Abstract
Polyploidy is very common within angiosperms, and several studies are in progress to ascertain the effects of early polyploidization at the molecular, physiological, and phenotypic level. Extensive studies are available only in synthetic allopolyploids. By contrast, less is known about the consequences of autopolyploidization. The current study aimed to assess the occurrence and extent of genetic, epigenetic, and anatomical changes occurring after oryzaline-induced polyploidization of Solanum commersonii Dunal and Solanum bulbocastanum Dunal, two diploid (2n=2×=24) potato species widely used in breeding programmes. Microsatellite analysis showed no polymorphisms between synthetic tetraploids and diploid progenitors. By contrast, analysis of DNA methylation levels indicated that subtle alterations at CG and CHG sites were present in tetraploids of both species. However, no change occurred concurrently in all tetraploids analysed with respect to their diploid parent, revealing a stochastic trend in the changes observed. The morpho-anatomical consequences of polyploidization were studied in leaf main veins and stomata. With only a few exceptions, analyses showed no clear superiority of tetraploids in terms of leaf thickness and area, vessel number, lumen size and vessel wall thickness, stomata pore length and width, guard cell width, and stomatal density compared with their diploid progenitors. These results are consistent with the hypothesis that there are no traits systematically associated with autopolyploidy.
Key words: chromosome doubling, methylation, microsatellite, stomata, vessels, potato.
Introduction
Polyploidy is widespread in higher plants and represents a major evolutionary force (Ramsey and Schemske, 2002). Genome sequencing has provided evidence that even a number of species traditionally considered diploid (e.g. corn, Arabidopsis, rice) have experienced genome duplication events during their evolution. Polyploidization entails a reorganization of the full genome to enable harmonious cohabitation in the same nucleus of genes of different origin (in the case of allopolyploids) or of duplicated genes from identical or very similar genomes (in the case of autopolyploids). The structural and functional modifications induced by polyploidization are probably the key to the success of polyploid plants in nature. Indeed, it may greatly affect the morphology, physiology, and secondary metabolism of newly formed offspring. For a specific trait, the phenotype could be similar to a progenitor, intermediate between the progenitors, or new (Comai et al., 2000). Possible morpho-anatomical and physiological polyploidy effects include, among others, changes in cell and organ size (Schranz and Osborn, 2004), leaf width:length ratio (Stanys et al., 2006), and transpiration and/or photosynthetic rate (Levin, 2002). Polyploidy can also cause the appearance of traits with a significant impact on crop production, such as resistance to stresses and a shift from sexual to apomictic reproduction (Quarin et al., 2001). Phenotypic changes caused by polyploidization are expected to be the result of genetic modifications. The first study demonstrating whole-genome changes after chromosome doubling was carried out in Brassica spp. by Song et al. (1995), who reported nuclear modifications in newly synthesized polyploids on the basis of restriction fragment length polymorphism patterns. Since then, similar phenomena have been documented in a wide range of plant species (reviewed by Yang et al., 2011). Ploidy-induced structural genomic changes are strongly associated with epigenetic alterations. The latter refer to heritable changes in gene expression that occur without alteration of the DNA sequence, such as DNA (cytosine) methylation, histone methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation (Lauria and Rossi, 2011). Epigenetic changes often occur during genome merger and doubling as demonstrated, for instance, in wheat and Arabidopsis allopolyploids (reviewed by Chen, 2007).
Extensive molecular and phenotypic studies are available in several synthetic allopolyploids, including the wheat group (Triticum–Aegilops) (Levy and Feldman, 2004), Arabidopsis thaliana (L.) Heynh. (Comai et al., 2000), Tragopogon miscellus G. B. Ownbey (Tate et al., 2006), and Nicotiana spp. (Lim et al., 2006). This type of polyploidy is generally subjected to conspicuous and rapid genetic and epigenetic changes during the first generations and offers a preferential model to study the effects of polyploidization on phenotype and gene expression. Alterations may be necessary to overcome incompatibility between divergent genomes and have been considered important for the so-called initial ‘revolutionary phase’ of polyploid formation (Levy and Feldman, 2004). As autopolyploids arise by doubling of similar homologous genomes, it is believed that substantial alterations might not be required after their formation (Comai, 2005; Parisod et al., 2010). According to most findings reported in the literature, changes are less dramatic than those induced by allopolyploidization. Stupar et al. (2007) reported no DNA polymorphism within a ploidy series of Solanum phureja Juz. & Bukasov. Similarly, Allario et al. (2011) showed that microsatellite profiles of diploid and tetraploid Citrus limonia Osbeck were identical. Transcriptome and proteome analysis have generally confirmed the presence of only subtle changes in cabbage (Albertin et al., 2005), potato (Stupar et al., 2007), maize (Riddle et al., 2010), and Rangpur lime (Allario et al., 2011). Studies in A. thaliana (L.) Heynh. have corroborated the findings that stable, non-stochastic, developmentally specific gene expression alterations occur following autopolyploidization (Yu et al., 2010). In the same species, microarray analysis did not allow the identification of a reliable group of ploidy-regulated genes (Pignatta et al., 2010). A recent paper by Li et al. (2012) reported a small but stable increase in the expression of cell-cycle genes (ICK1, ICK2, and ICK5) in autotetraploid seedlings of A. thaliana.
Given the evolutionary significance and practical breeding value of autopolyploidy and the scant coverage in the literature, the objective of this research was to assess the occurrence and extent of genetic, epigenetic, and anatomical changes occurring after artificial somatic polyploidization of two diploid (2n=2×=24) potato species, Solanum commersonii Dunal and Solanum bulbocastanum Dunal. Both species possess important resistance traits (to low temperatures and late blight, respectively) that can be exploited for breeding the tetraploid (2n=4×=48) cultivated potato. However, they are sexually isolated from Solanum tuberosum. Doubling their chromosome complement is an important prerequisite to sexually overcome incompatibility barriers (Carputo and Barone, 2005). Therefore, these species represent useful plant materials to compare the effects of early polyploidization at phenotypic and DNA levels.
Materials and methods
Plant material
Synthetic autotetraploids of S. bulbocastanum Dunal and S. commersonii Dunal were generated as described previously (Caruso et al., 2011). Briefly, a clone of diploid S. commersonii (PI 243503), coded cmm1t, and a clone of diploid S. bulbocastanum (PI 275190), coded blb1c, were subjected to oryzaline treatment to produce the autotetraploid genotypes used in this study. Those deriving from cmm1t were coded cmm15, cmm23, cmm24, cmm27, and cmm30. Those produced from blb1c were coded blb10, blb22, blb25, and blb26. To collect material for molecular and morpho-anatomical analyses, plants were micropropagated on MS medium (Murashige and Skoog, 1962) with sucrose (30g l–1) and microagar (9g l–1)–1, adjusted to pH 5.8, and then transferred into pots in a temperature-controlled greenhouse.
Molecular analysis
Fully developed young leaves of diploid progenitors and synthetic tetraploids were collected from individual plants on the same date. From each genotype, DNA was isolated from a leaf pool of three different plants using a DNeasy Plant mini kit (Qiagen, Valencia, USA) following the manufacturer’s instructions. DNA quality and integrity were checked by gel electrophoresis and spectrophotometric assay. simple sequence repeat (SSR) analyses were carried out with 12 nuclear microsatellite (SSR) primer pairs chosen from three sources: five primer pairs (STM1053, STM0031, STM1104, STM1052, and STM1106) from Milbourne et al. (1998), three (STM5114, STM5127, and STG0001) from Ghislain et al. (2009), and four (STI004, STI012, STI030, and STI032) from Feingold et al. (2005). These were recommended by the Centro Internacional de la Papa (http://www.cipotato.org) based on quality criteria, genome coverage, and locus-specific information. PCR reactions were performed in a 20 µl volume containing 1× reaction buffer with 2.5mM MgCl2, 0.2mM of each dNTP, 25 pM 6-carboxyfluorescein (FAM)-labelled forward SSR primer, 15 pM reverse SSR primer, 1U of GoTaq DNA polymerase (Promega, Madison, USA), and 40ng of genomic DNA. PCR was carried out using the following cycling profiles: 4min at 94°C, 31 cycles of 45 s at 94°C, 1min at annealing temperature, and 1min at 72°C, with a final extension step of 5min at 72°C. PCR products were separated on an ABI PRISM® 3130 DNA Analyzer. Size calibration was performed with the molecular weight ladder GenScanTM 500 ROXTM standard (Applied Biosystems, Foster City, USA). SSR alleles were detected and scored using GeneScan® Analysis software (Applied Biosystems).
To detect methylation patterns at the 5’-CCGG-3’ sites, the methylation-sensitive amplification polymorphism (MSAP) technique was employed. Two methylation-sensitive isoschizomer enzymes, HpaII and MspI, were used, and a double digestion was set up according to the protocol of Reyna-López et al. (1997) and adapted by Xiong et al. (1999). Fifteen selective amplifications were performed for S. commersonii diploid and tetraploid genotypes, using two FAM-labelled EcoRI primers (E-TCCA and E-TCAA) combined with different HpaII/MspI (HM) primers: E-TCCA/HM-AAC; E-TCCA/HM-AAG; E-TCCA/HM-AGG; E-TCCA/HM-ACG; E-TCCA/HM-ACA; E-TCCA/HM-ACC; E-TCCA/HM-ACT; E-TCCA/HM-AGC; E-TCAA/HM-AGG; E-TCAA/HM-AGC; E-TCAA/HM-ACT; E-TCAA/HM-ACC; E-TCAA/HM-ACA; E-TCAA/HM-AAG; and E-TCAA/HM-AAC). Seven selective amplifications were used for S. bulbocastanum diploid and tetraploid genotypes combining E-TCCA with the following HM primers: HM-AAC; HM-AAG; HM-ACA; HM-ACC; HM-ACT; HM-AGC; and HM-AGG). The GeneScan™ 500 ROX™ standard was used as size standard. SSR and MSAP fragments were separated electrophoretically using a capillary sequencer (ABI PRISM® 3130 Genetic Analyzer). A genetic profiler (GeneMapper®, Applied Biosystems) was used to display the results and to export data in text format. The intensity of the peaks was not taken into account, and fragments with identical mobility were considered to be monomorphic. For each genotype, polymorphic MSAP alleles were recorded as present (1) or absent (0). Methylation levels (hypo- and hypermethylation) were evaluated based on the different digestion patterns of the two methylation-sensitive isoschizomers, MspI and HpaII. All methylation changes in the CCGG site of diploid progenitors versus their tetraploid derivatives (including the appearance of new epi-alleles and the disappearance of parental epi-alleles) were calculated. For both SSR and MSAP analyses, DNA samples from each genotype were analysed in triplicate to provide technical replicates.
Morphological and anatomical analysis
Analyses were performed on diploid cmm1t and its tetraploids cmm23, cmm24, and cmm30, and on diploid blb1c and its tetraploids blb10, blb22, blb25, and blb26. Fully expanded leaves were collected at the same phenological stage (before flowering) on the same date. Digital images of the leaves were analysed using AnalySIS 3.2 (Olympus, Hamburg, Germany), a software program devised to quantify morpho-anatomical features. For anatomical analysis, from each genotype three leaves were selected and fixed in 40% formaldehyde:glacial acetic acid:50% ethanol: 5:5:90 by volume) for several days. Subsamples (5×5mm) containing the main vein were dissected, dehydrated in an ethanol series up to 90%, and embedded in JB4 acrylic resin (Polysciences Europe, Eppelheim, Germany). Semi-thin cross-sections (5 µm thick) were cut with a rotary microtome. Sections were stained with 0.5% toluidine blue in water (Feder and O’Brien, 1968), mounted with Canada balsam, and observed under a light microscope (BX60, Olympus). Digital photomicrographs of leaf sections were obtained with a digital camera (CAMEDIA C4040; Olympus) at various magnifications and analysed with AnalySIS 3.2. In addition to leaf area (LA) and lamina thickness (LT), the following parameters were measured with regard to the main veins: number of vessels in the main vein (VN), area and mean diameter of vessel lumen (VLA and VLD, respectively), and vessel wall thickness (VWT). VLA, VLD, and VWT were measured for a minimum of ten vessels per section. In measuring VWT, cell corners were avoided. Scanning electron microscopy (SEM) analysis was carried out using one leaf for each individual plant. A sample of 10mm2 of surface was removed from the inter-nerval zone at the point where the width of the leaf blade was at its maximum. Both leaf surfaces (abaxial and adaxial) were examined. Specimens were dried and then gold coated with a sputter coater K550 (Emitech, Ashford, UK). Images were obtained using a Zeiss SEM DSM 940A at 5–10kV. For measurements, photomicrographs were taken at ×500 magnification (standard area of 0.25mm2) and stomatal aperture was measured. In particular, stomata pore length (SL), stomata pore width (SW) and guard cell width (CW) were taken into account. Stomatal density (SD) was then measured on a standard area of 1mm2. About ten stomata were measured for each genotype. All morphological and anatomical data collected were subjected to statistical analysis using R software version 2.15.1 (http://www.r-project.org/). Means and standard errors were calculated. Post hoc mean separation was performed using Duncan’s multiple range test. Differences were considered significant when P ≤0.05.
Results
Microsatellite analysis
In diploid S. commersonii and its tetraploids, we were able to detect a total of 15 alleles (Table 1). Marker STM1052 did not amplify scorable alleles and was therefore not taken into account. Four loci (STG0001, STI004, STI012, and STM1053) were heterozygous, while the others were homozygous. On average, 1.4 alleles per locus were detected. For S. bulbocastanum, we were able to detect a total of 18 alleles by analysing 11 microsatellite loci. Marker STM0031 was ruled out from the analysis as it did not amplify scorable alleles. Seven loci were heterozygous: STM1052, STM1053, STM1104, STM1106, STG0001, STI004, and STI032. On average, we identified 1.4 alleles per locus. In neither species did the tested loci show polymorphisms between the tetraploids and the diploid progenitor they were derived from (data not shown), suggesting a lack of structural changes following polyploidization.
Table 1.
Description of the 12 SSR markers used to assess the genome integrity of S. bulbocastanum and S. commersonii synthetic tetraploids and the diploid parents (cmm1t and blb1C) they were derived from. For each marker, the respective name, source, repeat motifs, forward (Fw) and reverse (Rv) primer sequences, annealing temperature (T a), map location, and amplified allele sizes are reported.
| Name | Repeat motif | Primer sequences | T a (C°) | Map location | Allele size (nt)a |
|---|---|---|---|---|---|
| STG0001 | (CT)n | Fw_CAGCCAACATTTGTACCCCT | 55 | XI | C: 126, 158 |
| Rv_ACCCCCACTTGCCATATTTT | B: 130, 138 | ||||
| STI004 | (AAG)n | Fw_GCTGCTAAACACTCAAGCAGAA | 57 | VI | C: 66, 72 |
| Rv_CAACTACAAGATTCCATCCACAG | B: 78, 81 | ||||
| STI0012 | (ATT)n | Fw_GAAGCGACTTCCAAAATCAGA | 53 | IV | C: 179, 188 |
| Rv_AAAGGGAGGAATAGAAACCAAAA | B: 167 | ||||
| STI0030 | (ATT)n | Fw_TGACCCTCCAACTATAGATTCTTC | 56 | XII | C: 133 |
| Rv_TGACAACTTTAAAGCATATGTCAGC | B: 79 | ||||
| STI0032 | (GGA)n | Fw_TGGGAAGAATCCTGAAATGG | 54 | V | C: 118 |
| Rv_TGCTCTACCAATTAACGGCA | B: 115, 118 | ||||
| STM0031 | (AC)n…(AC)n GCAC (AC)n (GCAC)n | Fw_CATACGCACGCACGTACAC | 52 | VII | C: 63 |
| Rv_TTCAACCTATCATTTTGTGAGTCG | B: ND | ||||
| STM1052 | (AT)n GT (AT)n (GT)n | Fw_CAATTTCGTTTTTTCATGTGACAC | 54 | IX | C: ND |
| Rv_ATGGCGTAATTTGATTTAATACGTAA | B: 207, 224 | ||||
| STM1053 | (TA)n (ATC)n | Fw_TCTCCCCATCTTAATGTTTC | 55 | III | C: 120, 172 |
| Rv_CAACACAGCATACAGATCATC | B: 120, 172 | ||||
| STM1104 | (TCT)n | Fw_TGATTCTCTTGCCTACTGTAATCG | 57 | VIII | C: 195 |
| Rv_CAAAGTGGTGTGAAGCTGTGA | B: 165, 177 | ||||
| STM1106 | (ATT)n | Fw_TCCAGCTGATTGGTTAGGTTG | 56 | X | C: 174 |
| Rv_ATGCGAATCTACTCGTCATGG | B: 141, 150 | ||||
| STM5114 | (ACC)n | Fw_AATGGCTCTCTCTGTATGCT | 53 | II | C: 289 |
| Rv_GCTGTCCCAACTATCTTTGA | B: 286 | ||||
| STM5127 | (TCT)n | Fw_TTCAAGAATAGGCAAAACCA | 55 | I | C: 246 |
| Rv_CTTTTTCTGACTGAGTTGCCTC | B: 243 |
a C, S. commersonii; B, S. bulbocastanum; ND, not detected.
MSAP analysis
As reported in Table 2, due to the different sensitivities exhibited by the isoschizomers (McClelland et al., 1994), methylation of the internal cytosine (C5mCGG, or CG sites) or the hemi- (single-strand) methylation of the external cytosine (5mCCGG, or CHG sites) was unequivocally distinguished (for details, see Supplementary Fig. S1 at JXB online). In particular, from the comparison of EcoRI/HpaII and EcoRI/MspI restrictions, four types of alteration in cytosine methylation at the CCGG sites of random genomic loci were identified: CG hypermethylation (hereafter coded as CG hyper), CG hypomethylation (coded as CG hypo), CHG hypermethylation (coded as CHG hyper), and CHG hypomethylation (coded as CHG hypo) (see Supplementary Fig. S2 at JXB online). In agreement with Ochogavía et al. (2009), to rule out confounding polymorphic bands due to mutations or methylation events, only ‘genuine epigenetic polymorphic markers’ (called unambiguous) that appeared in one but not in both HpaII and MspI digestions were scored for a given genotype. Differences detected between the diploid and synthetic tetraploids appeared to be stochastic (Table 2). In S. commersonii, out of 2545 epialleles, 87% did not change. To estimate better the predominant event between methylation and demethylation, a parameter termed methylation ratio (MR) was calculated as the ratio between the hyper- and hypomethylation frequencies according to the following formula: MR=(CG hyper+CHG hyper)/(CG hypo+CHG hypo). Hypermethylation frequencies were slightly higher than those of hypomethylation in cmm15 and cmm27 (MR=1.5 and 2.1, respectively). By contrast, in cmm23 and cmm24, the two events occurred to similar extents (MR=0.9 and 1.0, respectively). Cmm15 and cmm30 showed the highest CG and CHG hyper incidences (5.2 for both). On average, the CHG were the sites changing the most (7.4 vs 6.0% of CG sites), with the exception of cmm15. MSAP analysis of S. bulbocastanum genotypes revealed no change for 94% of loci examined (Table 2). Also, in this species, all differences detected between the diploid blb1c and its synthetic tetraploids were stochastic. In blb10 and blb22, hypermethylation events were higher than hypomethylations (MR=1.2 and 2.3, respectively), whereas in blb25 and blb26 the opposite was true (MR=0.7 for both). On average, CG sites changed more than CHG (4.2% vs 2.2%), although in blb25, hyper- and hypomethylation at CHG sites occurred at the same frequency (2.3%). For both species, no change occurred concurrently in all tetraploids analysed with respect to their diploid parent, revealing a stochastic trend in the changes observed.
Table 2.
Results from MSAP fingerprinting analysis carried out on diploid S. commersonii (cmm1t) and S. bulbocastanum (blb1c) progenitors and their related autotetraploids. Values refer to the total number of scored fragments and the genuine epigenetic polymorphic markers (called unambiguous, see Results), and the number (%) of hyper- and hypomethylation events at CG and CHG sites occurring in each tetraploid with respect to its diploid progenitor. The methylation ratio (MR) is also shown.
| Genotype | Scored, n | Unambiguous, n | Hyper, n (%) | Hypo, n (%) | MRa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CG | CHG | CG | CHG | ||||||||
| S. commersonii | |||||||||||
| cmm15 | 749 | 460 | 24 | (5.22) | 20 | (4.35) | 16 | (3.48) | 13 | (2.83) | 1.52 |
| cmm23 | 837 | 594 | 22 | (3.70) | 16 | (2.69) | 15 | (2.53) | 26 | (4.38) | 0.93 |
| cmm24 | 720 | 494 | 14 | (2.83) | 22 | (4.45) | 15 | (3.04) | 20 | (4.05) | 1.03 |
| cmm27 | 789 | 518 | 14 | (2.70) | 21 | (4.05) | 7 | (1.35) | 10 | (1.93) | 2.06 |
| cmm30 | 789 | 479 | 15 | (3.13) | 25 | (5.22) | 10 | (2.09) | 17 | (3.55) | 1.48 |
| Average | 776.80 | 509.00 | 17.80 | (3.50) | 20.80 | (4.13) | 12.60 | (2.48) | 17.20 | (3.32) | 1.32 |
| S. bulbocastanum | |||||||||||
| blb10 | 366 | 215 | 5 | (2.33) | 2 | (0.93) | 4 | (1.86) | 2 | (0.93) | 1.17 |
| blb22 | 455 | 324 | 15 | (4.63) | 3 | (0.93) | 5 | (1.54) | 3 | (0.93) | 2.25 |
| blb25 | 389 | 212 | 2 | (0.94) | 2 | (0.94) | 3 | (1.42) | 3 | (1.42) | 0.66 |
| blb26 | 451 | 283 | 4 | (1.41) | 4 | (1.41) | 8 | (2.83) | 4 | (1.41) | 0.67 |
| Average | 415.25 | 258.50 | 6.50 | (2.33) | 2.75 | (1.05) | 5.00 | (1.91) | 3.00 | (1.17) | 1.10 |
a Ratio between hyper- and hypomethylation frequencies (CG hyper+CHG hyper)/(CG hypo+CHG hypo).
Leaf morpho-anatomical analyses
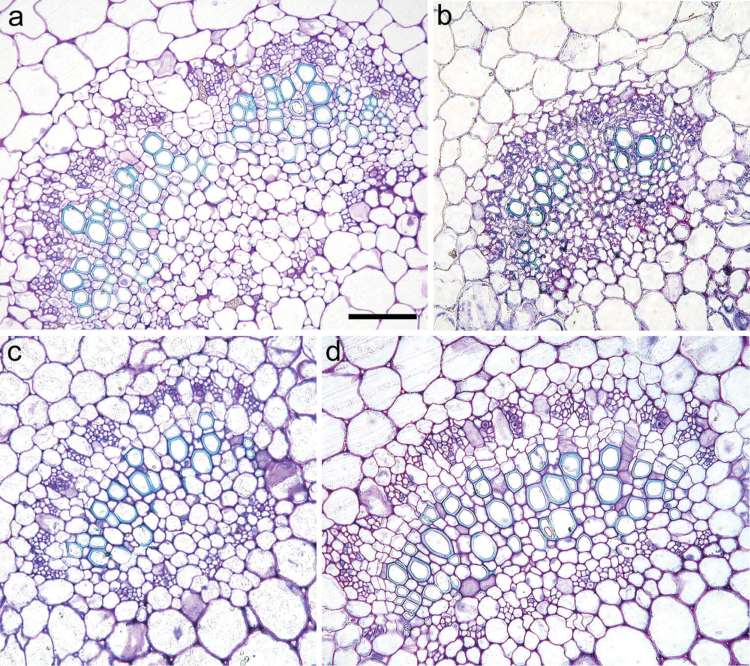
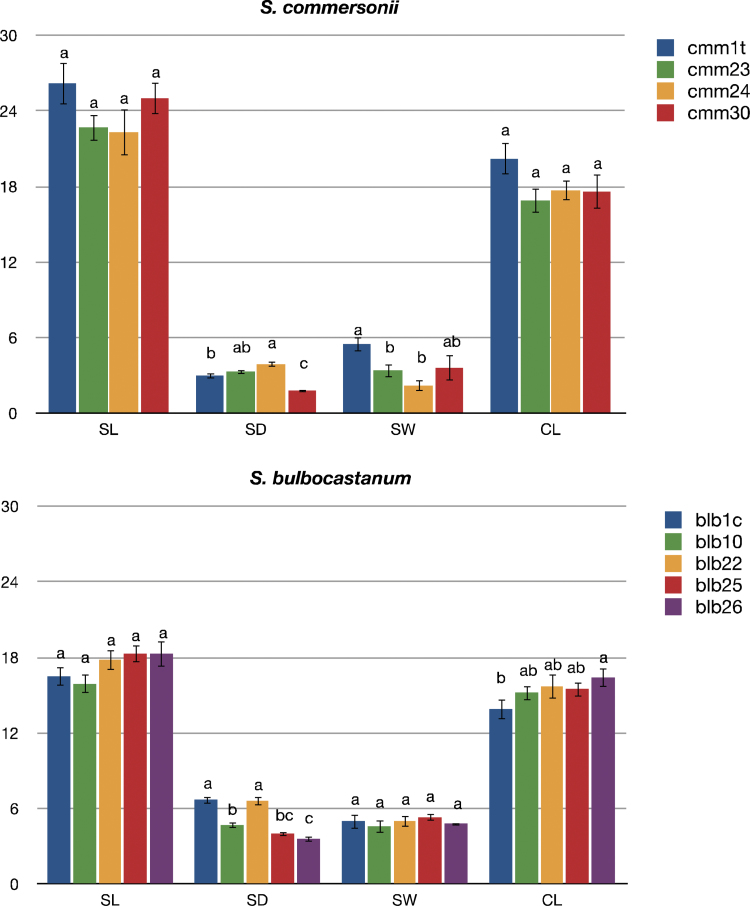
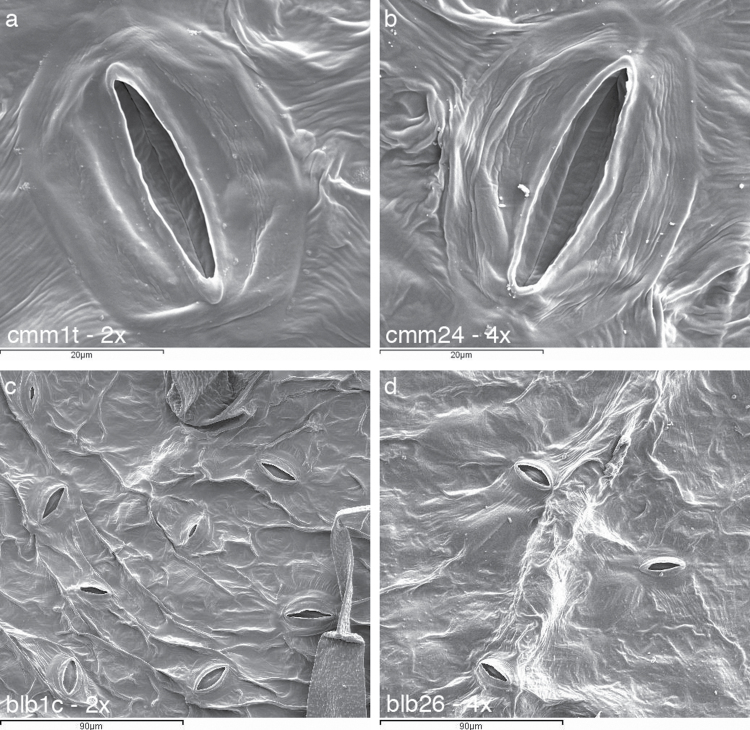
The quantitative leaf anatomical traits are summarized in Table 3. Both diploids and tetraploids had leaves with the typical dorsoventral structure with no evident qualitative modifications induced by polyploidization. In neither species did LT change significantly following polyploidization. The only exception was cmm30, whose LT increased significantly compared with diploid cmm1t. For LA, in S. commersonii cmm23 and cmm30 this was similar to that in diploid cmm1t. By contrast, cmm24 showed a significantly lower LA. The same parameter was more variable in tetraploids of S. bulbocastanum. Indeed, compared with the diploid parent blb1c, blb22 showed a larger LA, blb10 and blb25 a similar LA, and blb26 a lower LA. Diploid cmm1t generally performed differently from its synthetic tetraploids in terms of xylem characteristics. Indeed, VN, VLA, VLD, and VWT in diploid S. commersonii were 51, 44, 25, and 15% higher than the mean values of its tetraploids, respectively (Fig. 1a, b; Table 3). A different trend was observed in S. bulbocastanum for VN and VLA. Indeed, the main veins of tetraploids generally comprised a greater number of larger vessels than their diploid counterparts (Fig. 1c, d; Table 3). VLD and VWT of diploid S. bulbocastanum did not deviate much from the mean value of its tetraploids. SEM analysis data are shown in Figs 2 and 3. In both species, SL did not change significantly following polyploidization. By contrast, SD underwent changes following genome doubling. They were stochastic in S. commersonii tetraploids with respect to cmm1t, whereas in S. bulbocastanum, the SD changes were more consistent, with all tetraploids showing lower values compared with blb1c (Fig. 3a, b). Diploid cmm1t behaved differently from its synthetic tetraploids in terms of SW (Fig. 3c, d) but not in terms of CW. By contrast, diploid blb1c was similar to its tetraploids in terms of SW but not in terms of CW.
Table 3.
Results (mean ±standard error) on leaf lamina thickness (LT) and area (LA ), number of vessels per main vein (VN), vessel lumen area (VLA), and diameter (VLD), and vessel wall thickness (VWT) of S. commersonii and S. bulbocastanum synthetic tetraploids and the diploid parents they derived from (cmm1t and blb1c).
| Genotype | LT (µm)a,b | LA (cm2) | VN (n) | VLA (µm2) | VLD (µm) | VWT (µm) |
|---|---|---|---|---|---|---|
| S. commersonii | ||||||
| cmm1t (2n=2×) | 255.6±8.0 a | 64.87±5.48 b | 88.6±8.4 b | 376.0±23.3 c | 22.6±0.1 d | 2.7±0.05 c |
| cmm23 (2n=4×) | 245.1±4.7 a | 59.47±6.37 b | 45.3±5.20 a | 267.7±16.0 b | 19.9±0.0 c | 2.7±0.06 c |
| cmm24 (2n=4×) | 237.0±6.6 a | 32.74±1.74 a | 47.6±2.6 a | 114.2±6.3 a | 12.7±0.0 a | 2.0±0.03 a |
| cmm30 (2n=4×) | 281.8±8.5 b | 58.47±6.88 b | 39.0±4.2 a | 246.1±18.5 b | 18.3±0.0 b | 2.2±0.05 b |
| S. bulbocastanum | ||||||
| blb1c (2n=2×) | 243.0±7.9 ab | 27.84±0.92 bc | 33.3±2.3 a | 390.9±24.1 a | 23.5±0.1 a | 3.1±0.07 b |
| blb10 (2n=4×) | 222.8±9.2 ab | 29.97±0.73 c | 53.3±0.6 b | 499.3±44.5 ab | 27.1±0.1 a | 2.7±0.06 a |
| blb22 (2n=4×) | 256.1±10.1 b | 34.00±1.51 d | 52.0±1.5 b | 483.0±47.0 ab | 26.8±0.1 a | 3.0±0.07 b |
| blb25 (2n=4×) | 212.6±6.8 a | 25.23±1.31 ab | 46.0±1.2 b | 445.0±27.8 ab | 25.3±0.1 a | 2.9±0.05 b |
| blb26 (2n=4×) | 229.6±12.5 ab | 22.81±0.64 a | 36.0±5.6 a | 512.0±42.2 b | 26.4±0.0 a | 3.1±0.08 b |
a Each value of all measured parameters represents the mean of triplicate determinations.
b In each column, within each species, means denoted by the same letter did not differ significantly at P ≤0.05 according to Duncan’s multiple range test.
Fig. 1.
Light microscopy views of cross-sections of main veins in leaves of the diploid (a) and tetraploid (b) S. commersonii, and diploid (c) and tetraploid (d) S. bulbocastanum. Bar, 100μm.
Fig. 2.
Results of stomata pore length (SL, µm), stomata density (SD, pores mm–2), stomata pore width (SW, µm), guard cell width (CW, μm) of S. commersonii and S. bulbocastanum synthetic tetraploids and the diploid parents they were derived from (cmm1t and blb1c). Each value represents the average number of ten determinations (mean ±standard error). Means denoted by the same letter did not differ significantly at P ≤0.05 according to Duncan’s multiple range test.
Fig. 3.
Scanning electron microscopy results showing 2× versus 4× differences in terms of stomata pore width (a, b) and density (c, d) in S. commersonii (cmm) and S. bulbocastanum (blb), respectively.
Discussion
Studies on allopolyploids have contributed considerably to gaining insights into polyploidy-related processes. However, such polyploids inherently change two parameters at once, and hybridization rather than polyploidization per se may give rise to genome modifications (Doyle et al., 2008). By contrast, autopolyploids have the advantage of restricting the changes to one parameter, namely chromosome number, ruling out whole-genome alterations primarily connected with incompatible genomes. In this study, to understand better the chromosome doubling effects on genome stability, we tested the behaviour of SSR loci after polyploidization. None of the studied SSR loci demonstrated changes in microsatellite length. Our data not only confirmed the doubled diploid nature of our tetraploid genotypes but also revealed that structural changes due to polyploidization did not affect the genome of the two species analysed. Similar findings have also been reported by Ozkan et al. (2006) in A. thaliana, Stupar et al. (2007) in Solanum spp., and Allario et al. (2011) in Citrus ssp. Genome modifications have rarely been reported. A rapid elimination of DNA sequences was detected in induced autopolyploid of Phlox drummondii Hook by Raina et al. (1994). In a recent review, Parisod et al. (2010) suggested that, in synthetic autopolyploids, rapid elimination of DNA sequences may be possible, but this phenomenon is not so extensive during the first generations following genome doubling.
While genetic mutations can explain the cause of variability mainly over evolutionary time, epigenetic changes, which are potentially reversible, offer a flexible expedient for a polyploid cell to respond to genomic shock, especially in the early stages of polyploid formation, leading to various genetic variations. Alterations in cytosine methylation level were observed in our S. bulbocastanum and S. commersonii tetraploids versus their diploid parents (Table 2). In particular, various types of change were identified, including both hypo- and hypermethylation at CG and CHG sites, known to be the most affected targets of DNA methyltransferase (Law and Jacobsen, 2010). On average, the trend we observed was a subtle increase in methylation, although in all genotypes we detected both phenomena. Our data suggest that the S. commersonii and S. bulbocastanum epigenome landscape responds to autopolyploidization, probably to handle the consequences deriving from ploidy alterations. Ploidy-driven DNA methylation changes are likely to be more extensive than those we considered. Indeed, the real number of random loci undergoing methylation changes is probably underestimated, because, as suggested by Ochogavía et al. (2009), only a subset of MSAP markers (those called ‘genuine epigenetic polymorphic markers’) were prudentially analysed. Although our first-generation polyploids represent the plant materials to use in potato breeding, the DNA methylation changes occurring at the observed genetic loci have the potential to be inherited. Indeed, several studies suggest that epigenetic alterations generally persist through meiosis, showing a stable inheritance over many generations (reviewed by Hauser et al., 2011). In autotetraploid A. thaliana, Mittelsten Scheid et al. (2003) reported that epialleles lead to heritable gene silencing persisting after segregation from the inactivating allele. Investigating the mechanisms beyond polyploidy-associated transcriptional gene silencing in A. thaliana, Baubec et al. (2010) found that two epigenetic marks, namely DNA methylation and histone methylation, cooperate to give rise to a ‘double lock’ on transcriptional silencing, thus generating an extremely stable epigenetic state. For this reason, epialleles in polyploid plants might be highly relevant for long-term adaptation of gene expression patterns (Baubec et al., 2010). In our polyploids, the Alexander’s stain revealed a high pollen viability (>70% in all tetraploids; not shown), suggesting that it may be possible to obtain subsequent generations to study the effects of meiosis on the stability of epialleles.
For both species, no change occurred concurrently in all tetraploids analysed with respect to their diploid parent, revealing a stochastic trend in the changes observed. The occurrence of stochastic methylation alterations in our genetically identical autopolyploids confirms results previously reported in maize (Zhao et al., 2007). Such a phenomenon demands additional studies on subsequent generations to verify whether the identified methylation variations are artefacts of the lack of meiosis. Ploidy-induced DNA methylation changes were reported by Ochogavía et al. (2009) in Eragrostis curvula Schrad. Nees and Lavania et al. (2012) in Cymbopogon spp. The authors reported enhanced cytosine methylation as a result of genome duplication. By contrast, in Paspalum notatum Flügge (Martelotto et al., 2007) and watermelon (Wang et al., 2009), DNA methylation sites were similar between diploids and autotetraploids. These reports, along with our results, suggest that epigenetic modifications are probably induced by genome doubling per se but appear to be linked to different taxa and might be related to (i) the plastic nature of the epigenetic landscape of the species, which tends to respond more or less promptly to external and internal stimuli; and (ii) the CG/CHG content of a given species, as those rich in either CG or CHG may depend on methylation more heavily than species with lower CG/CHG content to regulate gene expression following polyploidization.
Despite their importance, the morphological and anatomical effects of alterations in ploidy level are not well understood (Riddle et al., 2006), and how ploidy and cell size are linked is still unclear (Tsukaya, 2008). In general, polyploidy results in significant cell enlargement, probably to maintain a constant balance between the nuclear and cytoplasm volume (Cavalieri and Smith, 1985). Phenotypic analysis at the cellular level has demonstrated that synthetic polyploidy resulted in larger epidermal and guard cells in Cymbopogon spp. (Lavania et al., 2012) and Nicotiana spp. (Anssour et al., 2009), larger mesophyll cells in P. drummondii Hook (Vyas et al., 2007), and thicker leaves and larger mesophyll cells in Citrus spp. (Romero-Aranda et al., 1997). Polyploidization has also been considered responsible for changes at the vascular level in Manihot esculenta Crantz, including the formation of smaller xylem elements characterized by thinner cell walls (Nassar et al., 2008). In our research, we focused on characterizing the main veins and stomatal parameters in leaves, as they are related to water conductivity, photosynthetic capacity, and hydraulic vulnerability under drought conditions (Blackman et al., 2010). In general, our analyses did not show a consistent superiority of tetraploids compared with their diploid progenitors (Table 3, Figs 1a, b, 2, 3c, d). Exceptions were observed in S. commersonii for leaf LT and in S. bulbocastanum for VN and VLA (Fig. 1c, d) and CW (Fig. 1a, b). Lack of consistent phenotypic superiority of synthetic tetraploids on their diploid parents has been reported for a number of anatomic traits in potato (Stupar et al., 2007), Citrus limonia Osbeck (Allario et al., 2011), Artemisia annua L. (Banyai et al., 2010), and Atriplex confertifolia Torr. et Frém (Warner and Edwards, 1989). We hypothesize that the occasional tetraploid superiority we found may be due to genetic variation on the phenotypic effects of autopolyploidization. To explain the lack of phenotypic superiority of several tetraploids over their diploid parents, at least two hypotheses can be formulated. Firstly, plants with a higher ploidy may experience fitness costs due to elevated DNA content and replication, defective mitosis, and increased cell size (Stupar et al., 2007). To support this hypothesis further, a recent treatment by Tsukaya (2008) reported that an increase in ploidy might alter the ratio between components of the cell with different dimensions, adding stress to polyploid cell activity. Secondly, the epigenetic changes detected in our synthetic polyploids, albeit subtle, may have caused modifications in the expression of relevant genes. The chance that small methylation changes are likely to exert an effect on the phenotype has been described previously (Gemma et al., 2009; Einstein et al., 2010). Therefore, it would be interesting to study the whole-genome expression patterns of our diploid and tetraploid materials as well as other mechanisms regulating gene expression, such as histone modifications and chromatin states.
In conclusion, our results on genome reorganization following chromosome doubling indicated that structural changes did not occur in the wild potato genotypes analysed here. By contrast, methylation modifications were found, with possible effects on gene silencing or activation that deserve further investigation. In terms of leaf anatomical characteristics, polyploidization did not result in a consistent and clear phenotypic superiority of tetraploids. Although larger tetraploid/diploid combinations should confirm these data, they support the idea that studies on autopolyploids should assume the null hypothesis that there are not traits systematically associated with autopolyploidy and, based on this assumption, that genome multiplication per se may be an evolutionary advantage (Parisod et al., 2010).
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Possible methylation status at 5’-CCGG sites, and the corresponding banding patterns produced by the isoschizomers HpaII and MspI.
Supplementary Fig. S2. Examples of MSAP electropherograms and banding patterns on 2× and 4× genotypes of S. commersonii.
Acknowledgements
We thankfully acknowledge Clara Conicella and Federica Consiglio for critically reading the manuscript. We are grateful to the CSIM (Centro Servizi Interdipartimentali di Microscopia, University of Molise, Campobasso, Italy), to Lucia Maiuro (Dipartimento Agricoltura, Ambiente, Alimenti) and Maria Teresa Scarano (CNR-IGV, Institute of Plant Genetics, Res. Div. Portici) for carrying out the SEM analysis. This paper received the contribution number 279 by the Department of Soil, Plant, Environmental and Animal Production Sciences. This research was carried out in the frame of the PRIN project ‘Effects of the ploidy level on gene expression and genome structure in alfalfa and potato’ funded by MiUR.
Glossary
Abbreviations:
- 2n
chromosome number of somatic cells
- 2x
diploid
- 4x
tetraploid
- CW
guard cell length
- FAM
6-carboxyfluorescein
- LA
leaf area
- LT
lamina thickness
- MSAP
methylation-sensitive amplification polymorphism
- SD
stomatal density
- SEM
scanning electron microscopy
- SL
stomata pore length
- SSR
simple sequence repeats
- SW
stomata pore length
- VLA
area of vessel lumen
- VLD
mean diameter of vessel lumen
- VN
number of vessels in the main vein
- VWT
vessel wall thickness.
References
- Albertin W, Brabant P, Catrice O, Eber F, Jenczewski E, Chèvre AM, Thiellement H. 2005. Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics 5, 2131–2139 [DOI] [PubMed] [Google Scholar]
- Allario T, Brumos J, Colmenero-Flores JM, Tadeo F, Froelicher Y, Talon M, Navarro L, Ollitrault P, Morillon R. 2011. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. Journal of Experimental Botany 62, 2507–2519 [DOI] [PubMed] [Google Scholar]
- Anssour S, Krügel T, Sharbel TF, Saluz HP, Bonaventure G, Baldwin IT. 2009. Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia . Annals of Botany 103, 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai W, Sangthong R, Karaket N, Inthima P, Mii M, Supaibulwatana K. 2010. Overproduction of artemisinin in tetraploid Artemisia annua L. Plant Biotechnology 27, 427–433 [Google Scholar]
- Baubec T, Dinh HQ, Pecinka A, Rakic B, Rozhon W, Wohlrab B, Haeseler von A, Mittelsten Scheid O. 2010. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis . The Plant Cell 22, 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman CJ, Brodribb TJ, Jordan GJ. 2010. Leaf hydraulic vulnerability is related to conduit dimensions and drought resistance across a diverse range of woody angiosperms. New Phytologist 188, 1113–1123 [DOI] [PubMed] [Google Scholar]
- Carputo D, Barone A. 2005. Ploidy manipulation through sexual hybridization. Annals of Applied Biology 146, 71–79 [Google Scholar]
- Caruso I, Lepore L, De Tommasi N, Dal Piaz F, Aversano R, Garramone R, Carputo D. 2011. Secondary metabolite profile in induced tetraploids of wild Solanum commersonii Dun. Chemistry & Biodiversity 8, 2226–2237 [DOI] [PubMed] [Google Scholar]
- Cavalieri AJ, Smith OS. 1985. Grain filling and field drying of a set of maize hybrids released from 1930 to 1982. Crop Science 25, 856–860 [Google Scholar]
- Chen ZJ. 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology 58, 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploids. Nature Reviews Genetics 6, 836–846 [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B. 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. The Plant Cell 12, 1551–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis D, Soltis P, Wendel JF. 2008. Evolutionary genetics of genome merger and doubling in plants. Annual Review of Genetics 42, 443–461 [DOI] [PubMed] [Google Scholar]
- Einstein F, Thompson RF, Bhagat TD, Fazzari MJ, Verma A, Barzilai N, Greally JM. 2010. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS ONE 5, e8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder N, O’Brien TP. 1968. Plant microtechnique: some principles and new methods. American Journal of Botany 55, 123–142 [Google Scholar]
- Feingold S, Lloyd J, Norero N, Bonierbale M, Lorenzen J. 2005. Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theoretical and Applied Genetics 111, 456–466 [DOI] [PubMed] [Google Scholar]
- Gemma C, Sookoian S, Alvariñas J, García SI, Quintana L, Kanevsky D, González CD, Pirola CJ. 2009. Maternal pregestational BMI is associated with methylation of the PPARGC1A promoter in newborns. Obesity 17, 1032–1039 [DOI] [PubMed] [Google Scholar]
- Ghislain M, Nunez J, Herrera MDR, Pignataro J, Guzman F, Bonierbale M, Spooner DM. 2009. Robust and highly informative microsatellite-based genetic identity kit for potato. Molecular Breeding 23, 377–388 [Google Scholar]
- Hauser MT, Aufsatz W, Jonak C, Luschnig C. 2011. Transgenerational epigenetic inheritance in plants. Biochimica et Biophysica Acta 1809, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M, Rossi V. 2011. Epigenetic control of gene regulation in plants. Biochimica et Biophysica Acta-Gene Regulatory Mechanisms 1809, 369–378 [DOI] [PubMed] [Google Scholar]
- Lavania UC, Srivastava S, Lavania S, Basu S, Misra NK, Mukai Y. 2012. Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. The Plant Journal 71, 539–549 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics 3, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution. New York: Oxford University Press; [Google Scholar]
- Levy AA, Feldman M. 2004. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society 82, 607–614 [Google Scholar]
- Li X, Yu E, Fan C, Zhang C, Fu T, Zhou Y. 2012. Developmental, cytological and transcriptional analysis of autotetraploid Arabidopsis . Planta 236, 579–596 [DOI] [PubMed] [Google Scholar]
- Lim KY, Souckova-Skalicka K, Sarasan V, Clarkson JJ, Chase MW, Kovarik A, Leitch AR. 2006. A genetic appraisal of a new synthetic Nicotiana tabacum (Solanaceae) and the Kostoff synthetic tobacco. American Journal of Botany 93, 875–883 [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Ortiz JPA, Stein J, Espinoza F, Quarin CL, Pessino SC. 2007. Genome rearrangements derived from autopolyploidization in Paspalum sp. Plant Science 172, 970–977 [Google Scholar]
- McClelland M, Nelson M, Raschke E. 1994. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Research 22, 3640–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbourne D, Meyer RC, Collins AJ, Ramsay LD, Gebhardt C, Waugh R. 1998. Isolation, characterization and mapping of simple sequence repeat loci in potato. Molecular Genetics and Genomics 259, 233–245 [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Afsar K, Paszkowski J. 2003. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana . Nature Genetics 34, 450–454 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497 [Google Scholar]
- Nassar NMA, Graciano Ribeiro D, Fernandes SDC, Araujo PC. 2008. Anatomical alterations due to polyploidy in cassava, Manihot esculenta Crantz. Genetics and Molecular Research 7, 276–283 [DOI] [PubMed] [Google Scholar]
- Ochogavía AC, Cervigni G, Selva JP, Echenique VC, Pessino SV. 2009. Variation in cytosine methylation patterns during ploidy level conversions in Eragrostis curvula . Plant Molecular Biology 70, 17–29 [DOI] [PubMed] [Google Scholar]
- Ozkan H, Tuna M, Galbraith DW. 2006. No DNA loss in autotetraploids of Arabidopsis thaliana . Plant Breeding 125, 288–291 [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytologist 186, 5–17 [DOI] [PubMed] [Google Scholar]
- Pignatta D, Dilkes BP, Yoo SY, Henry IM, Madlung A, Doerge RW, Jeffrey Chen Z, Comai L. 2010. Differential sensitivity of the Arabidopsis thaliana transcriptome and enhancers to the effects of genome doubling. The New Phytologist 186, 194–206 [DOI] [PubMed] [Google Scholar]
- Quarin CL, Espinoza F, Martinez EJ, Pessino SC, Bovo OA. 2001. A rise of ploidy level induces the expression of apomixis in Paspalum notatum . Sexual Plant Reproduction 13, 243–249 [Google Scholar]
- Raina SN, Parida A, Koul KK, Salimath SS, Bisht MS, Raja V, Khoshoo TN. 1994. Associated DNA changes in polyploids. Genome 37, 560–564 [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. 2002. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics 33, 589–639 [Google Scholar]
- Reyna-López GE, Simpson J, Ruiz-Herrera J. 1997. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Molecular Genetics and Genomics 253, 703–710 [DOI] [PubMed] [Google Scholar]
- Riddle NC, Hongmei J, An L, Doerge RW, Birchler JA. 2010. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theoretical and Applied Genetics 120, 341–353 [DOI] [PubMed] [Google Scholar]
- Riddle NC, Kato A, Birchler JA. 2006. Genetic variation for the response to ploidy change in Zea mays L. Theoretical and Applied Genetics 114, 101–111 [DOI] [PubMed] [Google Scholar]
- Romero-Aranda R, Bondada BR, Syvertsen JP, Grosser JW. 1997. Leaf characteristics and net gas exchange of diploid and autotetraploid Citrus . Annals of Botany 79, 153–160 [Google Scholar]
- Schranz ME, Osborn TC. 2004. De novo variation in life history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae). American Journal of Botany 91, 174–183 [DOI] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proceedings of the National Academy of Sciences, U S A 92, 7719–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanys V, Weckman A, Staniene G, Duchovskis P. 2006. In vitro induction of polyploidy in Japanese quince (Chaenomeles japonica). Plant Cell, Tissue and Organ Culture 84, 263–268 [Google Scholar]
- Stupar RM, Bhaskar PB, Yandell BS, et al. 2007. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 176, 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JA, Ni Z, Scheen AC, Koh J, Gilbert CA, Lefkowitz D, Jeffrey CZ, Soltis PS, Soltis DE. 2006. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173, 1599–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. 2008. Controlling size in multicellular organs: focus on the leaf. PLoS Biology 6, e174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P, Bisht MS, Miyazawa SI, Yano S, Noguchi K, Terashima I, Funayama-Noguchi S. 2007. Effects of polyploidy on photosynthetic properties and anatomy in leaves of Phlox drummondii . Functional Plant Biology 34, 673–682 [DOI] [PubMed] [Google Scholar]
- Wang CG, Hui L, Xue ZY, Chen CB, Gu Y, Sun DL, Song WQ. 2009. Marker-based analysis of genome structure and DNA methylation in a watermelon (Citrullus lanatus) ploidy series. Botanical Studies 50, 389–402 [Google Scholar]
- Warner DA, Edwards GE. 1989. Effects of polyploidy on photosynthetic rates, photosynthetic enzymes, contents of DNA, chlorophyll, and sizes and numbers of photosynthetic cells in the C4 dicot Atriplex confertifolia . Plant Physiology 91, 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong LZ, Xu CG, Saghai Maroof MA, Zhang QF. 1999. Patterns of cytosine methylation in an elite rice hybrid and its parental lines by a methylation-sensitive amplification polymorphism technique. Molecular Genetics and Genomics 261, 439–446 [DOI] [PubMed] [Google Scholar]
- Yang X, Ye CY, Cheng ZM, Tschaplinski TJ, Wullschleger SD, Yin W, Xia X, Tuskan GAA. 2011. Genomic aspects of research involving polyploid plants. Plant Cell, Tissue and Organ Culture 3, 387–397 [Google Scholar]
- Yu Z, Haberer G, Matthes M, Rattei T, Mayer KFX, Gierl A, Torres-Ruiz RA. 2010. Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana . Proceedings of the National Academy of Sciences, U S A 107, 17809–17814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chai Y, Liu B. 2007. Epigenetic inheritance and variation of DNA methylation level and pattern in maize intra-specific hybrids. Plant Science 172, 930–938 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.