Abstract
Tomato is a model and economically important crop plant with little information available about gene expression in roots. Currently, there have only been a few studies that examine hormonal responses in tomato roots and none at a genome-wide level. This study examined the transcriptome atlas of tomato root regions (root tip, lateral roots, and whole roots) and the transcriptional regulation of each root region in response to the plant hormones cytokinin and auxin using Illumina RNA sequencing. More than 165 million 1×54 base pair reads were mapped onto the Solanum lycopersicum reference genome and differential expression patterns in each root region in response to each hormone were assessed. Many novel cytokinin- and auxin-induced and -repressed genes were identified as significantly differentially expressed and the expression levels of several were confirmed by qPCR. A number of these regulated genes represent tomato orthologues of cytokinin- or auxin-regulated genes identified in other species, including CKXs, type-A RRs, Aux/IAAs, and ARFs. Additionally, the data confirm some of the hormone regulation studies for recently examined genes in tomato such as SlIAAs and SlGH3s. Moreover, genes expressed abundantly in each root region were identified which provide a spatial distribution of many classes of genes, including plant defence, secondary metabolite production, and general metabolism across the root. Overall this study presents the first global expression patterns of hormone-regulated transcripts in tomato roots, which will be functionally relevant for future studies directed towards tomato root growth and development.
Key words: Auxin, cytokinin, lateral root, RNA sequencing, root, root tip, tomato.
Introduction
Tomato (Solanum lycopersicum) is one of the main crops grown throughout the world. Many studies toward the improvement and increase in yield of this plant have focused on flower (Goméz et al., 1999; Molinero-Rosales et al., 2003) and fruit development (de Freitas et al., 2012; Pattison and Catalá, 2012). Roots in tomato, on the other hand, which constitute an important system to carry out water conduction, mineral uptake, anchorage, synthesis of bio-active substances, gravitropic responses, and sensing stress have not been studied in detail. This is particularly true for gene expression patterns in tomato roots that will be essential for any future studies focused on growing, breeding, or engineering plants with more desirable root traits.
The root system comprises different regions that are involved in specific functions for the overall plant sustenance. Two essential regions are the root tips and lateral roots. The root tip encompasses the root apical meristem, which provides all the cells and base organization for the growing root (Scheres et al., 2002). Moreover, this is a region of active cell division, elongation (Dolan et al., 1993), and is also the region where synthesis of important hormones (Feldman,1979; Muller et al., 1998) occurs which are transported to different locations in the plant. Lateral roots on the other hand extend from the primary root in horizontal manner to provide support and increased water and nutrient uptake.
Two key hormones that play pivotal roles in root growth and development are cytokinin and auxin. Both of these hormones have been implicated to have roles in several processes such as in root vascular development, initiation of lateral roots, and root gravitropism (Aloni et al., 2006; Vanneste and Friml, 2009). Quite often, cytokinin displays crosstalk with auxin during root development, such as in the establishment of root stem-cell niche (Müller and Sheen, 2008), meristem size and root growth (Dello Ioio et al., 2008; Zhang et al., 2011), and lateral root organogenesis (Marhavý et al., 2011), although sometimes in an antagonistic manner. Through several studies, the regulatory roles of hormones in plant development have been elucidated. This includes several cytokinin-induced genes, such as the CRE1/AHK4 cytokinin receptor (Che et al., 2002), CRFs that are involved in Arabidopsis cotyledon and leaf development (Rashotte et al., 2006), and a O-glucosyltransferase in rice that catalyses the inactivation of zeatin-type CKs by O-glucosylation (Hirose et al., 2007). Numerous auxin-regulated genes have similar important roles, such as SAURs that are strongly expressed in epidermal and cortical cells (Vanneste and Friml, 2009) and AUF1 that is involved in crosstalk between cytokinin and auxin during Arabidopsis root growth (Zheng et al., 2011).
With the availability of the complete tomato genome (The Tomato Genome Consortium, 2012), it has become possible to perform genome-wide transcriptome analysis to study gene expression patterns across different plant tissues under different conditions without de novo assembly. Next-generation high-throughput RNA sequencing technology (RNA-seq) using massively parallel sequencing has revolutionized transcriptome analysis and, when compared to microarrays, RNA-seq can detect all expressed genes without the generation of an array of probes, with reduced background noise and large dynamic range. This is particularly important in species such as tomato, where publicly available microarrays cover only one-third of the complete genome.
This study utilized Illumina RNA sequencing on 15-day-old hydroponically grown Micro-Tom plants to analyse the transcriptome of tomato roots with the main focus on the spatial patterning and regulation of genes in the root by the hormones cytokinin and auxin. This study is one of only a few tomato genome-wide expression profiles and generates a transcriptome atlas of tomato root regions: root tip, lateral roots, and whole roots. Additionally, this transcriptome analysis of hormone regulation in tomato root reveals novel genes regulated by each of these hormones. This comprehensive analysis of tomato root transcriptome can further be utilized as a reference to conduct future research on tomato roots.
Materials and methods
Plant materials, growth conditions, and hormone treatment
The tomato dwarf cultivar Micro-Tom plants were grown hydroponically in CYG germination pouches from Mega International under 26/22 °C, 16/8h light/dark photoperiod at 150 µE. Seedlings at 14 days after sowing were treated with exogenous 5 µM cytokinin (N6-benzyladenine, BA) or 5 µM auxin (naphthalene acetic acid, NAA) dissolved in dimethylsulphoxide (DMSO) for 24h by directly adding the hormones or control DMSO to the growth pouches, such that 15-day-old plant tissues were collected.
RNA extraction, library preparation, and Illumina GAIIX sequencing
Root tips (RT, encompassing the meristem and the elongation zone), lateral roots (LR), and whole roots (WR, including RT and LR) were collected from 15-day-old Micro-Tom plants from seedlings with or without treatment with cytokinin, auxin, and DMSO and immediately flash-frozen in liquid nitrogen and ground to obtain fine powder. Total RNA was extracted from the tissue using a Qiagen RNeasy Kit as per the manufacturer’s instructions and used for messenger RNA isolation with polyA selection and subsequent library construction with the TruSeq RNA sample preparation protocol from Illumina (San Diego, CA). Two biological replicates were sequenced and analysed for each of the nine tissue–treatment combinations. Single-end sequencing was performed on the 18 samples by the Illumina GAIIX platform, generating 165,894,496 1×54bp reads, totalling 8.99 Gbp. Raw sequence data is available for download at NCBI Sequence Read Archive under the number SRA058709.
Differential expression analysis with S. lycopersicum (ITAG2.3) reference
Reads for each of the 18 samples were aligned to the S. lycopersicum genome reference with GSNAP (Wu and Nacu, 2010). The high throughput sequencing data was managed using the Alpheus pipeline and database resource (Miller, 2008). Gene expression was quantified as the total number of reads for each sample that uniquely aligned to the reference, binned by gene coordinate. On average, each sample had 7.6 million uniquely aligning reads.
Differential expression analysis was performed with the negative binomial test of DESeq (Anders, 2010) on three partitions of the dataset: three tissues DMSO-only treatment, three tissues DMSO versus cytokinin treatment, and three tissues DMSO versus auxin treatment. To perform robust analyses, each set was normalized, filtered, and tested within DESeq separately with recommended settings. The first experiment interrogated tissue-specific signals with the DMSO-treated samples from root tip, lateral root, and whole root (n = 6). The second experiment investigated the effect of cytokinin treatment in each root tissue in reference to the corresponding DMSO control (n = 12). The final experiment looked at the effect of auxin treatment in each root tissue in reference to the corresponding DMSO control (n = 12). A multiple testing correction (the Benjamini–Hochberg false discovery rate method; Benjamini and Hochberg, 1995) was applied and genes with an adjusted P-value (P adj) ≤0.05 were considered to be significantly differentially expressed. For multiple tissue differential expression, this study shows comparisons with a log2 fold-change that were significant (P adj ≤ 0.05) in one or more of the root regions, indicating P adj in all regions.
Verification of mRNA-seq results by quantitative real-time PCR
A 500ng aliquot of the total RNA for each tissue type (see above) was used for reverse transcription using Quanta qScript cDNA supermix and the cDNA was diluted 20 times before it was used for analysis by quantitative real-time PCR (qPCR). Each reaction consisted of 2 µl 20-fold cDNA, 9 µl SYBR-Green supermix, 1 µl of 6 µM forward and reverse primers, and 7 µl sterile water. qPCR was performed in a Eppendorf Mastercycler ep realplex with the following parameters: one cycle of 2min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 45 s at 57 °C, and 35 s at 68 °C. The last step for each reaction was melting curve generation to test the amplicon specificity. All qPCR reactions were performed in two technical and two biological replicates. All samples were compared with the control gene TIP41 (Expósito-Rodríguez et al., 2008). The primer sequences for the genes which were verified through qPCR are presented in Supplementary Table S1 (available at JXB online).
Results and discussion
Tomato root transcriptome analysis
To obtain a global view of genes expressed across the tomato root, single-end Illumina GAIIX RNA sequencing was performed on samples derived from RT (encompassing the meristem and elongation zone), LR, and WR (including RT and LR) after 24h of treatment with cytokinin (5 µM BA) or auxin (5 µM NAA) versus the vehicle control DMSO. This generated a total of 165,894,496 1×54bp reads from all root tissue samples that were aligned to the S. lycopersicum reference genome. On average, samples generated 7.6 million uniquely aligning reads from which gene expression was quantified as the total number of reads for each sample that uniquely aligned to the reference, binned by gene.
Based on reads and filtering, ~17,300 genes were robust enough to be used for differential expression analysis on three subsets of the data – tissue-level expression patterns by utilizing untreated (DMSO) RT, LR, and WR samples – and the hormone expression in these tissues of cytokinin as well as auxin treatment in reference to DMSO. Since a large number of genes were found to be significantly regulated (P adj ≤ 0.05), study focused the differential expression analysis on genes with >2.0 log2 fold-change, which were designated as hormone or tissue regulated (Table 1). The discussion of these results is concentrated on genes regulated commonly across the different tissues examined, as they represent some of the most highly regulated hormone-responsive genes with important known gene function in each root region. Furthermore, qPCR was performed in order to confirm the results of RNA sequencing on 12 differential expression-regulated genes affected by cytokinin and auxin treatments. This comparison yielded very similar expression directionality and level of regulation for all genes examined across all root tissues, indicating that the log2 fold-change values obtained from RNA sequencing were accurate (Table 2).
Table 1.
Hormone-regulated genes commonly induced in all root tissues. Shown are the log2 fold-change values (log2FC) for genes commonly induced (>2.0) by cytokinin and auxin among root tip (RT), lateral root (LR), and whole root (WR) with adjusted P-values ≤0.05 in one or more tissue.
| GeneID | Description | RT | LR | WR | |||
|---|---|---|---|---|---|---|---|
| log2FC | P adj | log2FC | P adj | log2FC | P adj | ||
| Induced by cytokinin | |||||||
| Solyc01g088160 | SlCKX2 | 2.58 | 0.046321 | 3.14 | 0.00771 | 3.32 | 0.003426 |
| Solyc04g080820 | SlCKX4 | 2.90 | 8.56E-06 | 3.04 | 2.12E-06 | 2.18 | 0.000725 |
| Solyc12g008900 | SlCKX6 | 5.39 | 0.027127 | 7.31 | 0.00434 | 7.32 | 0.002308 |
| Solyc02g071220 | SlRRA2 | 2.31 | 3.48E-05 | 3.18 | 7.98E-09 | 3.03 | 2.01E-08 |
| Solyc06g048930 | SlRRA6 | 2.53 | 0.117332 | 2.90 | 0.05133 | 3.85 | 0.007115 |
| Solyc06g048600 | SlRRA7 | 2.55 | 0.000164 | 2.88 | 2.00E-05 | 3.17 | 2.25E-06 |
| Solyc09g092470 | TPS14 | 6.22 | 2.07E-26 | 5.21 | 1.26E-22 | 3.85 | 2.75E-14 |
| Solyc01g105960 | (E)-beta-ocimene synthase | 4.42 | 9.02E-14 | 3.61 | 2.66E-11 | 3.08 | 3.86E-09 |
| Solyc02g080790 | SlDHS | 3.45 | 0.000293 | 6.05 | 4.94E-10 | 5.69 | 2.61E-09 |
| Solyc06g076760 | Laccase 1a | 3.75 | 8.39E-07 | 2.27 | 0.002648 | 2.63 | 0.000348 |
| Solyc06g073580 | 1-aminocyclopropane-1-carboxylate oxidase 1 | 2.68 | 0.016622 | 2.81 | 0.014623 | 2.47 | 0.022694 |
| Solyc12g008740 | 1-aminocyclopropane- 1-carboxylate synthase | 3.02 | 8.96E-12 | 4.26 | 1.15E-14 | 3.21 | 1.38E-08 |
| Solyc04g010330 | Auxin-regulated protein | 2.18 | 0.351047 | 2.73 | 0.163185 | 3.78 | 0.02978 |
| Solyc02g080120 | Gibberellin 2-beta- dioxygenase 7 | 3.06 | 6.76E-05 | 3.16 | 5.34E-06 | 2.37 | 0.000459 |
| Solyc01g059950 | GAI-like protein 1 | 4.87 | 2.53E-27 | 3.50 | 2.87E-14 | 2.37 | 1.25E-08 |
| Solyc06g082770 | LOB domain family protein | 2.60 | 1.16E-06 | 3.34 | 4.00E-18 | 2.45 | 7.70E-10 |
| Solyc12g100150 | LOB domain protein 4 | 4.34 | 1.24E-05 | 4.84 | 2.39E-07 | 5.99 | 5.71E-09 |
| Induced by auxin | |||||||
| Solyc01g107400 | Indole-3-acetic acid (IAA)-amido synthetase | 3.15 | 1.24E-06 | 2.20 | 0.001216 | 2.11 | 0.003209 |
| Solyc02g064830 | SlGH3–3 | 7.31 | 8.65E-107 | 6.61 | 4.49E-98 | 8.22 | 1.03E-108 |
| Solyc02g092820 | SlGH3–4 | 6.56 | 1.56E-55 | 6.76 | 2.66E-57 | 7.16 | 3.70E-59 |
| Solyc07g063850 | SlGH3–9 | 4.95 | 7.98E-34 | 4.93 | 7.20E-33 | 4.51 | 2.70E-28 |
| Solyc06g084070 | SlIAA2 | 2.57 | 3.34E-11 | 3.64 | 7.49E-20 | 3.31 | 9.31E-16 |
| Solyc12g096980 | SlIAA5 | 3.99 | 3.01E-45 | 3.53 | 1.50E-38 | 3.45 | 1.85E-36 |
| Solyc06g008590 | SlIAA10 | 3.78 | 7.08E-05 | 3.64 | 0.000265 | 2.99 | 0.004369 |
| Solyc03g120380 | SlIAA11 | 4.91 | 1.47E-32 | 5.69 | 8.86E-37 | 5.60 | 3.68E-29 |
| Solyc07g008020 | SlIAA19 | 7.58 | 1.41E-50 | 6.92 | 7.03E-46 | 7.17 | 2.95E-41 |
| Solyc08g021820 | SlIAA21 | 5.47 | 9.70E-09 | 5.06 | 2.01E-07 | 4.26 | 1.09E-05 |
| Solyc09g083280 | SlIAA23 | 2.22 | 2.62E-25 | 2.50 | 7.22E-31 | 2.45 | 6.08E-29 |
| Solyc01g010970 | SlAGO7 | 2.35 | 1.05E-16 | 2.24 | 5.98E-15 | 2.02 | 1.23E-12 |
| Solyc12g096110 | Protein BREVIS RADIX | 4.19 | 9.56E-33 | 3.67 | 1.42E-29 | 3.52 | 1.92E-20 |
| Solyc07g048000 | Phototropic-responsive NPH3 | 3.29 | 4.54E-15 | 2.97 | 2.15E-11 | 2.27 | 6.34E-07 |
| Solyc03g044090 | Phototropic-responsive NPH3 family protein | 2.44 | 1.70E-10 | 2.74 | 3.04E-12 | 2.43 | 1.67E-09 |
| Solyc04g072300 | Ethylene-responsive transcription factor 4 | 3.49 | 5.57E-05 | 4.52 | 9.39E-07 | 3.72 | 0.000107 |
| Solyc05g050830 | Ethylene-responsive transcription factor 4 | 5.99 | 3.07E-09 | 6.17 | 4.36E-09 | 4.82 | 3.81E-06 |
| Solyc03g007460 | SlCRF4 | 2.63 | 5.25E-21 | 2.70 | 1.31E-22 | 2.36 | 1.04E-15 |
| Solyc01g079260 | WRKY transcription factor 4 | 2.22 | 3.11E-23 | 2.07 | 5.77E-20 | 2.21 | 2.02E-21 |
| Solyc11g017100 | GRAS family transcription factor | 3.27 | 0.000112 | 3.54 | 5.35E-05 | 2.12 | 0.026739 |
Table 2.
Experimental validation of a subset of cytokinin/auxin-regulated transcripts. Shown are the log2 fold-change values from qPCR versus RNA sequencing in root tip (RT), lateral root (LR), and whole root (WR) for genes induced by cytokinin (A) or auxin (B) and repressed by cytokinin and auxin (C).
| Gene ID | Description | Tissue | log2 fold-change | |
|---|---|---|---|---|
| mRNA sequencing | qPCR | |||
| (A) Genes induced by cytokinin | ||||
| Solyc04g078900 | Cytochrome P450 | RT | 4.67 | 4.71 |
| LR | 5.66 | 3.56 | ||
| WR | 7.00 | 2.65 | ||
| Solyc12g008900 | SlCKX6 | RT | 5.39 | 4.37 |
| LR | 7.30 | 3.67 | ||
| WR | 7.31 | 3.39 | ||
| Solyc01g111900 | O-Methyltransferase | RT | 6.69 | 5.54 |
| LR | 4.78 | 4.88 | ||
| WR | 4.12 | 3.83 | ||
| Solyc01g090810 | Expansin | RT | 3.88 | 4.22 |
| LR | 3.76 | 3.12 | ||
| WR | 5.97 | 5.51 | ||
| Solyc12g100150 | LOB domain | RT | 4.34 | 4.61 |
| LR | 4.84 | 4.28 | ||
| WR | 5.98 | 3.99 | ||
| (B) Genes induced by auxin | ||||
| Solyc01g090810 | Expansin | RT | 7.27 | 6.54 |
| LR | 7.32 | 6.39 | ||
| WR | 5.16 | 4.74 | ||
| Solyc03g120380 | SlIAA11 | RT | 4.91 | 4.76 |
| LR | 5.68 | 5.08 | ||
| WR | 5.60 | 4.65 | ||
| Solyc07g008020 | SlIAA19 | RT | 7.58 | 7.41 |
| LR | 6.92 | 7.62 | ||
| WR | 7.16 | 6.87 | ||
| Solyc03g120060 | Cytochrome P450 | RT | 7.85 | 7.61 |
| LR | 6.80 | 7.65 | ||
| WR | 5.70 | 5.68 | ||
| Solyc02g064830 | SlGH3-3 | RT | 7.30 | 8.11 |
| LR | 6.60 | 8.06 | ||
| WR | 8.22 | 6.88 | ||
| (C) Genes repressed by cytokinin and auxin | Cytokinin | Auxin | ||||
|---|---|---|---|---|---|---|
| mRNA sequencing | qPCR | mRNA sequencing | qPCR | |||
| Solyc12g009650 | Proline-rich protein | RT | –2.98 | –3.69 | –7.32 | –4.98 |
| LR | –2.46 | –2.17 | –4.81 | –2.60 | ||
| WR | –3.26 | –2.75 | –3.51 | –2.47 | ||
| Solyc01g094910 | Ferric reductase oxidase | RT | –4.17 | –2.15 | –5.72 | –3.24 |
| LR | –5.50 | –2.71 | –6.30 | –2.83 | ||
| WR | –6.54 | –3.12 | –5.05 | –3.50 |
Principal component analysis and variance decomposition (both as implemented in SAS JMP Genomics 5.1) of the overall, full transcriptome dataset (n = 18) showed distinct differences between the auxin and cytokinin treatments, responsible for 68.6% of the variance (Supplementary Fig. S1). With regards to tissue differences, 19.5% of the variance, there was greater similarity between LR and WR samples, while the RT has a more distinct profile.
Cytokinin-induced genes
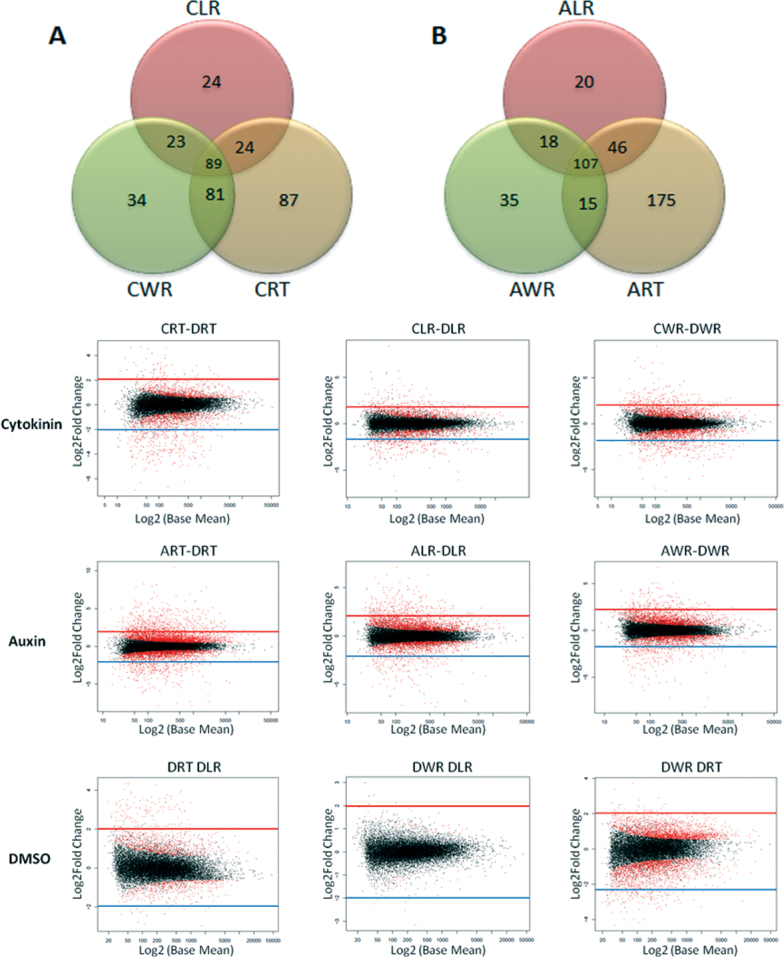
Genes were determined to be cytokinin regulated if they had >2.0 log2 fold-change in at least one root region sampled. This study identified 89 genes that were cytokinin regulated in all root tissues, while 87, 24, and 34 were specifically regulated in RT, LR, and WR tissues, respectively. Additionally, there were 24 genes commonly regulated in RT and LR tissues, 23 between LR and WR tissues, and 81 between RT and WR tissues (Fig. 1 and Supplementary Table S2).
Fig. 1.
Differential expression of genes in tomato root tip (RT), lateral root (LR), and whole root (WR). Treatment of root regions are noted as prefixes (C, cytokinin, BA; A, auxin, NAA; D, DMSO). (A, B) Venn diagrams illustrating differentially expressed and significantly induced genes in tomato root either between or unique to specific tissues; gene details are presented in Table 1 and Supplementary Tables 1 and 2. (C) MvA differential expression analysis plots indicating genes’ log2 fold-change versus the log2 base mean for root tissues treated with cytokinin, auxin, and DMSO control. Genes with >2.0 log2 fold-change values were identified as differentially regulated as noted in plots by lines (red, induced; blue, repressed). Significantly differentially expressed genes are red dots (P adj ≤ 0.05)
Differentially expressed cytokinin-induced genes found in this study appear to be mainly involved in metabolism, development, defence, transport, and tissue-specific related processes. Some of the commonly found highly cytokinin-regulated genes are the cytokinin oxidase/dehydrogenases (CKX) (Brenner et al., 2012). Three CKX genes (SlCKX2, SlCKX4, SlCKX6) were found to be induced in all root tissues (Table 1), while SlCKX1 was induced specifically between RT and LR and SlCKX5 between RT and WR (Supplementary Table S1). CKX genes across plant genera are involved in the breakdown and catabolism of cytokinin (Frébort et al., 2011), so it is not surprising to see several CKXs induced with the application of exogenous cytokinin. CKXs have also been linked to a number of cytokinin-related growth and developmental processes and recently it was shown that overexpression of CKX genes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco (Werner et al., 2010). Since this study has shown that CKXs can be highly induced in roots, it would be interesting to see if similar growth and developmental effects could be seen from CKX overexpression in tomato.
Seven type-A response regulator (RR) genes were found induced in this study, which are designated SlRRAs. SlRRA2, SlRRA6, and SlRRA7 were induced in all root tissues (Table 1), SlRRA4 specifically in LR, and SlRRA1, SlRRA3, and SlRRA5 were induced between LR and WR (Supplementary Table S2). Type-A RRs are commonly found to be highly induced by cytokinin treatment as they are negative regulators of cytokinin signalling (Brenner et al., 2012). It can be proposed that induction of these genes probably occurred to dampen the increased exogenous cytokinin levels added to the roots. Since cytokinin is a negative regulator of root growth, it would be interesting to know how the levels of cytokinin are maintained to appropriate amounts by the genes involved in cytokinin catabolism and the cytokinin signalling pathway components.
Defence genes, including those that produce volatile compounds in response to pathogens or predators, were among those induced by cytokinin, including the sesquiterpene synthase gene TPS14 and (E)-beta-ocimene synthase (Solyc01g105960) (Table 1; Paré and Tumlinson, 1999). Sesquiterpenes such as alpha-humulene are widely distributed in plants and have been shown to function as insect/pathogen repellent in direct defence (Suga et al., 1993). In support of the present results, TPS14 has been shown to be highly expressed in tomato roots (Bleeker et al., 2011). (E)-beta-Ocimene synthase produces a monoterpene released upon herbivory, mechanical wounding, or jasmonic acid treatment (Faldt et al., 2003). Cytokinin induction of these defence genes supports previous studies of cytokinin eliciting a defence response and the defence response being linked to the cytokinin signalling pathway (Choi et al., 2010; Naseem et al., 2012), although the underlying molecular mechanism behind this connection in tomato roots is obscure and needs further investigation.
Genes associated with abiotic environmental stresses were differentially induced, such as deoxyhypusine synthase SlDHS and six laccase genes: Solyc06g076760 among RT, LR, and WR; Solyc05g052360, Solyc05g052370, and Solyc06g082240 in RT; Solyc05g052400 in LR; and Solyc03g083900 in WR (Table 1 and Supplementary Table S2). DHS is an enzyme that catalyses enzymatic reactions leading to the activation of a eukaryotic translation initiation factor-5A (eIF-5A) (Park et al., 1993, 1997). One study has shown that transcript levels of SlDHS increased due to osmotic stress and chilling injury in tomato leaves (Wang et al., 2001) and the induction of SlDHS by cytokinin as reflected in the present data suggests that the hormone possibly has a role in abiotic stress response mediated by DHS.
Laccases are multi-copper-containing glycoproteins that have been proposed to be involved in lignin synthesis in plants (O’Malley et al., 1993). In maize roots, ZmLAC1, was shown to be induced by NaCl (Liang et al., 2006). A similar increased transcript level was observed for a tomato laccase gene by salt that were even more abundantly produced when treated with both salt and ABA, likely suggesting an involvement in stress response (Wei et al., 2000). Interestingly, in maize, it has been proposed that apoplastic laccases may have a role in cytokinin breakdown (Galuszka et al., 2005). How this group of genes mediates cytokinin response in responding to stress conditions remains unclear.
Several genes involved in different hormone biosynthesis, degradation, and signalling pathways were also induced. This includes the ethylene biosynthesis enzymes 1-aminocyclopropane-1-carboxylate (ACC) synthase (Solyc12g008740) and ACC oxidase 1 (Solyc06g073580) (Lin et al., 2009), an auxin-regulated protein (Solyc04g010330), and two gibberellic acid (GA)-related genes in involved in GA degradation, gibberellin 2-beta-dioxygenase 7 (Solyc02g080120) (Thomas et al., 1999), and GA signalling, a GAI-like protein 1 (Solyc01g059950) (Table 1; Thomas and Sun, 2004). Induction of these other hormone genes by cytokinin indicates the existence of a complex hormonal crosstalk pathway in tomato roots. Additionally two genes of the LATERAL ORGAN BOUNDARIES family (Solyc12g100150 and Solyc06g082770) (Table 1) were found to be induced by cytokinin. This gene family has been linked to a range of developmental processes; including lateral root formation, as well as regulation by auxin and even cytokinin in one case, further supporting hormone connections to root development (Majer and Hochholdinger, 2011).
Cytokinin-repressed genes
More than 50 genes were repressed by cytokinin treatment and can be broadly categorized into processes related to metabolism, development, and transport (Supplementary Table S3). There is a wide range of metabolic processes linked to Cytochrome P450 genes of which four were found to be repressed in the root (Nelson and Werck-Reichhart, 2011). There were several different types of transporters or channel proteins that were repressed including, two aquaporins and proteins associated with potassium, manganese, and malate (Supplementary Table S3). While these transporters all facilitate the movement of water or important solutes into the root, it is unclear why they would be repressed in the presence of additional cytokinin. One additionally interesting class of genes is the peroxidases, several of which were found as repressed while others were found to be induced by cytokinin. Peroxidases are known to have a variety of roles in developmental- and defence-related processes and the altered regulation of different peroxidases indicates a complex role for cytokinin in their regulation (Passardi et al., 2005).
Auxin-induced genes
From a total of 17,299 differentially expressed genes, there were 107 genes induced by auxin among all root tissues with >2.0 log2 fold-change, whereas 175, 20, and 35 genes were specifically regulated in RT, LR, and WR tissues respectively (Fig. 1). Also, there were 46 commonly regulated genes in RT and LR tissues, 18 between LR and WR tissues, and 15 between RT and WR tissues (Supplementary Table S4). Overall, the number of auxin-induced genes outnumbered the cytokinin-induced genes in the root tip. Interestingly, several genes induced by auxin were also induced by cytokinin, including cytokinin oxidase/dehydrogenase, ACC synthase, gibberellin 2-oxidase (same IDs as mentioned above). This supports a range of findings that indicates there is a complex crosstalk among hormones during root growth and development (Depuydt and Hardtke, 2011).
Among the differentially expressed auxin-induced genes were four GH3 family genes involved in indole-3-acetic acid (IAA)-amido synthetase (Solyc01g107400, SlGH3-3, SlGH3-4, and SlGH3-9) with log2 fold-change ranging from 2.10–8.22 (Table 1). Of these, SlGH3-3, SlGH3-4, and SlGH3-9 were recently found as being group III members of the GH3 family and were three of the five strongest auxin-induced GH3 family members in etiolated tomato seedlings treated with hormone for 1 or 3h (Kumar et al., 2012), supporting the present finding of the strong auxin induction of these genes. As these genes are involved in controlling auxin homeostasis levels in the plant, conjugating excess active IAA to an inactive form, their induction from the addition of exogenous auxin is not surprising, although their endogenous role in the in root is not well known (Kumar et al., 2012).
Auxin induction was also seen in a number of genes belonging to two major auxin-responsive transcriptional regulator families: auxin/indole-3-acetic acid (Aux/IAA) and auxin response factor (ARF). These gene family members are essential players in auxin signalling, with ARFs acting to activate auxin-regulated targets and Aux/IAA functioning as a transcriptional repressor by binding to ARFs and preventing their transcription factor activity, of which numerous examples have been identified in tomato (Kumar et al., 2011; Wu et al., 2012). In the present study, the Aux/IAA genes SlIAA2, SlIAA5, SlIAA10, SlIAA11, SlIAA19, SlIAA21, and SlIAA23 were induced among all root tissues (Table 1), while three ARFs were specifically induced in RT: SlARF2, SlARF3, and SlARF9 (Supplementary Table S4). The induction of these SlIAA genes would also be expected in response to the addition of exogenous auxin, in a manner parallel to the induction of SlRRAs by cytokinin, as both are acting to block excess hormone pathway signalling. Interestingly a recent report revealed that SlIAA genes can also be transcriptionally regulated by ethylene in tomato seedlings (Audran-Delalande et al., 2012) suggesting they may function in other signalling pathways and contribute towards complex hormonal crosstalk. Several genetic approaches have been utilized that implicate the importance of Aux/IAA and ARFs in lateral root formation (De Smet, 2012). While the present study has found that these genes are induced by auxin in the root, they are not found uniquely induced in LR tissues; potentially because LR tissues were not sampled during LR initiation when they are often required.
Auxin also induced SlAGO7, a member of the recently described tomato argonaute (AGO) gene family (Table 1; Bai et al., 2012). Argonaute proteins are components of the RNA-induced silencing complex, which is involved in small RNA-mediated silencing to regulate gene expression. In tomato it was shown that several members of this group were differentially regulated in response to abiotic and viral stresses. Although, SlAGO7 in particular was not observed to be differentially regulated by abiotic or viral stress conditions, its induction by auxin provides a new basis for further research. Interestingly miRNAs that are utilized by the RNA-induced silencing complex have been shown to be involved in root cap and LR formation by targeting specific ARFs (Wang et al., 2005), and can be regulated by auxin in LR development (Yoon et al., 2010) suggesting a potential role for SlAGO7 in this process.
Another gene, BREVIS RADIX (BRX, Solyc12g096110), which has a role in root development, was induced by auxin (Table 1). BRX was first implicated as a regulator of cell proliferation and elongation in the growth zone of root tip (Mouchel et al., 2004), whose protein function is unknown, as far as is known. It has been shown in Arabidopsis that BRX has a role in cytokinin-mediated inhibition of lateral root initiation which occurred due to loss of auxin response in presumptive founder cells in brx-2 with indirect involvement of brassinosteroid in the process (Li et al., 2009). How auxin regulation of BRX is involved in root development needs to be further explored.
Interestingly, two NPH3 (non-phototropic hypocotyl 3, Solyc03g044090, and Solyc07g048000) genes were also induced by auxin (Table 1). The function of most NPH3 family genes is still being investigated; however, recently this group has been linked to the proper localization of PIN protein auxin transporter (Furutani et al., 2011). The addition of exogenous auxin might result in a change in the endogenous auxin transport throughout the root, although this finding does further connect NPHs to auxin-regulated processes.
A range of different transcription factor family genes, which are involved in diverse plant metabolic and developmental roles, were induced by auxin in all root tissues. These include two ERFs (Solyc05g050830 and Solyc04g072300), SlCRF4, a WRKY (Solyc01g079260), and a GRAS family transcription factor (Solyc11g017100) (Table 1). In addition, there was induction by auxin of some transcription factor genes in a root tissue-specific manner. Root tip-specific transcription factors include six MYB family members, an AP2/ERF (Solyc02g092050), heat stress transcription factor A3 (Solyc09g082670), and CYCLOIDEA (Supplementary Table S4). There were two lateral root-specific auxin-induced transcription factors: GATA transcription factor 22 (Solyc01g100220) and SlCRF6. The transcription factor bHLH151 (Solyc06g009510) was induced in WR, but not in the other specific tissues, suggesting that it may be induced in the non-RT or non-LR parts of the primary root (Supplementary Table S4).
Auxin-repressed genes
Auxin was found to repress a larger number of genes, more than 110, compared to the ~50 in cytokinin (Supplementary Table S5). Five different 2-oxoglutarate-dependent dioxygenase (ODDs) class genes with a range of functions from catalysing the production of alkaloids in roots to regulating GA biosynthesis were repressed. A previous report has shown that auxin can inhibit the production of alkaloid production in Hyoscyamus niger (Hashimoto et al., 1986) and the present finding would support that link. There are also numerous instances connecting auxin and GA in hormone crosstalk, and since ODDs can function to both activate and inactivate GAs in the biosynthesis pathway, these genes could also support this relationship (Yamaguchi, 2008).
A number of other genes repressed by auxin belong to several of the same gene families or categories that were also repressed by cytokinin, such as CYP p450s, peroxidases, hormone regulation (ACC and GA), transporters (aquaporins and solute/nutrient transporters), genes involved in the biosynthesis of secondary metabolites, and other metabolic and developmental processes (Supplementary Table S5). Other auxin-repressed genes belong to various classes of transcription factors that are involved in protein turnover and stress.
Common genes induced/repressed by both cytokinin and auxin among all root tissues
This study identified several genes that were commonly induced (5) or repressed (28) by cytokinin and auxin in all root three tissues (Supplementary Table S6), which could be broadly categorized into metabolic and developmental processes. One of the commonly auxin- and cytokinin-induced genes is an expansin (Solyc01g090810), which was further verified as induced by both hormones using qPCR (Table 2). Interestingly this gene is highly related to the beta-expansin CIM1 from soybean that has previously been shown to be induced by both auxin and cytokinin, indicating that the two hormones can be involved in regulating expansin genes and potentially function to control cell growth in the root (Downes et al., 2001). The present study also confirmed by qPCR the common repression by auxin and cytokinin of ferric reductase oxidase (Solyc01g094910) involved in iron uptake by roots and a proline-rich protein (Solyc12g009650) involved in multiple processes including root nodule formation and response to external stimuli (Sheng et al., 1991; Table 2). The joint regulation of these genes by these two hormones indicates that they likely have vital roles in the root connected to hormone crosstalk.
Transcriptome atlas of tomato root regions: root-tip, lateral root, and whole root
In addition to examining the auxin and cytokinin hormone response in root tissues, this study used differential expression analysis on non-hormone-treated (DMSO) root samples to identify genes expressed in specific regions of the root. Identified were RT, LR, or WR abundantly expressed genes using differential expression comparisons between RT and WR, LR and WR, and RT and LR with the criteria >2.0 log2 fold-change and P adj ≤ 0.05 (Supplementary Table S7).
Among the differentially expressed genes in RT, there were several classes of genes involved in defence response, pectin modification, CYP p450, transcription factors (MYB, GATA, and BHLH), root cap proteins, an expansin, nodulin, cell division, and genes involved in other metabolic processes. The differential expression of many of these genes in the RT is not surprising as they are involved in active processes that occur in the RT such as expansion and cell division, as well as the encountering of a variety of biotic/abiotic stresses.
Differentially expressed genes in LR revealed a number of genes that normally play roles in metabolism processes including decarboxylase family protein, UDP-glucosyltransferases, alcohol and aldehyde dehydrogenases, long-chain fatty-acid-CoA ligase, fatty acid elongase 3-ketoacyl-CoA synthase, and ER glycerol-phosphate acyltransferase. In addition to these metabolic genes, a number of auxin-related genes were also differentially expressed in LR, such as Aux/IAAs and SAURs that are known to be linked to LR initiation and development (De Smet, 2012).
Since WR in this study comprised RT and LR, there were common genes expressed between these regions that are involved in metabolism and development. Compared to RT, WR and LR displayed similar profile with more than 120 common genes expressed indicating similarity in functionality of these regions. In addition, there was overrepresentation of genes expressed in WRs related to a range of function, such as ACC oxidases, transport of solute/minerals, CYP p450s, protein turnover, receptor-like kinases, and different transcription factors related to ethylene, auxin, and cytokinin. Other interesting genes expressed in WRs were three class I heat shock proteins, three water-stress inducible proteins, and two phloem proteins. As these genes cover a wide range of functional classes, it is possible that they may work as individuals or have combinatorial effects for the overall root development in tomato.
In conclusion, in order to identify the genes expressed and/or regulated by the hormones cytokinin and auxin in the tomato root, transcriptome analysis was performed using RNA sequencing. These results present the first genome-wide root tissue analysis of gene response to these hormones. This study reveals numerous significantly novel differentially expressed genes as well as tomato orthologues of auxin- and cytokinin-regulated transcripts. In addition, this RNA sequencing analysis has confirmed hormone regulation of some previously identified tomato auxin- and cytokinin-regulated genes from other studies. Several interactions were identified between different plant hormones and have been discussed in the context of hormonal crosstalk in the root. This study has also generated an atlas of root tissue-specific transcripts and their auxin and cytokinin regulation in whole roots, root tips, and lateral roots. This expression atlas indicates several classes of genes that may have specific roles in different root tissues. Taken together, this study provides a solid foundation of gene expression in tomato roots, both in different tissues and in response to different hormones, which should allow future investigations to more easily study and work to improve tomato roots, plant efficiency, and fruit yield.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Primer sequences of genes used for validation of RNA-seq results by quantitative real-time PCR.
Supplementary Table S2. Differentially induced genes >2.0 log2 fold-change between and among RT, LR, and WR by cytokinin.
Supplementary Table S3. Differentially repressed genes >2.0 log2 fold-change between and among RT, LR, and WR by cytokinin.
Supplementary Table S4. Differentially induced genes >2.0 log2 fold-change between and among RT, LR, and WR by auxin.
Supplementary Table S5. Differentially repressed genes >2.0 log2 fold-change between and among RT, LR, and WR by auxin.
Supplementary Table S6. Differentially induced/repressed genes by both cytokinin and auxin.
Supplementary Table S7. Differentially expressed genes in RT, LR, and WR identified from control (DMSO) samples.
Supplementary Fig. S1. Principal component analysis.
Acknowledgements
The authors gratefully acknowledge all members of the Rashotte lab for critical reading of the manuscript. This work was funded by the USDA-NRI (grant 2008-35304-04457), the AAES-HATCH (grants 370220-310007-2055, and 103600-310010-2055), the NIH through the National Center for Research Resources (grant 5P20RR016480-12), and the National Institute of General Medical Sciences (grant 8P20GM103451-12). Support for this research was also provided to S.G. by an Auburn University Cellular and Molecular Biosciences Peaks of Excellence Research Fellowship.
References
- Aloni R, Aloni E, Langhans M, Ullrich CI. 2006. Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and gravitropism. Annals of Botany 97, 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11, R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. 2012. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant and Cell Physiology 53, 659–672 [DOI] [PubMed] [Google Scholar]
- Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH, Yang YH, Xie BY. 2012. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum . Gene 501, 52–62 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57, 289–300 [Google Scholar]
- Bleeker PM, Spyropoulou EA, Diergaarde PJ, et al. 2011. RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Molecular Biology 77, 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T. 2012. Gene regulation by cytokinin in Arabidopsis . Frontiers in Plant Sciences 3, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH. 2002. Global and hormone-induced gene expression changes during shoot development in Arabidopsis . The Plant Cell 14, 2771–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. 2010. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis . Developmental Cell 19, 284–295 [DOI] [PubMed] [Google Scholar]
- de Freitas ST, Handa AK, Wu Q, Park S, Mitcham EJ. 2012. Role of pectin methylesterases in cellular calcium distribution and blossom-end rot development in tomato fruit. The Plant Journal 71, 824–835 [DOI] [PubMed] [Google Scholar]
- De Smet I. 2012. Lateral root initiation: one step at a time. New Phytologist 193, 867–873 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS. 2011. Hormone signalling crosstalk in plant growth regulation. Current Biology 21, R365–R373 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84 [DOI] [PubMed] [Google Scholar]
- Downes BP, Steinbaker CR, Crowell DN. 2001. Expression and processing of a hormonally regulated β-expansin from soybean. Plant Physiology 126, 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito–Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. 2008. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology 8, 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faldt J, Arimura G, Gershenzon J, Takabayashi J, Bohlmann J. 2003. Functional identification of AtTPS03 as (E)-β-ocimene synthase: a monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana . Planta 216, 745–751 [DOI] [PubMed] [Google Scholar]
- Feldman LJ. 1979. Cytokinin biosynthesis in roots of corn. Planta 145, 315–321 [DOI] [PubMed] [Google Scholar]
- Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P. 2011. Evolution of cytokinin biosynthesis and degradation. Journal of Experimental Botany 62, 2431–2452 [DOI] [PubMed] [Google Scholar]
- Furutani M, Sakamoto N, Yoshida S, Kajiwara T, Robert HS, Friml J, Tasaka M. 2011. Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 138, 2069–2078 [DOI] [PubMed] [Google Scholar]
- Galuszka P, Frébortová J, Luhová L, Bilyeu KD, English JT, Frébort I. 2005. Tissue localization of cytokinin dehydrogenase in maize: possible involvement of quinine species generated from plant phenolics by other enzymatic systems in the catalytic reaction. Plant and Cell Physiology 46, 716–728 [DOI] [PubMed] [Google Scholar]
- Goméz P, Jamilena M, Capel J, Zurita S, Angosto T, Lozano R. 1999. Stamenless, a tomato mutant with homeotic conversions in petal and stamens. Planta 209, 172–179 [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Yukimune Y, Yamada Y. 1986. Tropane alkaloid production in Hyoscyamus root cultures. Journal of Plant Physiology 124, 61–75 [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. 2007. Overexpression of a type-A response regulator alters rice morphologyand cytokinin metabolism. Plant and Cell Physiology 48, 523–539 [DOI] [PubMed] [Google Scholar]
- Kumar R, Tyagi AK, Sharma AK. 2011. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Molecular Genetics and Genomics 285, 245–260 [DOI] [PubMed] [Google Scholar]
- Kumar R, Agarwal P, Tyagi AK, Sharma AK. 2012. Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Molecular Genetics and Genomics 287, 221–235 [DOI] [PubMed] [Google Scholar]
- Li J, Mo X, Wang J, Chen N, Fan H, Dai C, Wu P. 2009. BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis . Planta 229, 593–603 [DOI] [PubMed] [Google Scholar]
- Liang M, Haroldsen V, Cai X, Wu Y. 2006. Expression of a putative laccase gene, ZmLAC 1, in maize primary roots under stress. Plant, Cell and Environment 29, 746–753 [DOI] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. 2009. Recent advances in ethylene research. Journal of Experimental Botany 60, 3311–3336 [DOI] [PubMed] [Google Scholar]
- Majer C, Hochholdinger F. 2011. Defining the boundaries: structure and function of LOB domain proteins. Trends in Plant Sciences 16, 47–52 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, et al. 2011. Cytokinin modulates endocytic trafficking of pin1 auxin efflux carrier to control plant organogenesis. Developmental Cell 21, 796–804 [DOI] [PubMed] [Google Scholar]
- Miller NA, Kingsmore SF, Farmer AD, et al. 2008. Management of high-throughput DNA sequencing projects: Alpheus. Journal of Computer Science and Systems Biology 1, 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero-Rosales N, Latorre A, Jamilena M, Lozano R. 2003. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218, 427–434 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. 2004. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes and Development 18, 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Hillebrand H, Weiler EW. 1998. Indole-3-acetic acid is synthesized from l-tryptophan in roots of Arabidopsis thaliana . Planta 206, 362–369 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, Dandekar T. 2012. Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. The Plant Cell 24, 1793–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Werck-Reichhart D. 2011. A P450-centric view of plant evolution. The Plant Journal 66, 194–211 [DOI] [PubMed] [Google Scholar]
- O’Malley DM, Whetten R, Bao W, Chen CL, Sederoff R. 1993. The role of laccase in lignification. The Plant Journal 4, 751–757 [Google Scholar]
- Paré PW, Tumlinson JH. 1999. Plant volatiles as a defense against insect herbivores. Plant Physiology 121, 325–331 [PMC free article] [PubMed] [Google Scholar]
- Park MH, Lee YB, Joe YA. 1997. Hypusine is essential for eukaryotic cell proliferation. Biological Signals 6, 115–123 [DOI] [PubMed] [Google Scholar]
- Park MH, Wolff EC, Folk JE. 1993. Hypusine: its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors 4, 95–104 [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. 2005. Peroxidases have more functions than a Swiss army knife. Plant Cell Reports 24, 255–265 [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Catalá C. 2012. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. The Plant Journal 70, 585–598 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. 2006. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses inconcert with a two-component pathway. Proceedings of the National Academy of Sciences, USA 103, 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan 2002. Root development. In Somerville CR, Meyerowitz EM, eds, The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists, doi/10.1199/tab.0101 Available at: www.aspb.org/publications/arabidopsis [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, D’Ovidio R, Mehdy MC. 1991. Negative and positive regulation of a novel proline-rich protein mRNA by fungal elicitor and wounding. The Plant Journal 1, 345–354 [DOI] [PubMed] [Google Scholar]
- Suga T, Ohta S, Munesada K, Ide N, Kurokawa M, Shimizu M, Ohta E. 1993. Endogenous pine wood nematicidal substance in pines, Pinusmassoniana, P. strobus and P. Palustris . Phytochemistry 33, 1395–1401 [Google Scholar]
- The Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. 1999. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proceedings of the National Academy of Sciences, USA 96, 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Sun TP. 2004. Update on gibberellin signaling. A tale of the tall and the short. Plant Physiology 135, 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136, 1005–1016 [DOI] [PubMed] [Google Scholar]
- Wang TW, Lu L, Wang D, Thompson JE. 2001. Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eukaryotic translation initiation factor 5A from tomato. Journal of Biological Chemistry 276, 17541–17549 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. 2005. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis . The Plant Cell 17, 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JZ, Tirajoh A, Effendy J, Plant AL. 2000. Characterization of salt-induced changes in gene expression in tomato (Lycopersicon esculentum) roots and the role played by abscisic acid. Plant Science 159, 135–148 [DOI] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novak O, Strnad M, Krämer U, Schmülling T. 2010. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. The Plant Cell 22, 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Peng Z, Liu S, He Y, Cheng L, Kong F, Wang J, Lu G. 2012. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Molecular Genetics and Genomics 87, 295–311 [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation. Annual Review of Plant Biology 58, 225–251 [DOI] [PubMed] [Google Scholar]
- Yoon EK, Yang JH, Lim J, Kim SH, Kim SK, Lee WS. 2010. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Research 38, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, To JPC, Cheng CY, Schaller GE, Kieber JJ. 2011. Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. The Plant Journal 68, 1–10 [DOI] [PubMed] [Google Scholar]
- Zheng X, Miller ND, Lewis DR, Christians MJ, Lee KH, Muday GK, Spalding EP, Vierstra RD. 2011. AUXIN UPREGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiology 156, 1878–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]