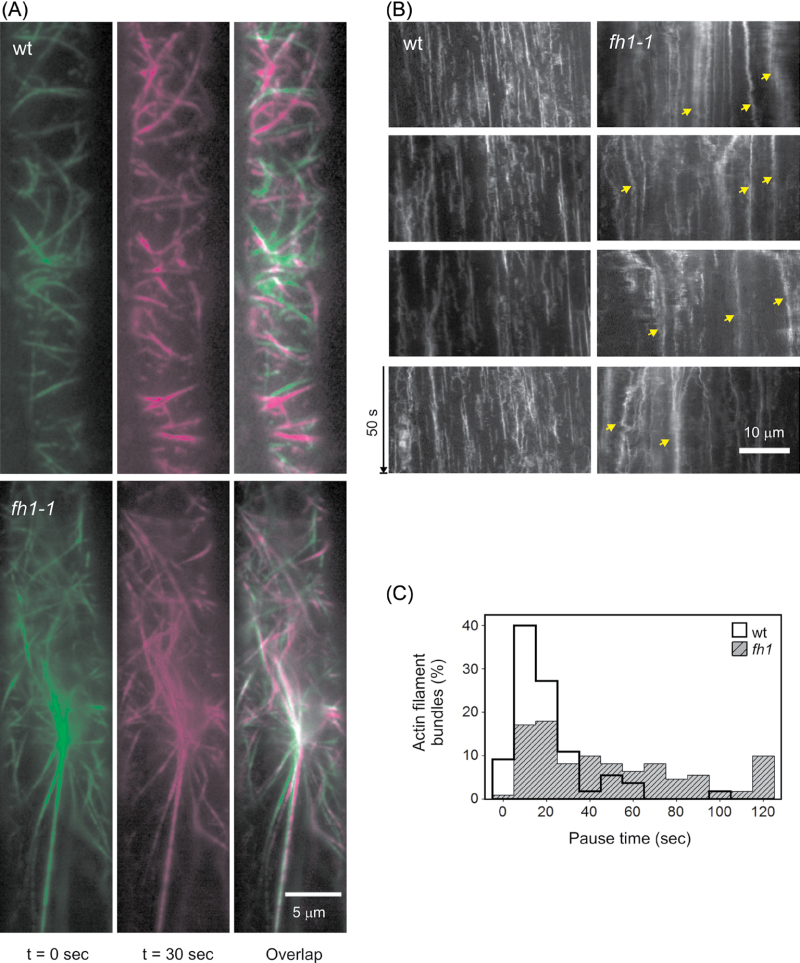
Differences in actin dynamics were observed between fh1 mutants and the wt (Fig. 5; Supplementary Video S1, S2 at JXB online). Mutant microfilament bundles were more abundant and less dynamic (in particular, they remained longer at pause) than those of wt seedlings, except a few rapidly moving bundles. This might reflect differences either in bundle size or in the degree of actin cross-linking.
Fig. 5.
GFP–FABD-tagged microfilament distribution and dynamics in the rhizodermis of fh1-1 and wt seedlings on standard medium. (A) VAEM images from two time points and their overlap showing growing or moving filaments in magenta, shrinking in green, and pausing and growing/shrinking in light green and light magenta, respectively. (B) Kymograph showing the static thick actin bundles in the mutant (arrows). (C) Distribution of actin bundle pause duration in mutant and wt.
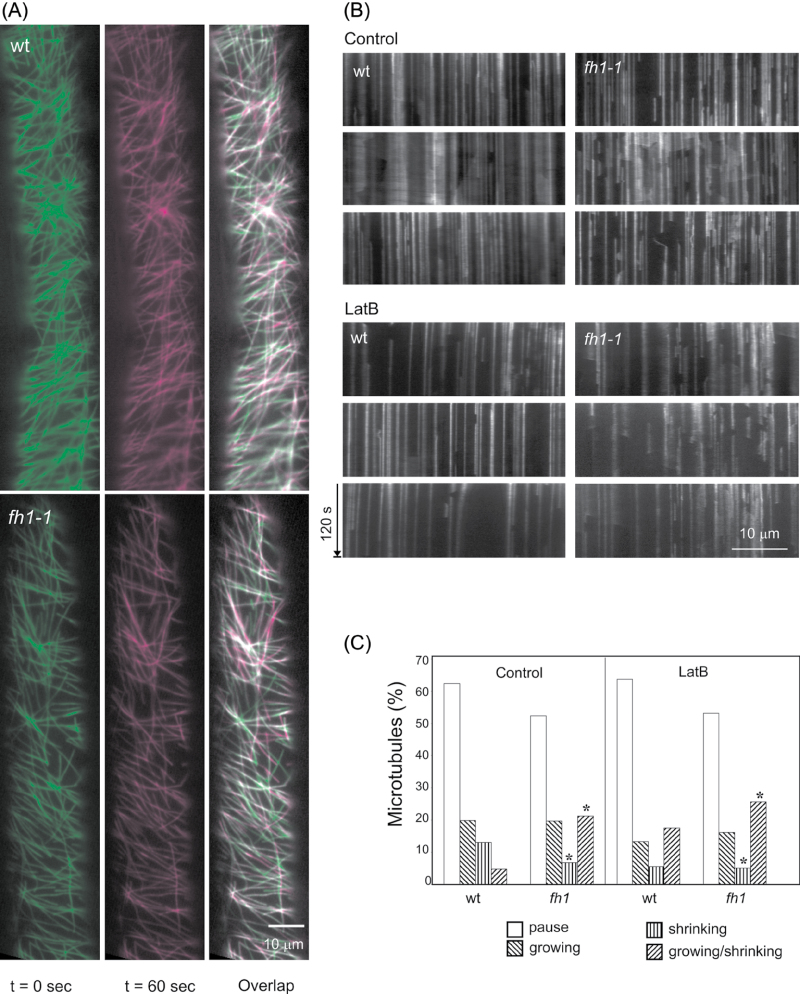
Differences between mutant and wt plants were also observed in microtubule dynamics (Fig. 6; Supplementary Video S3, S4 at JXB online). On the control medium, mutant microtubules exhibited increased dynamic instability compared with wt seedlings. LatB increased microtubule dynamics in both genotypes (Fig. 6A, B).
Fig. 6.
GFP–MAP4-tagged microtubule distribution and dynamics in fh1-1 mutant and wt rhizodermis. (A) VAEM images from two time points and their overlap showing growing microtubules in magenta, shrinking in green, pausing in light green, and growing/shrinking in light magenta. (B) Kymographs of microtubule dynamics under control conditions and on 0.1 µM LatB-supplemented medium. (C) Distribution of microtubule phases on control and 0.1 µM LatB-containing media. An asterisk indicates a significant difference between mutants and the wt (t-test P < 0.0001).
To quantify microtubule turnover, the distribution of microtubule phases was determined in images taken during the time span of 2min. Mutants had fewer shrinking or pausing microtubules but more microtubules undergoing stochastic transition (i.e. alternatively shrinking and growing) than the wt (Fig. 6C). LatB reduced the fraction of growing microtubules in both genotypes, and increased the fraction of growing/shrinking microtubules even in the wt (again, LatB-treated wt plants resembled fh1 mutants grown under control conditions). Oryz in both genotypes increased the percentage of pausing microtubules and reduced the growing, shrinking, and growing/shirinking fractions. The distribution of microtubule growth and shrinkage rates differed somewhat between fh1 and wt roots (Supplementary Fig. S4 at JXB online). Despite comparable average growth rates, a higher proportion of microtubules in fh1 cells grew more slowly than average; this difference persisted upon LatB treatment, while Oryz reduced the growth rate in both the fh1 mutant and the wt.
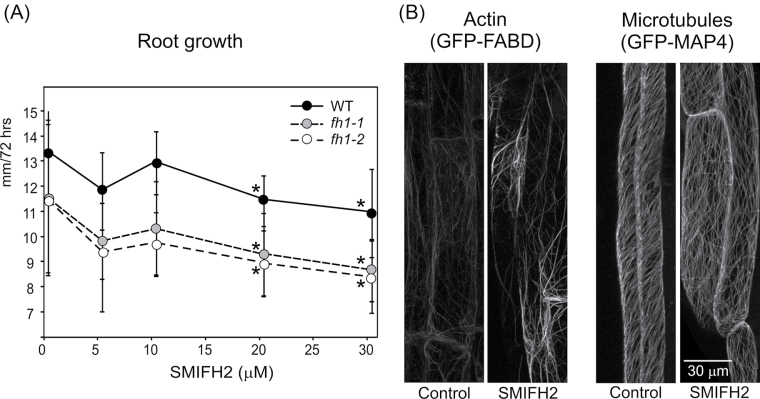
Effects of the formin inhibitor SMIFH2 mimic the fh1 mutation
To verify that the observed mutant phenotypes are due to disrupted formin function, the effects of a recently described inhibitor of formin-mediated actin assembly, SMIFH2 (Rizvi et al., 2009), were examined in wt seedlings. In the standard experimental set-up, significant reduction of root growth was observed at or above a concentration of 20 µM in both the wt and fh1 mutants (Fig. 7A). The effect of SMIFH2 was stronger when seedlings were exposed to the drug in the dark (possibly due to light sensitivity of the drug), and fh1-1 mutant roots were significantly more affected than those of the wt (Supplementary Fig. S5 at JXB online). SMIFH2-treated wt seedlings expressing GFP–FABD and GFP–MAP4 exhibited increased microfilament bundling and reduced microtubule density, especially after additional LatB treatment, again reminiscent of fh1 mutants (Fig. 7B).
Fig. 7.
Effects of SMIFH2 on longitudinal root growth and cytoskeletal organization. (A) Concentration-dependent growth inhibition; asterisks denote significant differences from non-treated seedlings of the same genotype (t-test P < 0.0001). (B) Rhizodermal microfilament and microtubule organization under control conditions and on 20 µM SMIFH2-supplemented medium.
Discussion
The first description is presented of a mutant phenotype in A. thaliana lacking the most expressed housekeeping class I formin, AtFH1. It is shown that AtFH1 affects actin and microtubule dynamics, processes central for cell expansion and development.
Using cytoskeletal inhibitors to uncover mutant phenotypes
Angiosperm FH2 proteins form a large family of paralogues: A. thaliana has 21 formin-encoding genes, 11 of them in class I. Ten of these (including AtFH1) share the characteristic clade-specific domain structure (Deeks et al., 2002; Grunt et al., 2008). Loss of a single formin gene thus rarely causes obvious phenotypic effects due to ‘functional redundancy’. Only subtle, if any, phenotypes have so far been documented for loss-of-function class I formin mutants. Such phenotypes are usually tissue specific, reflecting the pattern of gene expression. Loss of AtFH5 caused delayed endosperm cytokinesis (Ingouff et al., 2005), and pollen tube defects were elicited by RNA interference (RNAi) targeting the pollen formins AtFH3 in Arabidopsis or NtFH5 in tobacco (Ye et al., 2009; Cheung et al., 2010). Additional phenotypes were produced by overexpression, sometimes ectopic or heterologous, of wt or mutant proteins, such as AtFH1 (Cheung and Wu, 2004) or AtFH8 (Deeks et al., 2005; Yi et al., 2005).
Asymptomatic or mildly symptomatic doses of inhibitors of specific cellular functions may result in a ‘synthetic phenotype’ in mutants where the inhibitor’s target(s) are already weakened. In mutants of Arabidopsis formins AtFH8 (Xue et al., 2011) and AtFH12 (Cvrčková et al., 2012), LatB induced alterations in roots and/or root hairs. In the present report, the response of T-DNA mutants with insertions in AtFH1 to cytoskeletal inhibitors targeting either actin (LatB) or microtubules (Oryz) was examined, since no readily noticeable differences between fh1 mutants and the wt were observed under control conditions.
Low doses of LatB, inhibiting primary root growth and causing radial swelling in young seedlings, and enhancing the phenotype of some cytoskeletal mutations (Collings et al., 2006), affected the fh1 mutants more than the wt. However, the whole organ phenotype was subtle compared with effects on the level of individual cells or cytoskeletal structures, providing yet another example of organ- and tissue-level compensation of cell-level defects (see Breuninger and Lenhard, 2010). The shorter, thicker roots of LatB-treated mutants consisted of shorter and wider cells, suggesting altered cell expansion rather than cell division, consistent with previous observations (Baluška et al., 2001). LatB can also disrupt intracellular membrane trafficking (Zhang et al., 2010), crucial for polar auxin transport. Since auxin, in turn, affects actin, it is difficult to separate direct and auxin-mediated effects on root growth (Rahman et al., 2007).
LatB-treated fh1 mutants also exhibited malformed root hairs. Unlike pollen tubes ectopically overexpressing AtFH1, which have bulbous tips (Cheung and Wu, 2004), in the experiments presented here, mainly root hair bases were affected, resembling the phenotype of actin (act2) mutants (Gilliland et al., 2002; Nishimura et al., 2003) and suggesting defective focusing of exocytosis during the bulge stage.
Disruption of microtubules affected fh1 mutants and the wt similarly, consistent with AtFH1 functioning mainly through actin. However, mutants exhibit increased sensitivity to Oryz in the presence of LatB, suggesting that AtFH1 may participate in a cross-talk between microfilaments and microtubules, and that its loss might, under some circumstances, destabilize microtubules. Consistently, mutants are partially resistant towards the root growth inhibition, radial root swelling, and root twisting induced by the GFP–MAP4 marker and taxol, which can stabilize and bundle microtubules (Granger and Cyr, 2001; Hashimoto, 2002).
Formin inhibition mimics the mutant phenotype
The small molecule SMIFH2, a 2-thio-oxodihydropyrimidine-4,6-dione derivative, is an inhibitor of FH2 domain-mediated actin assembly, active in vitro against several formins, and eliciting actin-related phenotypes in yeast and mammalian cells (Rizvi et al., 2009). Its in vitro characterized targets represent sufficiently distant formin clades (see Grunt et al., 2008) to suggest that SMIFH2 should inhibit most or all formins.
In the present study, SMIFH2 reduced root growth, increased microfilament bundling, and decreased cortical microtubule density; that is, it mimicked some phenotypes observed in fh1 mutants (especially after LatB treatment). Consistent with SMIFH2 also targeting the remaining formins, fh1 mutants were still responding to the inhibitor. The stronger mutant allele, atfh1-1, was even somewhat more sensitive towards root growth inhibition in the dark, reminiscent of increased sensitivity of some cytoskeletal mutants to inhibitors (see above). While non-specific effects of SMIFH2 cannot be ruled out, as its reported inactive analogue (Rizvi et al., 2009) is not commercially available, the present observations support the notion that the mutant phenotypes are indeed due to perturbation of formin function.
Changes in actin and microtubule distribution and dynamics in fh1 mutants
Plant actin and microtubule networks undergo constant remodelling (Staiger et al., 2009; Blanchoin et al., 2010). They are mutually interdependent, and sometimes co-aligned; microtubule-disrupting drugs may affect actin organization, and vice versa (Collings et al., 2006; Smertenko et al., 2010; Sampathkumar et al., 2011). The actin–microtubule ‘cross-talk’ may be mediated by bifunctional proteins or protein complexes (see Petrášek and Schwarzerová, 2009).
The thicker, more compact actin bundles in the fh1 mutants are reminiscent of some Arabidopsis mutants with an altered balance between fine actin filaments and bundles, such as adf4 (Henty et al., 2011) or aip1 (Ketelaar et al., 2004). AtFH1 might stabilize microfilaments by bundling (Michelot et al., 2005, 2006), enhanced polymerization, or capping, as reported for its relative AtFH8 (Yi et al., 2005). Low doses of LatB also disrupt fine actin filaments, resulting in increased actin bundling and reduced stochastic dynamics (Staiger et al., 2009). It is thus not surprising that LatB enhanced the effects of the fh1 mutation and mimicked its phenotype in wt plants. Consistent with AtFH1 participating in actin–microtubule cross-talk, LatB also aggravated or phenocopied the presumably microtubule-related cell expansion phenotypes.
To gain insight into cytoskeletal dynamics in wt and mutant plants, VAEM, a fluorescence microscopy technique allowing time-lapse imaging of a thin cortical layer of the cytoplasm, recently also adopted in plants (Smertenko et al., 2010; Sparkes et al., 2011; Vizcay-Barrena et al., 2011; Wan et al., 2011), was used. Increased bundling and decreased dynamics of the cortical actin in fh1 mutants were observed, suggesting altered actin-bundling, capping, or severing activities. Indeed, some formins can sever actin (Harris et al., 2004; Yi et al., 2005), thereby contributing to overall actin mobility, and AtFH1 may also have this ability. AtFH1 also anchors actin filaments across the plasmalemma into the cell wall (Martiniere et al., 2011), which may effectively constrain bundling.
As suggested already by the root growth phenotypes discussed above, fh1 mutants exhibited increased microtubule dynamics (important for cell elongation; Shaw et al., 2003), although the plus-end growth rates were remarkably decreased. There are multiple documented cases of formins participating in actin–microtubule cross-talk or binding to microtubules (Bartolini and Gundersen, 2010; Chesarone et al., 2010).
Particular microtubule-binding motifs may be restricted to narrow formin lineages (Deeks et al., 2010; Li et al., 2010; Yang et al., 2011; Zhang et al., 2011). Formins might also bind microtubules indirectly via heterodimerization with tubulin-binding paralogues, though heterodimerization is so far documented only among closely related mammalian Diaphanous formins (Copeland et al., 2007). The microtubule-related effects may also be mediated by other microtubule-associated proteins; co-expression of AtFH1 with the At3g16060 kinesin (see data from http://string-db.org; Szklarczyk et al., 2011) is interesting in this respect. However, since AtFH1 is excluded from the areas of cell cortex occupied by microtubules (Martiniere et al., 2011), the effects on microtubule dynamics may be secondary to those on microfilaments.
In summary, phenotypic effects of loss of function of AtFH1, which altered root cell expansion, root hair morphogenesis, and cytoskeletal dynamics especially under conditions perturbing the actin cytoskeleton, were documented. Consistent effects were also elicited by the formin inhibitor SMIFH2. These results suggest the participation of AtFH1 in actin–microtubule cross-talk in vivo by an as yet unclear mechanism.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Trichoblast, atrichoblast, and root hair characteristics in inhibitor-treated wt and mutant seedlings.
Figure S1. Dose–response curves of wt and fh1-1 root growth parameters for varying concentrations of cytoskeletal inhibitors.
Figure S2. Effects of taxol and jasplakinolide on wt and fh1-1 root growth.
Figure S3. Effects of GFP–MAP4 and GFP–FABD on wt and fh1-1 root growth.
Figure S4. Distribution of microtubule growth and shrinkage rates in fh1-1 mutant and wt seedlings.
Figure S5. Effects of SMIFH2 on wt and fh1 root growth under dark conditions.
Video S1. Actin dynamics in wt rhizodermis under control conditions.
Video S2. Actin dynamics in fh-1 rhizodermis under control conditions.
Video S3. Microtubule dynamics in wt rhizodermis under control conditions.
Video S4. Microtubule dynamics in fh1-1 rhizodermis under control conditions.
Acknowledgements
This work was supported by the MSM 0021620858, GACR P305/10/0433, and SVV 265203 projects. We thank Aleš Soukup, Ondřej Šebesta, and Ondřej Horvath for help with microscopy and image analysis, Marta Čadyová for technical support, and two anonymous reviewers for helpful comments.
References
- Abe T, Hashimoto T. 2005. Altered microtubule dynamics by expresion of modified alfa-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. The Plant Journal 43, 191–204 [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11, 36–42 [Google Scholar]
- Alonso J, Stepanova A, Leisse T, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- Baluška F, Jasik J, Edelmann H, Salajova T, Volkmann D. 2001. Latrunculin B-induced plant dwarfism: plant cell elongation is F-actin-dependent. Developmental Biology 231, 113–124 [DOI] [PubMed] [Google Scholar]
- Bannigan A, Baskin T. 2005. Directional cell expansion—turning toward actin. Current Opinion in Plant Biology 8, 619–624 [DOI] [PubMed] [Google Scholar]
- Banno H, Chua N. 2000. Characterization of the Arabidopsis formin-like protein AFH1 and its interacting protein. Plant and Cell Physiology 41, 617–626 [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen G. 2010. Formins and microtubules. Biochimica et Biophysica Acta 1803, 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Henty J, Khurana P, Staiger C. 2010. Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Current Opinion in Plant Biology 13, 714–723 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Staiger C. 2010. Plant formins: diverse isoforms and unique molecular mechanism. Biochimica et Biophysica Acta 1803, 201–206 [DOI] [PubMed] [Google Scholar]
- Breuninger H, Lenhard M. 2010. Control of tissue and organ growth in plants. Current Topics in Developmental Biology 91, 185–220 [DOI] [PubMed] [Google Scholar]
- Chesarone M, DuPage A, Goode B. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nature Reviews. Molecular Cell Biology 11, 62–74 [DOI] [PubMed] [Google Scholar]
- Cheung A, Niroomand S, Zou Y, Wu H. 2010. A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proceedings of the National Academy of Sciences, USA 107, 16390–16395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A, Wu H. 2004. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. The Plant Cell 16, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R, Synek L, Žárský V, Fowler J. 2005. SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiology 138, 2005–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings D, Lill A, Himmelspach R, Wasteneys G. 2006. Hypersensitivity to cytoskeletal antagonists demonstrates microtubule–microfilament cross-talk in the control of root elongation in Arabidopsis thaliana. New Phytologist 170, 275–290 [DOI] [PubMed] [Google Scholar]
- Copeland S, Green B, Burchat S, Papalia G, Banner D, Copeland J. 2007. The diaphanous inhibitory domain/diaphanous autoregulatory domain interaction is able to mediate heterodimerization between mDia1 and mDia2. Journal of Biological Chemistry 282, 30120–30130 [DOI] [PubMed] [Google Scholar]
- Cvrčková F. 2000. Are plant formins integral membrane proteins?. Genome Biology 1,, RESEARCH001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrčková F, Grunt M, Žárský V. 2012. Expression of GFP–mTalin reveals an actin-related role for the arabidopsis class II formin AtFH12. Biologia Plantarum 3, 431–440 [Google Scholar]
- Deeks M, Cvrčková F, Machesky L, Mikitová V, Žárský V, Davies B, Hussey P. 2005. Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytologist 168, 529–540 [DOI] [PubMed] [Google Scholar]
- Deeks M, Fendrych M, Smertenko A, Bell K, Oparka K, Cvrčková F, Žárský V, Hussey P. 2010. The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. Journal of Cell Science 123, 1209–1215 [DOI] [PubMed] [Google Scholar]
- Deeks M, Hussey P, Davies B. 2002. Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends in Plant Science 7, 492–498 [DOI] [PubMed] [Google Scholar]
- Dvořáková L, Cvrčková F, Fischer L. 2007. Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genomics 8, 412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emons A, Hofte H, Mulder B. 2007. Microtubules and cellulose microfibrils: how intimate is their relationship?. Trends in Plant Science 12, 279–281 [DOI] [PubMed] [Google Scholar]
- Gilliland L, Kandasamy M, Pawloski L, Meagher R. 2002. Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiology 130, 2199–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland L, Pawloski L, Kandasamy M, Meagher R. 2003. Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. The Plant Journal 33, 319–328 [DOI] [PubMed] [Google Scholar]
- Granger CL, Cyr RJ. 2001. Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP–MBD. Protoplasma 216, 201–214 [DOI] [PubMed] [Google Scholar]
- Grunt M, Žárský V, Cvrčková F. 2008. Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing protein. BMC Evolutionary Biology 8, 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E, Li F, Higgs H. 2004. The mouse formin, FRL, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. Journal of Biological Chemistry 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- Hashimoto T. 2002. Molecular genetic analysis of left-right handedness in plants. Philosophical Transactions of the Royal Society B: Biological Sciences 357, 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty J, Bledsoe S, Khurana P, Meagher R, Day B, Blanchoin L, Staiger C. 2011. Arabidopsis actin depolymerizing factor 4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. The Plant Cell 23, 3711–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Kondo N, Hasezawa S. 2010. Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. The Plant Journal 61, 156–165 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Fitz J, Guerin C, Robert H, Sorensen M, Van Damme D, Geelen D, Blanchoin L, Berger F. 2005. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nature Cell Biology 7, 374–380 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Allwood E, Anthony R, Voigt B, Menzel D, Hussey P. 2004. The actin-interacting protein AIP is essential for actin organization and plant development. Current Biology 14, 145–149 [DOI] [PubMed] [Google Scholar]
- Knezevic S, Streibig J, Ritz C. 2007. Utilizing R software package for dose–response studies: the concept and data analysis. Weed Technology 21, 840–848 [Google Scholar]
- Li Y, Shen Y, Cai C, Zhong C, Zhu L, Yuan M, Ren H. 2010. The type II Arabidopsis formin 14 interacts with microtubules and microfilaments to regulate cell division. The Plant Cell 22, 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J, Granger C, Brincat J, Fisher D, Kao T, McCubbin A, Cyr R. 1998. A GFP–MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. The Plant Cell 10, 1927–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniere A, Gayral P, Hawes C, Runions J. 2011. Building bridges: formin 1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. The Plant Journal 66, 354–365 [DOI] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemma R, Guerin C, Huang S, Parcy F, Staiger C, Blanchoin L. 2006. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Current Biology 16, 1924–1930 [DOI] [PubMed] [Google Scholar]
- Michelot A, Guerin C, Huang S, Ingouff M, Richard S, Rodiuc N, Staiger C, Blanchoin L. 2005. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis Formin 1. The Plant Cell 17, 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Yokota E, Wada T, Shimmen T, Okada K. 2003. An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polimerization, reveals its distinctive function in root development. Plant and Cell Physiology 44, 1131–1140 [DOI] [PubMed] [Google Scholar]
- Peremyslov V, Prokhnevsky A, Dolja V. 2010. class XI myosins are required for development, cell expansion and F-actin organization in Arabidopsis. The Plant Cell 22, 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Schwarzerová K. 2009. Actin and microtubule cytoskeleton interactions. Current Opinion in Plant Biology 12, 728–734 [DOI] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor E, Baskin T. 2007. Auxin, actin and growth of the Arabidopsis thaliana primary root. The Plant Journal 50, 514–528 [DOI] [PubMed] [Google Scholar]
- Rizvi S, Neidt E, Cui J, Feiger Z, Skau C, Gardel M, Kozmin S, Kovar D. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chemistry and Biology 16, 1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Lindeboom J, Debolt S, Gutierrez R, Ehrhardt D, Ketelaar T, Persson S. 2011. Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. The Plant Cell 23, 2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S, Kamyar R, Ehrhardt D. 2003. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Deeks M, Hussey P. 2010. Strategies of actin reorganisation in plant cells. Journal of Cell Science 123, 3019–3028 [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. 2005. Spatial control of cell expansion by the plant cytoskeleton. Annual Review of Cell and Developmental Biology 21, 271–295 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Graumann K, Martiniere A, Schoberer J, Wang P, Osterrieder A. 2011. Bleach it, switch it, bounce it, pull it: using laser to reveal plant cell dynamics. Journal of Experimental Botany 62, 1–7 [DOI] [PubMed] [Google Scholar]
- Staiger C, Sheahan M, Khurana P, Wang X, McCurdy D, Blanchoin L. 2009. Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. Journal of Cell Biology 182, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, et al. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T. 2002. Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417, 193–196 [DOI] [PubMed] [Google Scholar]
- van der Honing H, Kieft H, Emons A, Ketelaar T. 2012. Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiology 58, 1426–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcay-Barrena G, Webb S, Martin-Fernandez M, Wilson Z. 2011. Subcellular and single-molecule imaging of plant fluorescent proteins using total internal reflection fluorescent microscopy (TIRFM). Journal of Experimental Botany 62, 5419–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Ash W, Fan L, Hao H, Kim M, Lin J. 2011. Variable-angle total internal reflection fluorescence microscopy of intact cells of Arabidopsis thaliana. Plant Methods 7, 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xue X, Ren H. 2012. New insights into the role of plant formins: regulating the organization of the actin and microtubule cytoskeleton. Protoplasma 249, Suppl. 2 S101–S107 [DOI] [PubMed] [Google Scholar]
- Xue X, Guo C, Du F, Lu Q, Zhang C, Ren H. 2011. AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Molecular Plant 4, 264–278 [DOI] [PubMed] [Google Scholar]
- Yang W, Ren S, Zhang X, et al. 2011. BENT UPPERMOST INTERNODE1 encodes the class II Formin FH5 crucial for actin organization and rice development. The Plant Cell 23, 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Zheng Y, Yan A, Chen N, Wang Z, Huang S, Yang Z. 2009. Arabidopsis Formin3 directs the formation of actin cables and polarized growth in pollen tubes. The Plant Cell 21, 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Guo C, Chen D, Zhao B, Yang B, Ren H. 2005. Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiology 138, 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, He J, McCormick S. 2010. Interdependence of endomembrane trafficking and actin dynamics during polarized growth of arabidopsis pollen tubes. Plant Physiology 152, 2200–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Tan H, Wan Y, Li G, Liang W, Yuan Z, Hu J, Ren H, Zhang D. 2011. RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. The Plant Cell 23, 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.