Abstract
Seed germination and flowering initiation are both transitions responding to similar seasonal cues. This study shows that ABSCISIC ACID-INSENSITIVE MUTANT 5 (ABI5), a bZIP transcription factor, which plays an important role in the abscisic acid (ABA)-arrested seed germination, is robustly associated with the floral transition in Arabidopsis. Under long-day conditions, overexpression of ABI5 could delay floral transition through upregulating FLOWERING LOCUS C (FLC) expression. In contrast, ectopically overexpressing FLC in an abi5 mutant reversed the earlier flowering phenotype. Further analysis indicated that transactivation of FLC could be promoted by ABI5 and/or other abscisic acid-responsive element (ABRE)-binding factors (ABFs). The expression of FLC that was promoted by ABI5 and/or other ABFs could be blocked in a triple SNF1-related protein kinase (SnRK) mutant, snrk2.2/2.3/2.6, despite the presence of ABA. In sharp contrast, when SnRK2.6 was coexpressed, the reduction of transactivity of FLC was reverted in mesophyll protoplasts of snrk2.2/2.3/2.6. Additional results from analysing transgenic plants carrying mutations of phosphoamino acids (ABI5 S42AS145AT201A), which are conserved in ABI5, suggested that SnRK2-mediated ABI5 and/or ABF phosphorylation may be crucial for promoting FLC expression. The transgenic plants ABI5 S42AS145AT201A were insensitive to ABA in seed germination, in addition to having an earlier flowering phenotype. Direct binding of ABI5 to the ABRE/G-box promoter elements existing in FLC was demonstrated by chromatin immunoprecipitation. Mutations at the ABRE/G-box regions in FLC promoter sequences abolished the ABI5-promoted transactivation of FLC. In summary, these results may decipher the inhibitory effect of ABA on floral transition in Arabidopsis.
Key words: ABA, ABFs, ABI5, chromatin immunoprecipitation, FLC, flowering time, SnRK2s.
Introduction
The transition to flowering initiation is one of the most important decisions in the plant life cycle (Simpson and Dean, 2002; Boss et al., 2004). Optimal timing for switching from vegetative growth to reproductive development is crucial to maximize the reproductive accomplishment. Accordingly, plants have evolved mechanisms to regulate the timing of floral initiation (Boss et al., 2004; Jack, 2004).
Four classic regulatory pathways involving the photoperiod, vernalization, autonomous pathways, and gibberellic acid (GA) have been identified in Arabidopsis (Mouradov et al., 2002). The environmental and endogenous signals can be collaboratively sensed by the plant in order to initiate transition of timely flowering (Moon et al., 2005; Liu et al., 2009). FLOWERING LOCUS C (FLC), one of the repressor integrators, tightly controls flowering signals (Michaels and Amasino, 1999, 2001). FLC expression can be repressed by vernalization and autonomous pathways via modulating the chromatin structure (Michaels, 2009); thus, flowering can be promoted through revoking the inhibitory effect of FLC on the expression of FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) (Helliwell et al., 2006; Searle et al., 2006).
Phytohormones have diverse effects in the growth and development of plants, which regulate multiple physiological, metabolic, and cellular processes. Regulations of phytohormones in the floral transition have been reported (Gray, 2004; Davis, 2009; Domagalska et al., 2010). The involvement of GA in the control of flowering in Arabidopsis has been demonstrated by applying GAs to a plant to stimulate bolting (Lang, 1956, 1957). Mutants deficient in GA biosynthesis exhibit dramatic delays in flowering under short-day conditions (Hisamatsu et al., 2000; Moon et al., 2003). The inhibitory effect of ABA in floral transition has been documented in an ABA-deficient mutant (Martinez-Zapater et al., 1994). Modulation of DELLA protein activity can instigate the inhibitory effect of ABA on flowering (Achard et al., 2006). Thus, ABA is considered as a floral repressor.
However, the mechanism of delayed flowering time by ABA in Arabidopsis is poorly understood, and whether ABA could undertake a convergent approach to sustain negative regulation on FLC is elusive. A study has shown that FLC-mediated seed germination proceeds through genes such as FT, SOC1, and APETALA1, making FLC a promising regulator of temperature-dependent seed germination (Chiang et al., 2009). In Arabidopsis, most abscisic acid-responsive element (ABRE)-binding factors (ABFs), which are classified as bZIP transcription factors, are involved in ABA signal transduction during seed germination and/or in vegetative growth (Choi et al., 2000; Jakoby et al., 2002). FD, another bZIP protein, mediates signals from FT at the shoot apex in Arabidopsis (Abe et al., 2005). ABF1, ABF3, and ABF4 are mainly expressed in vegetative tissues whereas ABSCISIC ACID-INSENSITIVE MUTANT 5 (ABI5), a bZIP transcription factor, is preferentially expressed during seed maturation and seed germination (Finkelstein and Lynch, 2000; Uno et al., 2000). Information about ABI5 in other developmental aspects is inadequate.
Seed germination and flowering initiation are both transitions that could result from responding to a similar seasonal cue and these two important life transitions might share common regulatory elements in genetic pathways. This study shows that, in addition to controlling seed germination, ABI5 may be functional in floral transition in Arabidopsis. The results indicate that overexpression of ABI5 could actually delay flowering initiation via upregulating FLC expression. Phosphorylation of ABI5 by sucrose nonfermenting 1-related protein kinase (SnRK) 2 directly influenced floral transition; without phosphorylation, the inhibitory effect of ABI5 on floral transition was abolished. Direct binding of ABI5 to FLC promoter regions could transactivate FLC expression. All data suggest a positive regulation by ABI5 on FLC activity for the control of floral transition in Arabidopsis.
Materials and methods
Plant materials, growth conditions, and measurement of flowering time
The abi5 mutant, abi5-4 (Lopez-Molina and Chua, 2000) and the wild type (Ws-0) were in the Wassilewskija background. Other plants used in this study were in the Columbia (Col-0) background. The snrk2.2/3/6+ mutant was kindly provided by Dr Jian-Kang Zhu (Purdue University) (Fujii et al., 2007; Fujii and Zhu, 2009). Primer sequences for identifying homozygous snrk2.2/2.3/2.6 are listed in Supplementary Table S1 (available at JXB online). Transgenic plants of ABI5-5, ABI5 S42A, ABI5 S145A, and ABI5 S42AS145AT201A were generated by Agrobacterium tumefaciens GV3101-mediated floral infiltration (Clough and Bent, 1998). To overexpress FLC in the abi5-4 mutant background, plasmid p35S::GFP-FLC was transformed into abi5-4 plants. To assess the expression pattern of FLC in planta, the plasmids pFLC::GUS and pFLC(m)::GUS were introduced into ABI5-5 plants, respectively. Plants were grown in soil or on MS medium (Phyto Technology, USA) containing 1% sucrose and 0.8% (w/v) agar at 23 °C in a growth room under long-day conditions (16h/8h light/dark) or short-day conditions (8h/16h light/dark). To detect the effect of ABA on flowering time, 2-week-old plants were sprayed with 100 µM ABA thrice a week until all plants start flowering. The stock solution of ABA (Sigma-Aldrich, USA) was dissolved in ethanol. The control treatment was performed with an equal amount of the solvent. Flowering time was scored as the number of total rosette leaves at bolting. At least 15 plants were scored in each group.
Plasmid construction
Primer sequences for cloning the constructs in this study are listed in Supplementary Table S2 and detailed information on the plasmids can be found in Supplementary Table S3. In brief, p35S::ABI5S42A-GFP, p35S::ABI5S145A-GFP, p35S::ABI5S42AS145A-GFP and p35S::ABI5S42AS145AT201A-GFP were made through site-directed mutagenesis by PCR amplification (Edelheit et al., 2009), based on plasmid p35S::ABI5-GFP. Plasmid p35S::ABI5-HA was constructed by inserting the ABI5 coding sequence into pBA002 at the XhoI/SacII site (Kost et al., 1998).
Transient expression assay
Mesophyll protoplasts were prepared from 4-week-old plants of Col-0 and snrk2.2/2.3/2.6 according to the methods described previously (Yoo et al., 2007). All plasmids were prepared through the purification with caesium chloride/ethidium bromide (Sambrook et al., 1989). Protein kinase inhibitor K252a (Sigma-Aldrich, USA) was dissolved in DMSO (Sigma-Aldrich, USA) and accordingly added to the culture. The relative activity of LUC/GUS was scored after transformation. The transformed protoplasts were incubated at 23 °C for 12 hours under darkness. Each experiment was repeated at least three times.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was essentially performed according to methods described in several reports (Lee et al., 2007; Xi et al., 2010). DNA was recovered as described by Wang et al. (2002). The precipitated DNA fragments were quantified by quantitative real-time PCR (qRT-PCR) amplification using specific primers (Supplementary Table S4).
Analysis of gene expression
Total RNA was isolated from 5-day-old seedlings. The protocols used for qRT-PCR experiments have been described previously (Wang et al., 2011). Primer sequences are available in Supplementary Table S5.
GUS histochemical staining
GUS signals were detected with the method described by Jefferson et al. (1987). Plants were pretreated with or without 100 µM ABA for 3 hours, and then immersed in 90% acetone for 30 minutes. After they were incubated in the GUS staining solution (0.5mg/ml X-Gluc, 50mM PBS, pH 7.0; 5.0mM potassium ferricyanide, 5.0mM potassium ferrocyanide, 0.1% Triton X-100) at 37 °C overnight, the stained plants were washed with 70% ethanol overnight. Images were taken with an inverted microscope (SMZ1500, Nikon).
Results
Overexpression of ABI5 in Col-0 influences flowering time
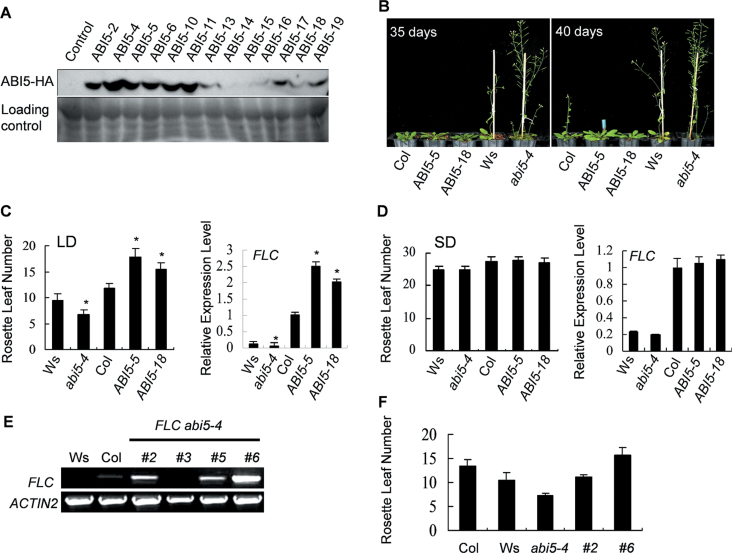
To assess the correlation between flowering time and ABI5 expression, overexpressing transgenic lines carrying plasmid p35S::ABI5-HA in Col-0 background were generated. A total of 43 lines were analysed by examining ABI5-HA protein level in individual transgenic plants (Fig. 1A). Quantifying the rosette leaf numbers indicated that flowering time of transgenic lines ABI5-5 and ABI5-18 was delayed under long-day conditions; however, abi5-4 showed slightly earlier flowering (Fig. 1B, C). Under short-day conditions, the flowering time was not much different in all examined lines (Fig. 1D). Subsequently, the microarray data available in public resources were analysed and the expression pattern of FLC was actually similar to that of ABI5 (Supplementary Fig. S1A; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). Thus, the present study examined the transcriptional correlation between ABI5 and FLC and found that both genes had decreased expression levels during the growth of seedlings (Supplementary Fig. S1B). Under long-day conditions, although the expression level of FLC in abi5-4 plants was downregulated, it was accordingly upregulated in ABI5-5 and ABI5-18 plants (Fig. 1C). This phenomenon was not observed in all of the examined lines under short-day conditions (Fig. 1D). To characterize the genetic relationship between ABI5 and FLC, p35S::FLC-GFP was introduced into abi5-4 plants. The expression level of FLC in transgenic hybrid FLC abi5-4 plants (T2) was then analysed (Fig. 1E) and the earlier flowering phenotype of abi5-4 was reverted (Fig. 1F). Thus, these results imply a positive role of ABI5 in regulating FLC expression under long-day conditions, in terms of disturbing floral transition in Arabidopsis.
Fig. 1.
Overexpression of ABI5 altered flowering time. (A) The expression level of ABI5-HA fusion protein in individual transgenic lines of p35S::ABI5-HA. (B) Flowering-time phenotypes in abi5-4 (Ws), ABI5-5 (Col) and ABI5-18 (Col) under long-day conditions. (C and D) Comparisons of total rosette leaf numbers (n > 15 for each experiment) and FLC expression under long-day (LD) (C) and short-day (SD) conditions (D) (*P < 0.05). (E) FLC expression in FLC abi5-4 transgenic lines (T2, homozygous) (#2, #3, #5, and #6 are individual lines). Total RNA was extracted from 10-day-old seedlings. ACTIN2 represents the loading control. (F) Comparisons of total rosette leaf numbers. Data are mean ± standard errors of three replicated experiments (n > 15 for each experiment).
ABI5 activates FLC transcription in an ABA-dependent manner
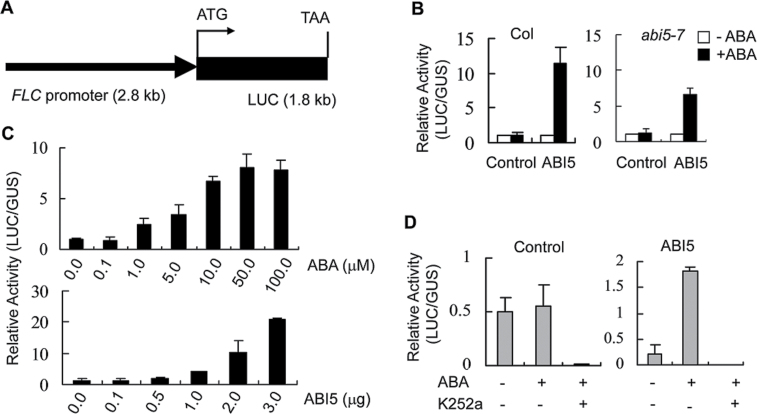
A FLC promoter fragment (2.8kb upstream of ATG) was cloned and fused to the luciferase (LUC) coding sequence (Fig. 2A), the resulting plasmid, pFLC::LUC, was used as the reporter construct. Plasmid pUBQ::GUS was used as the internal control (Wang et al., 2011). Plasmid p35S::ABI5-GFP (ABI5) and pFLC::LUC and pUBQ::GUS were cotransformed into mesophyll protoplasts of Col and abi5-7 (Col) respectively. The relative activity of LUC/GUS was significantly increased in coexpressing p35S::ABI5-GFP with ABA treatment (Fig. 2B). The fold-change of relative activity (LUC/GUS) was apparently greater in Col than that in abi5-7 (Fig. 2B). The relative activity (LUC/GUS) of FLC::LUC was further analysed under a range of ABA concentrations (0–100 µM). The relative activity (LUC/GUS) of FLC::LUC was enhanced simultaneously with increasing ABA concentrations when transformed with the same amount (5 µg) of p35S::ABI5-GFP plasmid DNA (Fig. 2C). Hence, the relative activity (LUC/GUS) of FLC::LUC with 50 µM ABA was compared with a gradient dose of p35S::ABI5-GFP plasmid DNA (0–3 µg). FLC::LUC activity gradually increased and was tightly linked to the amount of p35S::ABI5-GFP DNA (Fig. 2C). As ABI5 activity can be modulated by SnRK2s (Nakashima et al., 2009), the transfected protoplasts were treated with 50nM protein kinase inhibitor K252a. Application of K252a could completely abolish the activity of FLC::LUC despite of presence of ABA (Fig. 2D). In summary, the results suggest an effect of ABA on modulating flowering signalling, which may be through interactive communication between ABI5 and FLC.
Fig. 2.
ABI5-triggered FLC promoter activity is associated with abscisic acid (ABA) treatment. (A) Schematic diagram of pFLC::LUC construct. (B) Relative activity (LUC/GUS) of pFLC::LUC in coexpression with ABI5 with and without 50 µM ABA for 12 hours. (C) Relative activity (LUC/GUS) of pFLC::LUC with various concentrations of ABA, and various doses of p35S::ABI5-GFP DNA. (D) Relative activity (LUC/GUS) of pFLC::LUC was abolished when treated with 50nM protein kinase inhibitor (K252a) for 12 hours. Control, p35S::GFP; ABI5, p35S::ABI5-GFP. Data are mean ± standard errors of three replicated experiments (n > 5 for each experiment).
Phosphorylation of ABI5 is crucial in regulating flowering time
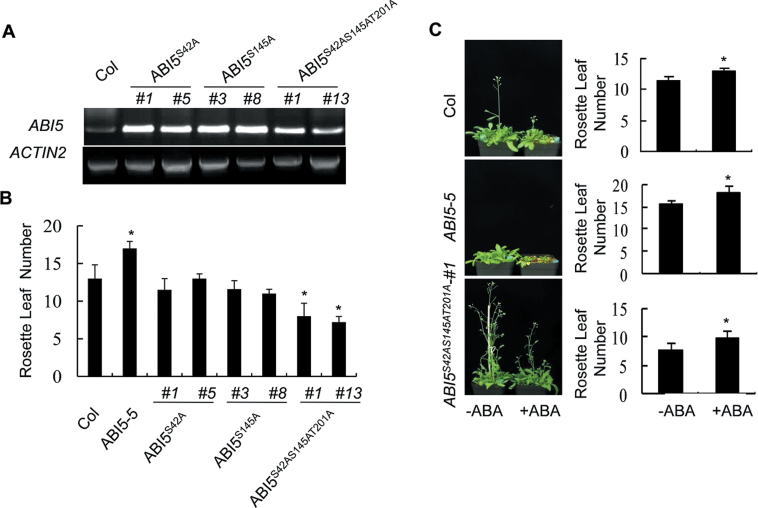
Four conserved phosphoamino acids (S41, S42, S145, T201) in ABI5 sequences have been identified (Lopez-Molina et al., 2002). To analyse the importance of ABI5 phosphorylation in the regulation of flowering time, three phosphoamino acids (S42, S145, and T201) were replaced by alanine, and transgenic plants expressing the mutated forms (ABI5 S42A, ABI5 S145A, and ABI5 S42AS145AT201A) were then generated. The expression level of ABI5 in the transgenic plants were evaluated by reverse-transcription PCR (Fig. 3A). Unlike ABI5-5 plants, flowering time was evidently earlier in the transgenic plants (Fig. 3B and Supplementary Fig. S2A, B). Moreover, the retarded flowering time with 50 µM ABA was also observed in plants carrying the triple mutation ABI5 S42AS145AT201A (Fig. 3C). To further untangle the correlation between the flowering time and ABA response, the ABA response of the ABI5 S42AS145AT201A line in seed germination was analysed. Higher germination rates were scored with the ABI5 S42AS145AT201A line on the plate containing 1 µM ABA (Supplementary Fig. 2C, D). Therefore, the data demonstrate that failure of ABI5 phosphorylation may lead to dysfunction of ABA signalling and suggest the importance of phosphorylation modification of ABI5 in the regulation of flowering time in Arabidopsis.
Fig. 3.
ABI5 phosphorylation is essential for flowering time regulation. (A) ABI5 expression was evaluated in independent transgenic lines, ABI5 S42A, ABI5 S145A, and ABI5 S42AS145AT201A. ACTIN2 represents the loading control. (B) Comparative analysis of total rosette leaf numbers. Data represent the means ± SEs of three replicated experiments (n > 20 for each experiment). (C) Application of exogenous abscisic acid (ABA) significantly delays flowering time in Col and ABI5-5 plants, but a partial effect is observed in ABI5 S42AS145AT201A plants. Data are mean ± standard errors of three independent experiments (n > 15 for each experiment).
Downstream targets in the flowering pathways can be altered by ABI5
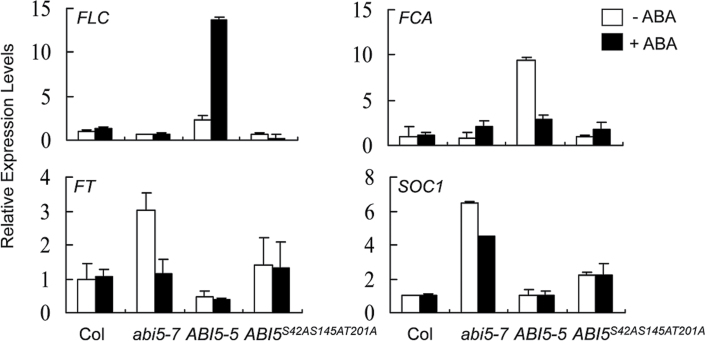
Several downstream targets whose expression can be regulated by FLC are key players in flowering signal transduction pathways. This study scored the expression of FLC in ABI5-5 plants under 50 µM ABA treatment (Fig. 4). In contrast, the level of FLC expression was reduced in abi5-7 and ABI5 S42AS145AT201A plants, which may explain their earlier flowering phenotypes (Figs. 1B and C and Supplementary Fig. S2A). Notably, the expression of FCA in ABI5-5 plants was inhibited; however, it was slightly increased in abi5-7 and ABI5 S42AS145AT201A plants after ABA treatment. Without ABA, FT expression was significantly upregulated in abi5-7 plants. Contrarily, with ABA treatment the level of FT expression was drastically decreased in abi5-7 but not in Col or ABI5 S42AS145AT201A plants. In addition, FT expression was sustained at lower level with or without ABA treatment in ABI5-5 plants. Similar expression patterns of SOC1 were also obtained (Fig. 4). Taken together, these data extend the suggestion that the negative effect of ABI5 on activation of FT and SOC1 may be associated with the ABI5 activity that is dependent on ABA signalling.
Fig. 4.
Relative expression levels of flowering-related genes. 10-day-old seedlings were treated with or without 50 µM abscisic acid (ABA) for 3 hours. Gene expression in Col without ABA treatment were taken as the reference level; then fold-changes in abi5-7 (Col), ABI5-5, and ABI5 S42AS145AT201A plants were normalized relative to the reference. Data are mean ± standard errors of three independent experiments (n > 4 for each experiment).
SnRK2s are required for ABI5-promoted FLC::LUC activity
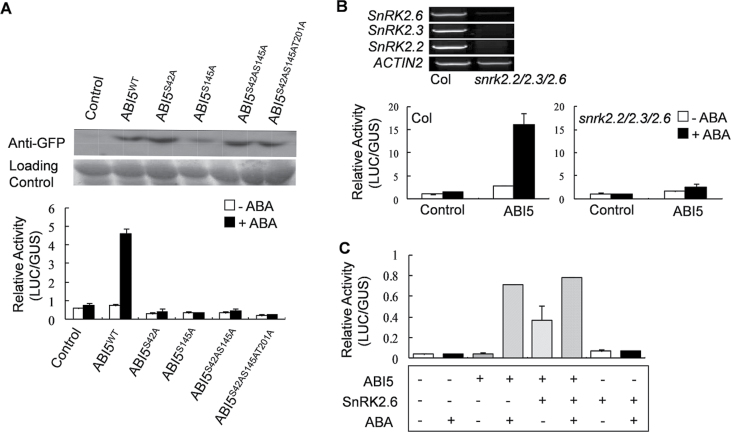
The relative activity (LUC/GUS) of FLC::LUC was abolished despite ABA treatment when p35S::ABI5S42A-GFP was coexpressed with pFLC::LUC. Similar results were obtained in coexpressions of p35S::ABI5S145A-GFP, p35S::ABI5S42AS145A-GFP, and p35S::ABI5S42AS145AT201A-GFP (Fig. 5A). Changing the phosphoamino acids to alanine did not affect the stability of ABI5s (Fig. 5A). A transient expression assay was conducted in mesophyll protoplasts isolated from homozygous plants of the triple mutant snrk2.2/2.3/2.6. ABA treatment failed to promote activation of FLC::LUC, even when ABI5 was coexpressed (Fig. 5B). When SnRK2.6 and ABI5 were coexpressed in Col protoplasts, the relative activity (LUC/GUS) of FLC::LUC could be detected; in addition, it was enhanced by 50 µM ABA (Fig. 5C). These data are consistent with the earlier flowering phenotype observed in heterozygous triple mutant snrk2.2/3/6+ plants (Supplementary Fig. S3A). Subsequently, the roles of other ABA-responsive ABFs in regulating FLC expression were analysed. With 50 µM ABA, all three tested ABFs (ABF1, ABF3, and ABF4) could trigger the relative activity of FLC::LUC in Col but not in snrk2.2/2.3/2.6 (Supplementary Fig. S3B). These data demonstrate that SnRK2s may play an important role in the ABI5- and/or ABFs-modulated activation of FLC at the transcriptional level.
Fig. 5.
SNF1-related protein kinases (SnRKs) may be involved in the positive regulation by ABI5 on FLC transcription. (A) Phosphoamino acid mutations of ABI5 inhibited the abscisic acid (ABA)-dependent FLC::LUC activity. Western blotting was used to detect ABI5. Data are mean ± standard errors of more than five independent experiments (n > 4 for each experiment). (B) The effect of ABI5 on FLC::LUC activity is significantly decreased in protoplasts of snrk2.2/2.3/2.6 mutants. Homozygous snrk2.2/2.3/2.6 plants were verified by semi-quantitative reverse-transcription PCR. Data are mean ± standard errors of three replicated experiments (n > 4 for each experiment). (C) Transient coexpression of p35S::SnRK2.6 (SnRK2.6) activates FLC promoter activity. Data are mean ± standard errors of more than five experiments (n > 4 for each experiment).
ABI5 regulates FLC expression via directly binding to FLC promoter elements
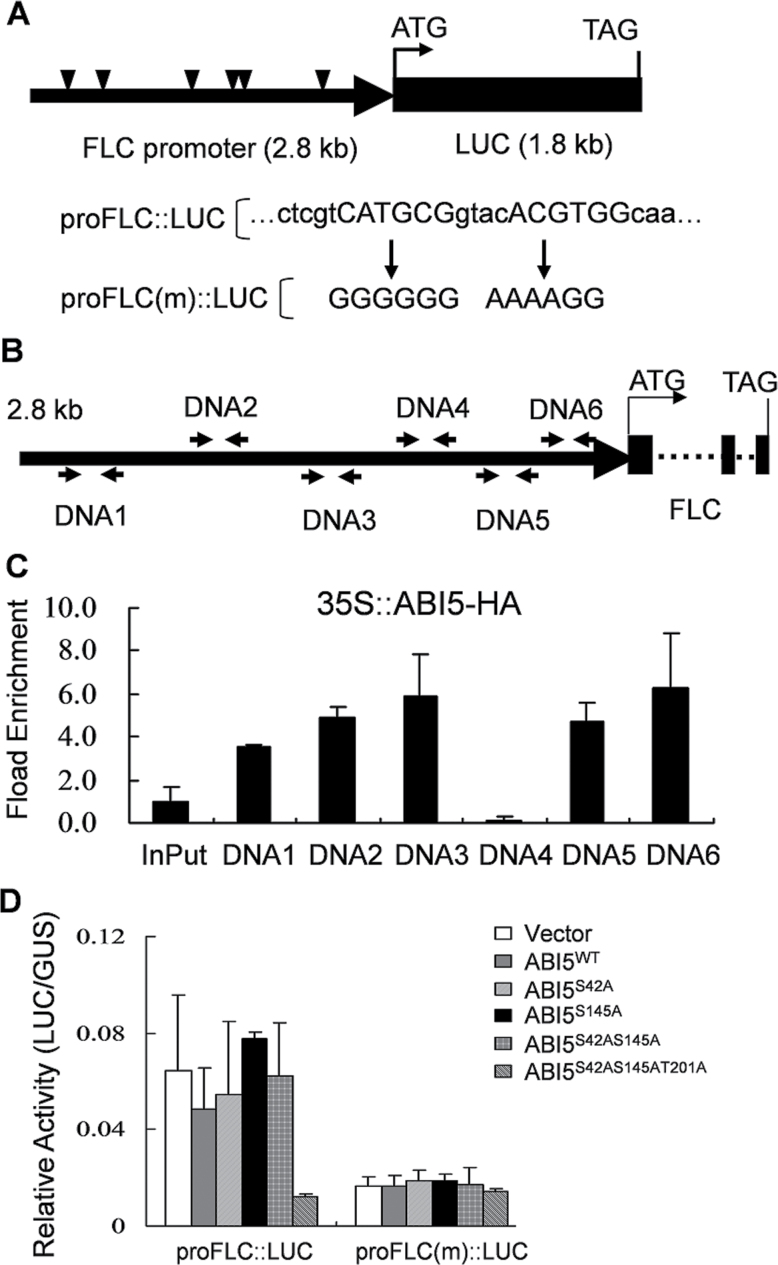
To determine the regulatory rule of ABI5 on FLC expression, this study analysed the 2.8-kb promoter sequences of FLC and found six putative variants of ABRE-like elements (PyACGTGG/TG) (Fig. 6A). In addition, a G-box (CACGTG) element in the promoter region might be classified as a ubiquitous regulatory DNA element, which might be bound by bZIP proteins. Thus, a chromatin immunoprecipitation assay was performed in ABI5-HA transgenic plants. Except for the DNA4 fragment, all other five DNA fragments (DNA1, DNA2, DNA3, DNA5, and DNA6) (Fig. 6B) were immunoprecipitated by ABI5-HA. The best binding efficiency was obtained with the proximal DNA6 fragment (Fig. 6C). The sequence information conserved in the DNA6 fragment was analysed further (Fig. 6B) and, not surprisingly, a putative G-box and a single ABRE-like motif were contained in DNA6 (Fig. 6A). Next, the elements of the putative G-box and ABRE-like motifs were mutated to evaluate their functions in modulating FLC transcription in a transient expression assay (Fig. 6D). When the ABRE-like motif was mutated alone, there was no obvious effect on FLC promoter activity (Supplementary Fig S4A). However, when mutations were created in both motifs, the relative activity of LUC/GUS, representing the expression of proFLC(m)::LUC, was impaired (Fig. 6D). Furthermore, the hybrid lines pFLC::GUS ABI5-5 and pFLC(m)::GUS ABI5-5 were generated to confirm the role of the ABRE/G-box elements in ABI5-regulated FLC promoter activity. Under 100 µM ABA treatment for 3 hours, a significantly enhanced GUS signal was detectable in all examined tissues of pFLC::GUS ABI5-5 plants, but no GUS signals were detected in tissues of pFLC(m)::GUS ABI5-5 plants (Supplementary Fig. S4C). Together with chromatin immunoprecipitation analysis, the results suggest that the ABA inhibitory effect on floral transition is most likely mediated by a bZIP transcription factor, such as ABI5 and/or other ABFs promoting FLC expression.
Fig. 6.
ABI5 directly binds to DNA elements in the FLC promoter. (A) Schematic diagram showing the fusion of reporter constructs of FLC::LUC; triangles indicate the positions of putative ABRE motifs (PyACGTGG/TG) located in the 2.8-kb FLC promoter segment; arrows indicate mutated sequences of ABRE-like and G-box motifs. (B) Schematic diagram to show FLC promoter elements; DNA1–6 indicate the DNA fragments; pairs of arrows represent the DNA fragments amplified in the chromatin immunoprecipitation assay. (C) Chromatin immunoprecipitation enrichment to show the binding ability of ABI5-HA to the DNA fragments of the FLC promoter, as shown by qRT-PCR. InPut, total input chromatin DNA. Data are mean ± standard error of one experiment (n > 3). Similar results were obtained from four independent experiments. (D) Mutated ABRE-like elements in the FLC promoter could partially abolish FLC promoter activity in a transient expression assay. Data are mean ± standard errors of three replicated experiments (n > 3 for each experiment).
Discussion
Negative effect of ABA on flowering time regulation in Arabidopsis
To control flowering time, plants have evolved a complex genetic network for responding to endogenous cues and environmental factors. The mechanism underlying ABA regulation on flowering time is poorly understood. This study showed that the bZIP transcription factors, including ABI5 and other ABFs (such as ABF1, ABF3, and ABF4) play negative roles in ABA-mediated inhibition of flowering time. Application of ABA may delay the flowering time in Arabidopsis (Fig. 3 and Supplementary Fig. S2; Jiang et al., 2012). ABI5 and/or other ABFs could distinctly promote the expression of FLC (Figs. 1–6 and Supplementary Figs. S3 and S4). The stimulation of FLC expression was dependent upon ABA treatment, which was mediated by SnRK2s regulation on ABI5 or other ABFs activity (Figs. 2 and 5 and Supplementary Fig. S3). Coupled with the genetic analysis of the hybrid lines such as FLC abi5-4 and pFLC::GUS ABI5-5 (Fig. 1 and Supplementary Fig. S4), it is most likely that the inhibitory effect of ABA on flowering time is achieved by activation of ABI5 and/or other ABFs and successive transactivation of FLC.
ABI5 phosphorylation is essential for promoting FLC expression
Protein kinases involved in ABA signal transduction have been identified, of which the SnRK2s have been widely studied (Anderberg and Walker-Simmons, 1992; Boudsocq et al., 2004). In Arabidopsis, there are 10 members in the SnRK2 family (SnRK2.1–SnRK2.10) (Hrabak et al., 2003). The triple mutant snrk2.2/2.3/2.6 plants show severe impairment in vegetative and reproductive growth, as well as in the control of transpiration water loss (Fujii and Zhu, 2009; Nakashima et al., 2009). The present study shows that the heterozygous triple mutant snrk2.2/3/6+ has an early flowering phenotype (Supplementary Fig. S3). The stimulation of FLC promoter activity by ABI5 and other ABFs was abolished in snrk2.2/2.3/2.6 plants (Fig. 5 and Supplementary Fig. S3), implying the correlation between SnRKs and ABI5-regulated FLC promoter activity (Fig. 5). On the contrary, coexpressing SnRK2.6 and ABI5 enhanced FLC promoter activity significantly (Fig. 5). Thus, the data not only pinpoint the role of SnRK2s in activating ABI5 but also indicate the importance of the phosphorylation state of ABI5 for transactivating FLC promoter activity. Transgenic plants carrying ABI5 mutations at phosphorylation sites recovered the early flowering phenotype (Fig. 3 and Supplementary Fig. S2), which clearly demonstrate the importance of ABI5 phosphorylation status in the control of flowering time. By analysing the expression of FLC in planta, this study found that FLC expression was upregulated by ABA in ABI5-5 plants and, in contrast, downregulated in abi5-7 and ABI5 S42AS145AT201A plants (Figs. 1 and 4). The role of individual consensus phosphoamino acids (such as S41, S42, S145, T201) in ABI5 activity has been evaluated in planta and it was suggested that none are essential for ABA-induced ABI5 function, based on the assessment of seed germination (Lopez-Molina et al., 2002). The present study found that each phosphoamino acid, S41, S42, S145, and T201, was actually critical for ABI5 to promote FLC expression (Figs. 3 and 5 and Supplementary Fig. S2). The possible explanation is that each individual consensus phosphoamino acid shares a redundant function in maintaining ABI5 activity during seed germination, because the prominent effect was obtained with the triple mutation ABI5 S42AS145AT201A (Figs. 3 and 5). It is speculated that the phosphorylation of ABI5 (including other ABFs) might be a limiting but essential step for facilitating ABI5 to interact with FLC promoter elements, which, in turn, triggers FLC transcription. Future experimental evidence is needed to clarify the differential roles of these consensus phosphoamino acids of bZIP transcription factors on the regulation of flowering time.
FLC transcription is directly activated by bZIP transcription factors
Previous studies have provided experimental evidence to support the notion that bZIP transcription factors, including ABI5 and ABFs, are involved in various developmental processes in plants (Lopez-Molina and Chua, 2000; Kang et al., 2002). The roles of bZIP transcription factors in the transition from vegetative growth to reproductive development are elusive. As for the control of flowering time, specific histone modifications at the FLC locus associated with the chromatin structure of the gene segment are indispensable for maintaining a regular expression level of FLC under various conditions (Dennis and Peacock, 2007). Many genes involved in the modification of FLC chromatin (i.e. methylation and acetylation) have been identified and they usually form a protein complex to collaboratively regulate FLC expression (Kim and Michaels, 2006; Kim et al., 2006, Deal et al., 2007). A feedback regulatory loop involving SOC1 (one of the targets of FLC) and many other transcription regulators (including bZIP transcription factors) has been suggested (Tao et al., 2012). This study found an interesting connection between floral transition and ABA signal transduction, which ABI5 and other ABFs can promote FLC transactivation in vivo (Figs. 2 and 5 and Supplementary Fig. S3). The correlation between ABI5 activity and FLC expression was verified by analysing the expression patterns of pFLC::GUS and pFLC(m)::GUS in ABI5-5 transgenic plants, providing genetic evidence of the requirement of ABRE/G-box elements for ABI5 to activate FLC expression (Fig. 4). The data tally the expression patterns of ABI5 and FLC in various tissues of Arabidopsis (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), suggesting that ABI5 and FLC may be commonly activated during seed germination and seedling growth.
Each bZIP transcription factor (ABF) possesses its preferential expression property in various tissues during Arabidopsis development and while responding to diverse environmental signals (Choi et al., 2000; Yamaguchi-Shinozaki and Shinozaki, 2006). The redundant or synergistic function of ABFs in regulating FLC expression may be remarkable during the plant life cycle. It is most likely that ABI5 (and/or other ABFs) is involved in the floral transition of Arabidopsis plants while responding to environmental cues. The fact that ABA concentrations more than 50 µM ABA did not increase the effect suggests that regulation of FLC expression by ABI5 (and/or other ABFs) is limited under stress conditions. In these analyses, ABI5 could directly bind to the putative ABRE/G-box elements in promoter segments of FLC in vivo (Fig. 6). An enriched binding efficiency of ABI5 to the proximal DNA6 element embedded in the FLC promoter was identified (Fig. 6), implicating the diversity of FLC promoter properties. The ABRE-element and G-box are common coupling partners and are functional as cis-elements in ABA signalling.
Conclusion
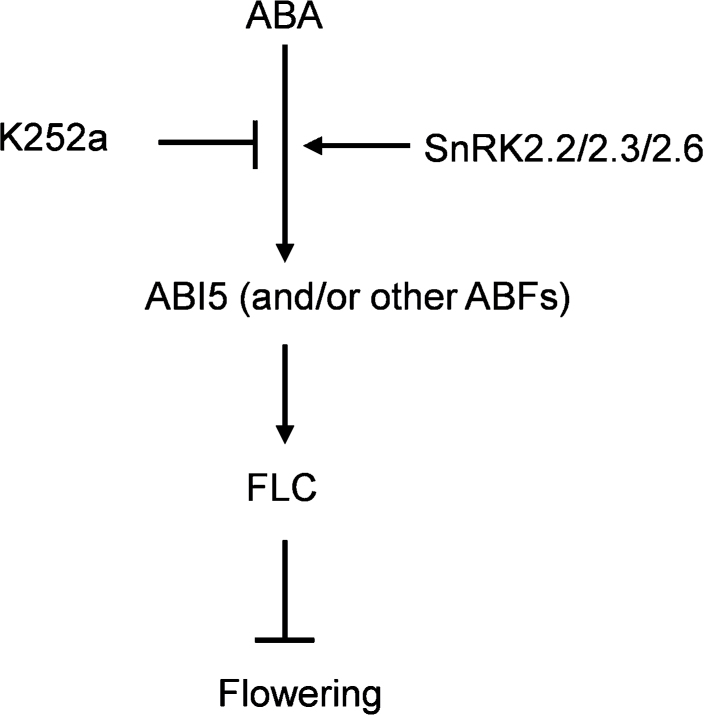
Overall, the existence of a previously unidentified mechanism underlying the transactivation of FLC expression by ABI5 (and/or other ABFs) is suggested in this study (Fig. 7). While plants are exposed to environmental stresses, the increased ABA content may stimulate the regulatory flexibility on floral transition through activating bZIP transcription factors whose activities can be modulated by SnRK2s. As such, activated ABI5 (and/or other ABFs) may trigger the expression of FLC in time; then the immediate adjustment in plants for responding to ambient changes may be initiated.
Fig. 7.
A working model to suggest the inhibitory effect of abscisic acid (ABA) on flowering time in Arabidopsis. Activation of ABI5 (or other ABFs) by ABA stimulation requires SnRK2.2/2.3/2.6. The direct binding of ABI5 and/or ABFs to the promoter elements of FLC is a critical step for promoting FLC transcription. Thereafter, flowering time may be postponed.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Comparisons of ABFs, ABI5, and FLC expression patterns in Arabidopsis.
Supplementary Fig. S2. Comparisons of flowering phenotypes, total rosette leaf numbers, and seed germination phenotypes in wild-type and transgenic lines.
Supplementary Fig. S3. SnRK2s are involved in regulation of FLC expression.
Supplementary Fig. S4. ABRE-like and G-box motifs in the FLC promoter are essential for FLC expression in responding to ABA.
Supplementary Table S1. Primer sequences for identification of snrk2.2/2.3/2.6 homozygous plants.
Supplementary Table S2. Primer sequences for plasmid construction.
Supplementary Table S3. Plasmids used in this study.
Supplementary Table S4. Primer sequences for analysis of enrichment of DNA fragments in chromatin immunoprecipitation assay.
Supplementary Table S5. Primer sequences for qRT-PCR experiments.
Acknowledgements
The authors are grateful to laboratory members for stimulating discussions and helpful comments on the manuscript. They thank Dr. Nam-Hai Chua (Rockefeller University) for kindly providing abi5-4 (Ws-0) mutant seeds, Dr. Ruth R. Finkelstein (University of California at Santa Cruz) for kindly providing abi5-7 (Col-0) mutant seeds, and Dr. Jian-Kang Zhu (Purdue University) for kindly providing snrk2.2/3/6+ mutant seeds. This work was supported by grants to Y. Wu from the Ministry of Science and Technology of China (2013CB126900) and the National Natural Science Foundation of China (31270333, 90817013), and by the Chinese 111 Project (B06018).
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 [DOI] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd PN. 2006. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 [DOI] [PubMed] [Google Scholar]
- Anderberg RJ, Walker-Simmons MK. 1992. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proceedings of the National Academy of Sciences, USA 89, 10183–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. 2004. Multiple pathways in the decisions to flower: enabling, promoting, and resetting. The Plant Cell 16, S18–S31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Laurière C. 2004. Identification of nine sucrose non-fermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana . Journal of Biology Chemistry 279, 41758–41766 [DOI] [PubMed] [Google Scholar]
- Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K. 2009. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 106, 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-i, Hong J-h, Ha J-o, Kang J-y, Kim SY. 2000. ABFs, a family of ABA-responsive element binding factors. Journal of Biology Chemistry 275, 1723–1730 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Davis SJ. 2009. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana . Plant, Cell and Environment 32, 1201–1210 [DOI] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB. 2007. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A. The Plant Cell 19, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ. 2007. Epigenetic regulation of flowering. Current Opinion in Plant Biology 10, 520–527 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Sarnowska E, Nagy F, Davis SJ. 2010. Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana . PLoS ONE 5, e14012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit O, Hanukoglu A, Hanukoglu I. 2009. Simple and efficient site-directed mutagenesis using two single-printer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnology 9, 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J-K. 2007. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis . The Plant Cell 19, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu J-K. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA 106, 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM. 2004. Hormonal regulation of plant growth and development. PLoS Biology 2, e311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. The Plant Journal 46, 183–192 [DOI] [PubMed] [Google Scholar]
- Hisamatsu T, Koshioka M, Kubota S, Fujime Y, King RW, Mander LN. 2000. The role of gibberellin biosynthesis in the control of growth and flowering in Matthiola incana . Physiologia Plantarum 109, 97–105 [Google Scholar]
- Hrabak EM, Chan CWM, Gribskov M, et al. 2003. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiology 132, 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T. 2004. Molecular and genetic mechanisms of floral control. The Plant Cell 16, S1–S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis . Trends in Plant Science 7, 106–111 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 20, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Kumar S, Eu YJ, Jami SK, Stasolla C, Hill RD. 2012. The Arabidopsis mutant, fy-1, has an ABA-insensitive germination phenotype. Journal of Experimental Botany 63, 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-y, Choi H-i, Im M-y, Kim SY. 2002. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. The Plant Cell 14, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I. 2006. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C . The Plant Cell 18, 2985–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD. 2006. SUPPRESSOR OF FRI4 encodes a nuclear-localized protein that required for delayed flowering in winter-annual Arabidopsis . Development 133, 4699–4707 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua N-H. 1998. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. The Plant Journal 16, 393–401 [DOI] [PubMed] [Google Scholar]
- Lang A. 1956. Induction of flower formation in biennial Hyoscyamus by treatment with gibberellin. Naturwissenschaften 43, 284–285 [Google Scholar]
- Lang A. 1957. The effect of gibberellin upon flower formation. Proceedings of the National Academy of Sciences, USA 43, 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. 2007. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis . Genes and Development 21, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Thong Z, Yu H. 2009. Coming into bloom: the specification of floral meristems. Development 136, 3379–3391 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua N-H. 2000. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana . Plant Cell Physiology 41, 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua N-H. 2002. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal 32, 317–328 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Coupland G, Dean C, Koornneef M. 1994. The transition to flowering in Arabidopsis . In: Meyerowitz EM, Somerville CR, eds, Arabidopsis. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, pp 403–433 [Google Scholar]
- Michaels SD. 2009. Flowering time regulation produces much fruit. Current Opinion in Plant Biology 12, 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 2001. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. The Plant Cell 13, 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I. 2005. Analysis of flowering pathway integrators in Arabidopsis . Plant Cell Physiology 46, 292–299 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek N-C, Kim S-G, Lee I. 2003. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis . The Plant Journal 35, 613–623 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. 2002. Control of flowering time: interacting pathway as a basis for diversity. The Plant Cell 14, S111-–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, et al. 2009. Three Arabidopsis SnRK2s protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiology 50, 1345–1363 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd edn Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis . Genes and Development 20, 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C. 2002. Arabidopsis, the rosetta stone of flowering time? Science 296, 285–289 [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. 2012. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis . The Plant Journal 70, 549–561 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. 2000. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences, USA 97, 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang W, Zhu C, Perry SE. 2002. A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, A MADS domain protein that preferentially accumulates in embryos. The Plant Journal 32, 831–843 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Zhao S, Liu Z, Feng YQ, Wu Y. 2011. Cytokinin antagonizes ABA-suppression to seed germination of Arabidopsis by down-regulating ABI5 expression. The Plant Journal 68, 249–261 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PloS ONE 2, e718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis . The Plant Cell 22, 1733–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57, 781–803 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocol 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.