Abstract
We have developed a porcine intestine epithelial cell line, designated SD-PJEC for the propagation of influenza viruses. The SD-PJEC cell line is a subclone of the IPEC-J2 cell line, which was originally derived from newborn piglet jejunum. Our results demonstrate that SD-PJEC is a cell line of epithelial origin that preferentially expresses receptors of oligosaccharides with Sia2-6Gal modification. This cell line is permissive to infection with human and swine influenza A viruses and some avian influenza viruses, but poorly support the growth of human-origin influenza B viruses. Propagation of swine-origin influenza viruses in these cells results in a rapid growth rate within the first 24 h post-infection and the titres ranged from 4 to 8 log10 TCID50 ml−1. The SD-PJEC cell line was further tested as a potential alternative cell line to Madin–Darby canine kidney (MDCK) cells in conjunction with 293T cells for rescue of swine-origin influenza viruses using the reverse genetics system. The recombinant viruses A/swine/North Carolina/18161/02 (H1N1) and A/swine/Texas/4199-2/98 (H3N2) were rescued with virus titres of 7 and 8.25 log10 TCID50 ml−1, respectively. The availability of this swine-specific cell line represents a more relevant substrate for studies and growth of swine-origin influenza viruses.
Introduction
The family Orthomyxoviridae contains three genera of influenza virus, which are identified by antigenic differences in their nucleoprotein (NP) and matrix protein. Influenza A virus infects humans, other mammals and birds, and causes all flu pandemics (Webster et al., 1992). Influenza B virus infects humans and seals (Osterhaus et al., 2000). Influenza C virus infects humans and pigs (Buonagurio et al., 1986). Viruses of the family Orthomyxoviridae contain six to eight segments of linear negative-sense ssRNA genome, influenza A and B viruses consists of eight RNA segments. The viral haemagglutinin protein is a major envelope glycoprotein encoded by one of these RNA segments. During the infection, viruses bind to a cell through interactions between the HA glycoprotein and sialic acid sugars on the surfaces of epithelial cells.
An important aspect of the influenza virus pathogenesis is the mechanism of cross-species transmission. Pigs are considered to be a mixing vessel, from which novel virus reassortants could emerge to cause pandemics (Scholtissek et al., 1983). Previous studies have demonstrated that HA receptor specificity can determine the infection of species (Maines et al., 2011; Shinya et al., 2006). Human influenza viruses preferentially attach to host cells expressing α2,6-linked sialic acids (Hatakeyama et al., 2005; Matrosovich et al., 2000; Webby & Webster, 2003), while avian influenza viruses prefer to attach to α2,3-linked sialic acids (Suzuki et al., 2000). It has been well documented that both α2,6- and α2,3-linked sialic acid receptors are present at the pig mucosal surfaces (Bateman et al., 2008; Chutinimitkul et al., 2010; Nicholls et al., 2008; Rogers et al., 1985; Takemae et al., 2010), allowing for the simultaneous infection of pigs with both avian- and human-origin influenza viruses (Trebbien et al., 2011).
The most common cellular model for influenza virus studies is the Madin–Darby canine kidney (MDCK) cells, and this cell line represents the most applicable alternative to egg-based virus isolation and propagation (Hussain et al., 2010; Roth et al., 2012). Other cell lines, including Vero, baby hamster kidney (BHK) and A549 also support the growth of influenza viruses (Govorkova et al., 1999a, 1999b; Rimmelzwaan et al., 2004). However, these cells may not represent the most relevant cell line for evaluation of influenza virus infection within pigs. Therefore, we developed the hypothesis that a more relevant swine-origin cell line would be very useful to study the host cell contributions to cross-species transmission and viral pathogenesis. To test this hypothesis, we developed a porcine epithelial cell line, SD-PJEC. The SD-PJEC is a subclone of the IPEC-J2 cell line, which is a non-transformed, non-tumorigenic small intestinal epithelial cell line originally derived from jejunal epithelia of a neonatal, unsuckled piglet (Berschneider, 1989; Rhoads et al., 1994). The capability of this cell line to support influenza virus replication was determined by using a panel of influenza A viruses of human, swine and avian origin, and influenza B viruses of human origin. We further explored the potential application of SD-PJEC cells in a reverse genetics system.
Results
SD-PJEC cells determined to be epithelial phenotype
The SD-PJEC cell line was generated from a subclone of IPEC-J2 cells (Berschneider, 1989; Rhoads et al., 1994). The cell line appears to be a more homogeneous cell population than that of the IPEC-J2 cell line. To determine the phenotype of the SD-PJEC cells, cells were stained with antibodies that recognized the marker proteins for different cell phenotypes, including cytokeratin (epithelial), vimentin (fibroblast), alpha smooth muscle actin (ASMA, smooth muscle) and desmin (smooth and striated muscles). As shown in Fig. 1, all SD-PJEC cells were positively stained for cytokeratin and few cells faintly stained for vimentin. This result indicates that the SD-PJEC cell line has an epithelial phenotype.
Fig. 1.
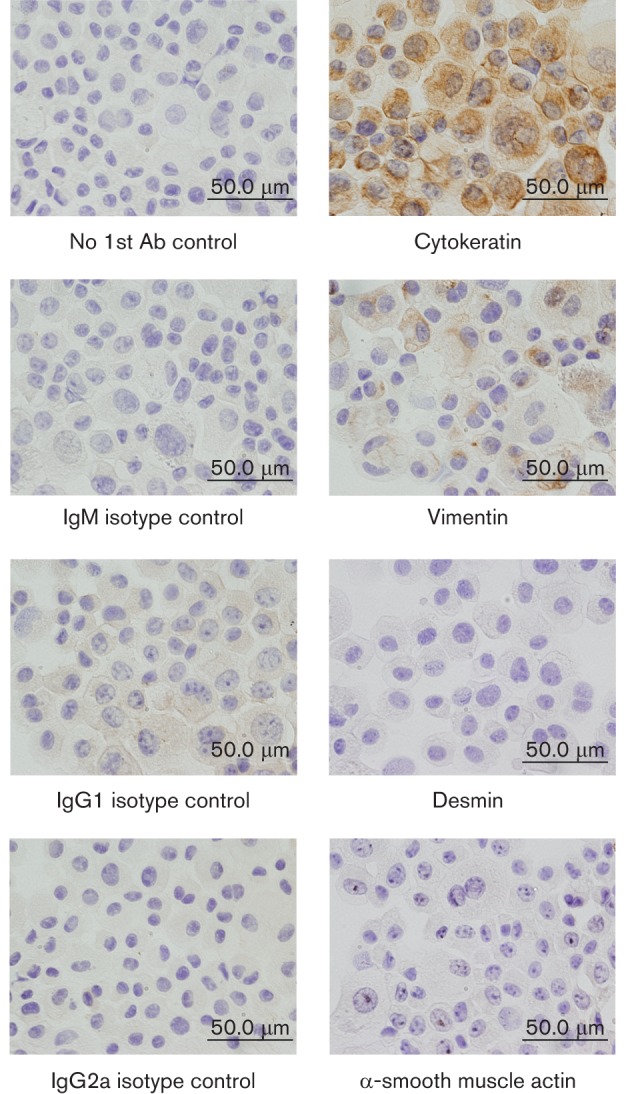
Staining of SD-PJEC cells for various epithelial, fibroblast and smooth muscle markers. Cytospins (1×105 SD-PJEC cells) were fixed in acetone. The presence of cytokeratin, vimentin, alpha smooth muscle actin (ASMA) and desmin proteins was detected by immunohistochemical (IHC) staining using mAbs specific for these proteins. Staining without primary antibody but only secondary antibody of mouse IgM, IgG1 and IgG2a were used as negative and isotype controls. After washing, cells were incubated with biotinylated goat anti-mouse isotype-specific Abs. This was followed by incubation of cells with the ABC solution. Then DAB substrate was added and cells were counterstained with haematoxylin. Note, all SD-PJEC cells were positively stained for cytokeratin and few cells faintly stained for vimentin, indicating their epithelial phenotype.
SD-PJEC cells express Sia2-6 galactose oligosaccharides receptor
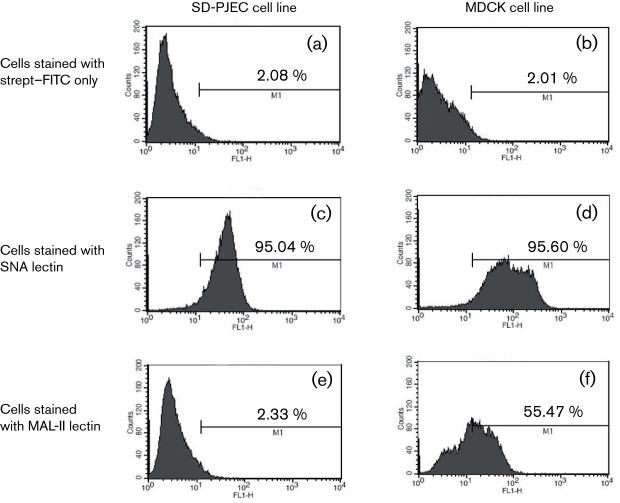
Previous studies determined that both influenza virus receptors, Sia2-6Gal and Sia2-3Gal, are present at the pig mucosal surfaces (Ito et al., 1998). We further determined whether these receptors were present on the SD-PJEC cells. As a comparison, MDCK cells were included in the analysis. Biotinylated Maackia amurensis lectin-2 (MAL-II) specific for Sia2-3Gal and Sambucus nigra agglutinin (SNA) specific for Sia2-6Gal were used to stain both cell lines as described previously (Meroz et al., 2011). Receptor specificity was evaluated by flow cytometric analysis. As shown in Fig. 2(a) and (b), only background fluorescent signals were detected in the negative control cells that were only stained with FITC-conjugated streptavidin. Both SD-PJEC and MDCK cells expressed Sia2-6Gal receptor showing positive SNA staining (Fig. 2c and d). However, the Sia2-3Gal receptors were not expressed on the surface of SD-PJEC cells, and only background fluorescent signal was detected for MAL-II lectin staining (Fig. 2e). In contrast, MDCK cells stained positive for MAL-II lectin (Fig. 2f). These results clearly indicate that the SD-PJEC cells mainly express Sia2-6Gal receptor, while the Sia2-3Gal receptor is expressed at minimal levels. The experiment was repeated four different times using SD-PJEC cells from passages 18 to 22. There were no significant changes in their staining pattern or percentages of cells stained for MAL-II (0.25–0.65 %) and SNA (84.16–92.96 %) across the passage 18–22 cells. This suggests that SD-PJEC cells were phenotypically stable over many cell passages tested in this study.
Fig. 2.
Influenza virus receptor expression in SD-PJEC and MDCK cell lines. Both SD-PJEC and MDCK cell lines were incubated with biotinylated MAL-II and SNA lectins followed by staining with streptavidin–FITC. The negative control cells for both cell lines were stained with streptavidin–FITC only. Stained samples were subjected to flow cytometric analysis. A representative experiment out of four experiments is shown here.
Replication efficiencies of human, avian and swine influenza viruses in SD-PJEC cells
To determine whether the SD-PJEC cells are permissive to influenza virus infection, we inoculated the cells with an influenza A virus, A/swine/Texas/4199-2/98 (H3N2). At 24 h post-infection (p.i.), cells were fixed and the expression of viral nucleocapsid protein (NP) was detected by immunofluorescence assay (IFA) using NP-specific antibody. As shown in Fig. 3, a specific fluorescent staining pattern was observed in nuclear and cytoplasm compartments of infected cells, while no fluorescent signal was detected in uninfected cells, suggesting that the cells were permissive for influenza A virus infection.
Fig. 3.
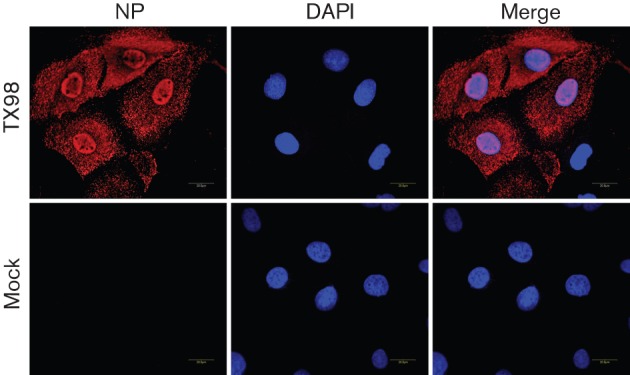
Immunofluorescence microscopy detection of NP expression in SD-PJEC-infected cells by A/swine/Texas/4199-2/98 (TX98). Confluent cells were infected with the influenza virus at an m.o.i. of 0.01. At 24 h p.i., cells were fixed and stained with a primary mAb to NP and Alexa Fluor 546-labelled goat anti-mouse antibody was used as secondary antibody. The nucleus was stained with DAPI. Mock-infected cells were used as a control. Specimens were visualized on a Zeiss LSM510 confocal microscope. A 0.8 mm slice through the nucleus is shown in each image.
We further compared the replication efficiency of influenza viruses in SD-PJEC cells with that in MDCK cells. A panel of human, swine and avian-origin influenza virus isolates were titrated on both MDCK and SD-PJEC cells in order to quantify replication efficiency in the newly developed SD-PJEC cells. Some of the influenza A viruses of swine origin grew to higher titres in SD-PJEC cells (Table 1). The titres ranged from 4 to 8 log10 TCID50 ml−1, with most of the viruses having titres of 6–7 log10 TCID50 ml−1. The replication of two H7 subtype avian influenza viruses was also assessed. Contemporary North American lineage influenza H7 viruses were reported to possess human receptor specificity (Belser et al., 2008) and we therefore predicted that these viruses would replicate well in the SD-PJEC cells. The results showed that both A/mallard/Alberta/177/04 (H7N9) and A/shorebird/Delaware/22/06 (H7N3) avian influenza A viruses replicated well in the SD-PJEC cells with less than 10-fold difference in virus titre between MDCK and SD-PJEC cells. Influenza B viruses have been reported to only naturally infect humans and seals (Osterhaus et al., 2000), and within humans, two distinct genetic lineages are co-circulating (McCullers et al., 2004). Using influenza B viruses that represent the genotypes of two human lineages, B/Memphis/13/03 (Victoria87 lineage) and B/Yamanashi/166/98 (Yamagata88 lineage), no CPE was observed in SD-PJEC cells. Influenza B viruses appeared to grow poorly in these cells, lack of cell-to-cell spreading, and very low virus titres were detected in the cell culture supernatant at 72 h p.i. (Table 1). In contrast, these two viruses replicated well in MDCK cells, with the virus titres reaching 6.5 and 7.4 log10 TCID50 ml−1, respectively.
Table 1. Comparison of propagation of influenza A and B viruses within MDCK and SD-PJEC cells.
| Virus isolate | Species | MDCK* | SD-PJEC |
| A/New Jersey/11/76-H1N1 | Human | 6.00 | 6.40 |
| A/Brisbane/59/07-H1N1 | Human | 7.00 | 6.60 |
| A/California/4/09-H1N1 | Human | 5.00 | 4.00 |
| A/swine/South Dakota/01/09-H1N1 | Swine | 7.40 | 6.40 |
| A/swine/South Dakota/02/09-H1N1 | Swine | 7.50 | 6.40 |
| A/swine/Minnesota/4390/11-H1N1 | Swine | 3.50 | 4.50 |
| A/swine/North Carolina/31/12-H1N1 | Swine | 5.00 | 8.00 |
| A/swine/Texas/042995-27/07-H1N2 | Swine | 9.00 | 8.40 |
| A/swine/Missouri/4460/11-H1N2 | Swine | 6.50 | 6.50 |
| A/swine/North Carolina/4478/11-H1N2 | Swine | 4.00 | 6.00 |
| A/swine/Minnesota/4579/11-H1N2 | Swine | 4.50 | 6.50 |
| A/swine/Minnesota/21/12-H1N2 | Swine | 2.50 | 3.50 |
| A/swine/North Carolina/28/12-H1N2 | Swine | 6.50 | 8.00 |
| A/swine/North Carolina/29/12-H1N2 | Swine | 8.33 | 6.75 |
| A/swine/Oklahoma/52/12-H1N2 | Swine | 7.50 | 7.00 |
| A/swine/Minnesota/4393/11-H3N2 | Swine | 7.00 | 6.00 |
| A/swine/Minnesota/4395/11-H3N2 | Swine | 3.50 | 5.50 |
| A/swine/Texas/2/98-H3N2 | Swine | 5.00 | 5.50 |
| A/swine/Illinois/26/12-H3N2 | Swine | 3.50 | 4.50 |
| A/swine/Minnesota/50/12-H3N2 | Swine | 4.00 | 5.00 |
| A/swine/Ohio/51/12-H3N2 | Swine | 7.50 | 5.50 |
| A/mallard/Alberta/177/04 (H7N9) | Avian | 7.70 | 6.80 |
| A/shorebird/Delaware/22/06 (H7N3) | Avian | 8.50 | 6.70 |
| B/Yamanashi/166/98-Yam88 | Human | 7.40 | 4.00 |
| B/Memphis/13/03-Vic87 | Human | 6.50 | 4.00 |
Values reported as log10 TCID50 ml−1.
To determine the kinetics of influenza virus replication in SD-PJEC cells, we compared the growth curves of a representative virus (A/swine/Texas/4199-2/98) in SD-PJEC cells and MDCK cells. Supernatants were harvested at different time points p.i., and virus titre was determined by titration in MDCK cells. The results showed that both viruses reach their peak titre at 36 h p.i. However, there was an approximately 100-fold higher peak viral titre in SD-PJEC cells, with a peak titre of 9.3 log10 TCID50 ml−1 in SD-PJEC cells in comparison to the peak titre of 7.3 log10 TCID50 ml−1 in MDCK cells (Fig. 4). Stability of the viruses in SD-PJEC cells was further investigated by serial passage of A/swine/Texas/4199-2/98 virus 10 times in these cells. We observed no mutations in HA gene of passage 10 virus, and the virus titre remains at a similar level to that of passage 1 virus (P1 = 5.5 log10 TCID50 ml−1; P10 = 6.5 log10 TCID50 ml−1).
Fig. 4.
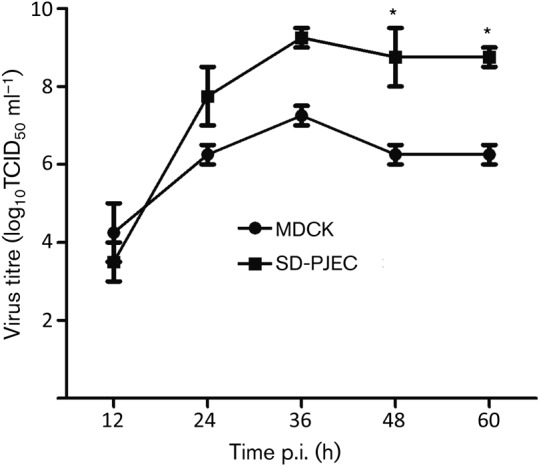
Comparison of growth kinetics of A/swine/Texas/4199-2/98 (H3N2) in MDCK and SD-PJEC cells. Confluent MDCK and SD-PJEC cells were infected with the influenza virus at an m.o.i. of 0.01. Virus titres were determined at 12 h intervals. The results shown are mean values from three replicates. Error bars show sem. Asterisks indicate that mean virus titres from different cell types differ significantly (P<0.05).
Application of the SD-PJEC cell line in reverse genetics
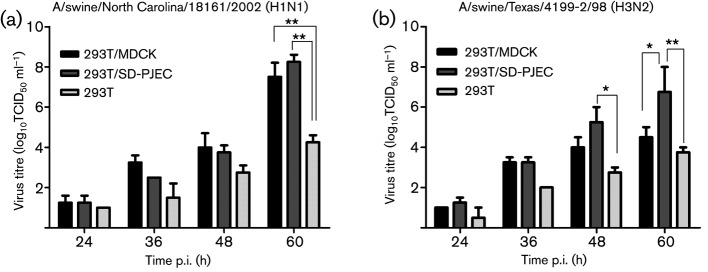
Reverse genetics systems allow for the production of influenza viruses from cloned viral cDNA (Hoffmann et al., 2000, 2002; Hoffmann & Webster, 2000). The commonly used cell lines to obtain influenza viruses from cDNA are 293T and MDCK cells. These two types of cells are co-cultured for initial transfection and virus rescue in order to achieve the maximal viral yield. However, MDCK cells may not be the optimum cell line for rescue of viruses that contain the genome segment(s) of swine origin. In this study, we explored the possibility of using the SD-PJEC cell line as an alternative to MDCK cells in the co-culture system. Two sets of eight-plasmid cDNA reverse genetics system for swine-origin influenza viruses, A/swine/North Carolina/18161/02 (H1N1) and A/swine/Texas/4199-2/98 (H3N2), were tested (Meroz et al., 2011; Solórzano et al., 2005). The virus rescue efficiency was compared between 293T/MDCK and 293T/SD-PJEC co-culture systems. As a control, we also rescued the virus on 293T cell alone. At 12, 24, 36, 48 and 60 h after addition of trypsin, the titres of virus in the cell culture supernatant were determined. For A/swine/North Carolina/18161/02, there was no significant difference (P>0.05) in virus titres in the supernatant from both co-culture systems, whereas virus titres in 293T cells alone were reduced about 3–4 log10 TCID50 ml−1 (Fig. 5a). In contrast, a significantly higher amount of A/swine/Texas/4199-2/98 viruses (P<0.05) was rescued from 293T/SD-PJEC co-culture system at 60 h p.i. The peak titre reached 7 log10 TCID50 ml−1 (60 h p.i.), which is 2.5 log higher than that rescued from the 293T/MDCK co-culture system. Again, the 293T cells alone recovered the lowest amount of recombinant viruses in comparison to the other two co-culture systems (Fig. 5b).
Fig. 5.
Rescue of recombinant influenza viruses from 293T/SD-PJEC co-culture system. Eight plasmids containing individual gene segments of A/swine/North Carolina/18161/2002 (a) or A/swine/Texas/4199-2/98 (b) were used to transfect 293T/SD-PJEC, 293T/MDCK or 293T cells alone. The amount of virus produced in culture supernatants was determined at 12 h intervals. The results shown at each time point are mean values from three independent experiments conducted with each virus. Error bars show sem. Asterisks indicate that mean virus titres from different culture methods differ significantly (*P<0.05; **P<0.01).
Discussion
In this study, we characterized a porcine small intestinal epithelial cell line (SD-PJEC) that supports the productive replication of certain influenza viruses. The SD-PJEC cell line represents an in vitro model system for evaluation of influenza virus infection within porcine cells. The recent emergence and pandemic classification of a triple reassortant virus containing swine, human and avian genetic components have raised greater concerns over the swine-origin viruses (Smith et al., 2009). There are significant concerns that these novel viruses would undergo further reassortment events within either the human or swine populations to yield viruses with increased virulence (Smith et al., 2009). Based on our current knowledge of influenza virus reassortment within the swine population (Khiabanian et al., 2010), the pandemic potential for a given influenza virus reassortant depends on the fitness of these reassortants within the swine host.
While studies aim to evaluate influenza virus virulence and pathogenesis require animal hosts (Ozawa et al., 2011), in vitro studies to determine the interaction between the virus and host that yield optimal virus replication and fitness can be performed using cell lines. To date, the majority of in vitro influenza virus experiments have been performed using MDCK cells (Sidorenko & Reichl, 2004; Heynisch et al., 2010), which may differ significantly from either human (Chakrabarti et al., 2010) or swine cells. Most studies for influenza pathogenesis in swine have been performed in animal infection models. Elucidation of the in vitro infection mechanisms depends on a reliable, continuous porcine cell line, and the SD-PJEC cell line represents such a model.
Since the SD-PJEC cells were generated from small intestine, this cell line is particularly suitable to study the pathogenic mechanism of influenza virus in digestive systems. Influenza infection normally causes symptoms, including coughing, sore throat, headache, fever, weakness, muscle aches, diarrhoea and vomiting. The seasonal influenza usually attacks the respiratory system, and is rarely associated with gastrointestinal symptoms such as vomiting or diarrhoea. However, when 2009 pandemic virus emerged, patients showed disease not only in the respiratory tract, but also within the digestive system, with vomiting and/or diarrhoea reported by nearly 30 % of people with laboratory-confirmed influenza infection (Shinde et al., 2009). This suggests that the influenza virus might be able to replicate in intestine cells, but to date, the knowledge concerning the pathogenic mechanism of influenza virus within the digestive system is limited due to a lack of a good in vitro cellular system. We believe that the intestinal epithelial cell line we present here would make an appropriate cellular model for evaluation of pathogenic mechanism of influenza viruses, especially for those viruses containing genome segment(s) of swine-origin influenza virus.
SD-PJEC cells were determined to have an epithelial phenotype with strong staining for epithelial marker cytokeratin. However, they were also weakly positive for vimentin staining. In a recent study, it was clearly shown that cultured bovine intestinal epithelial cells co-expressed the epithelial marker cytokeratin and the mesenchymal marker vimentin. It is known that vimentin protein is not expressed in intestinal epithelial cells in vivo although these cells can carry vimentin mRNA. In contrast, a post-transcriptional inhibition of vimentin synthesis observed in vivo is suppressed in vitro (Rusu et al., 2005). Thus, epithelial cells could express lower levels of vimentin along with cytokeratin in vitro. Another study on pig intestinal cells also reported similar findings (Kaeffer et al., 1993). Our results were consistent with these studies, and SD-PJEC cells strongly stained for cytokeratin but also weakly stained for vimentin.
It is intriguing that the SD-PJEC cells predominantly express the α2,6-galactose receptor. It is well-known that both α2,6- and α2,3-galactose receptors are expressed on the cell surface of the pig respiratory tract (Ito et al., 1998). Receptor expression levels differ in respiratory and intestine cells; therefore it would be useful to study the difference of influenza pathogenic mechanisms between the two different compartments (respiratory tract versus intestine). Since SD-PJEC cells lack the α2,3-galactose receptor, these cells could also be useful in viral receptor-based analyses, including those associated with the development of novel therapies based on sialic acid binding properties (Malakhov et al., 2006).
Our results further demonstrate that SD-PJEC could be used as an alternative cell line in a reverse genetics system. Application of 293T/SD-PJEC co-cultures would be useful for situations where 293T/MDCK co-cultures are unsuccessful for creation of swine-origin influenza viruses (Wanitchang et al., 2010). In addition, targeted mutations could be created to elucidate the molecular basis of the subsequent growth adaptation of a specific influenza virus in host cells using the 293T/SD-PJEC co-cultures and the reverse genetics system.
Besides its application in influenza pathogenesis studies, the SD-PJEC cell line could be a potential candidate to use in vaccine production. There is a great demand for the development of improved cell culture systems for human vaccine production (Lee & Hu, 2012), since the traditional embryonic egg-based method for virus propagation can result in antigenic changes (Fedson, 2008). Several influenza virus permissive cell lines have been explored previously (Hussain et al., 2010). For example, baby hamster kidney (BHK) cells were capable of supporting influenza virus propagation, but like eggs, receptor-binding variants of human influenza viruses were generated when growing in this cell line (Govorkova et al., 1999b). Also, while African green monkey kidney (Vero) cells were reported to fully support the replication of influenza A and B viruses, the viruses have to be adapted before they can be grown in these cells (Govorkova et al., 1999a). MDCK cells are still considered the best cell-based alternative to eggs for isolation and propagation of influenza viruses, but concerns over the tumorigenic potential of by-products from this cell line make it less suitable for vaccine production (Gregersen et al., 2011). Since SD-PJEC cells represent a non-transformed, non-tumorigenic cell line, they could be tested as a candidate cell line for use in vaccine production. To support this notion, we present similar growth characteristics, if not improved, when propagation of viruses within SD-PJEC cells compared to MDCK cells; and have observed no mutations in the HA gene upon sequential passage of the A/swine/Texas/4199-2/1998 virus (10 passages) within these cells.
In conclusion, we characterized a swine epithelial cell line, SD-PJEC that is permissive to infection with human and swine influenza A viruses and some avian influenza viruses, but poorly support the growth of human-origin influenza B viruses. The availability of this cell line provides an additional cellular model system for elucidation of the mechanism of influenza virus pathogenesis, especially for those associated with successful reassortment events within the intermediate swine host. Further evaluation of the interactions between this cell line and influenza viruses will allow for identification of relevant virulence factors and, eventually, the development of effective strategies to prevent influenza virus infection.
Methods
Cell lines and culture conditions.
The SD-PJEC cell line was generated by a subclone of IPEC-J2 cells, which appears to be a more homogeneous cell population. For subcloning the IPEC-J2 cells, a confluent cell monolayer was trypsinized with 0.25 % trypsin-EDTA and 100 representative cells were obtained based on haemacytometer counts. These 100 cells were seeded in a 96-well plate to obtain a cell density at approximately 1 cell per well in 250 µl cell culture medium. Cells were allowed to adhere for 24 h, and then washed and replaced with fresh medium. Cell culture medium was changed every other day to allow the cell to grow into a single clone in each well. Each clone of cells grown into a confluent monolayer in 96-well plate was expended into 24-well plates and a proportion of cells in each well were tested further for their permissiveness to influenza virus infection.
Both IPEC-J2 and SD-PJEC cells were grown in Dulbecco’s modified Eagle’s medium (DMEM): Ham’s F-12 (1 : 1) medium (DMEM/F12) (Invitrogen). The DMEM/F12 medium was supplemented with 5 % FBS (Hyclone), 1 % insulin-transferring selenium supplements (Invitrogen), 5 ng epidermal growth factor (Invitrogen) ml−1, 1 % penicillin–streptomycin (penicillin 10 000 U ml−1 and streptomycin 100 mg ml−1; Invitrogen) and 15 mM HEPES. Cell culture media were changed every other day. In addition, MDCK and HEK293T cells were also used in this study, and they were maintained in minimal essential medium (MEM) supplemented with 5 % FBS.
Influenza virus isolates.
Two sets of influenza viruses were used in this study. Table 1 lists a total of 25 influenza virus isolates of human, swine or avian origin. Nine of these isolates were obtained from the repository at the St. Jude Children’s Research Hospital (Memphis, TN); and two swine-origin influenza viruses were obtained from South Dakota Animal Disease Diagnostic Laboratory (Brookings, SD). The rest of the viruses were isolated from field samples submitted to Newport Laboratories (Worthington, MN). Virus stocks were grown on either SD-PJEC or MDCK cells in MEM supplemented with 0.3 % FBS and l-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-trypsin (1 µg ml−1 for MDCK and 0.1 µg ml−1 for SD-PJEC). Infected cells were incubated at 37 °C for 72 h, and observed daily for cytopathic effect (CPE). To confirm the presence of virus, haemagglutination assay with 0.5 % chicken erythrocytes was performed.
Phenotyping of SD-PJEC cells.
SD-PJEC cells were stained with antibodies which recognized various epithelial, fibroblast and smooth muscle markers using the protocol as described previously (Kaushik et al., 2008; Rhoads et al., 1994). Briefly, SD-PJEC cell cultures were trypsinized and washed with PBS. Cytospins (1×105 SD-PJEC cells) were prepared using a cytofuge (Cytospin 3; Thermo Shandon Inc.), air-dried, fixed in acetone and stored at 4 °C until they are ready for immunoassays. For antibody staining, slides were equilibrated at room temperature, rehydrated in PBS and then incubated with PBS containing 1 % goat serum to block non-specific protein binding. The presence of cytokeratin, vimentin, ASMA and desmin proteins was detected by immunohistochemical (IHC) staining using anti-cytokertain mAb C6909 (IgG2a isotype), anti-vimentin mAb V5255 (IgM isotype), anti-ASMA mAb A2547 (IgG2a isotype) and anti-desmin mAb D1033 (IgG1). mAbs M9144 (IgG2a isotype), M9269 (IgG1 isotype) and M5170 (IgM isotype) were used as irrelevant isotype-matched controls. Cells without primary antibody staining were used as negative control. All mAbs were purchased from Sigma and used at 1 µg ml−1 concentration with 1 h incubation. Slides were washed three times (3×) in PBS and then incubated with isotype-specific, biotinylated goat anti-mouse IgG2a, IgG1 or IgM antisera (1 : 2000 dilution; Caltag laboratories) for 30 min. Slides were washed 3× and then incubated in PBS containing 0.3 % hydrogen peroxide to block endogenous peroxidase activity. Antibody labelling was visualized by adding ready-to-use (RTU) HRP–streptavidin solution for 30 min followed by the addition of RTU diaminobenzene (DAB) substrate (Vector Laboratories). Cytospins were counterstained with haematoxylin, dried overnight, contained with a coverslip and examined under the light microscope. Pictures were taken under ×40 magnification using an Olympus AX70 microscope.
Flow cytometric analysis of sialic acid receptor expression.
Biotinylated MAL-II specific for Sia2-3Gal and SNA (Vector laboratories) specific for Sia2-6Gal were used to stain both SD-PJEC and MDCK cell lines as described previously (Meroz et al., 2011). Briefly, 5×105 cells of each cell type were incubated with biotinylated MAL-II and SNA lectins (final concentration 10 µg ml−1) followed by staining with streptavidin–FITC (1 : 200 dilution). The negative control cells for both cell lines were stained with streptavidin–FITC only. Stained samples were subjected to flow cytometric analysis.
Immunofluorescence microscopy.
SD-PJEC cells were infected with influenza virus A/swine/Texas/4199-2/1998 at an m.o.i. of 0.01 for 1 h and virus supernatant was replaced with the growth medium containing 1 µg TPCK-trypsin ml−1. Infected cells were incubated at 37 °C for 24 h and then fixed with methanol–acetone (1 : 1 ratio in volume) at −20 °C for 20 min. The fixed cells were stained with a primary mAb 42-100 to the NP at 37 °C for 1 h. The Alexa Fluor 549-labelled goat anti-mouse antibody (Kirkegaard & Perry Laboratories) was used as a secondary antibody and incubated for another hour. Nuclear staining with DAPI was performed as recommended by the manufacturer (Molecular Probes). Specimens were imaged using a Zeiss LSM510 confocal microscope with a ×63 objective, and images were processed with NIH ImageJ and Adobe Photoshop 6.0 software.
Viral growth kinetics analysis.
Growth kinetics of influenza viruses in SD-PJEC cells was compared with those in MDCK cells. Confluent cell monolayers were infected with influenza A/swine/Texas/4199-2/1998 virus at an m.o.i. of 0.01 and incubated at 37 °C for 1 h. The virus suspension was then removed, and the MEM containing 1 µg TPCK-treated trypsin ml−1 was added (1 µg ml−1 for MDCK and 0.1 µg ml−1 for SD-PJEC). Cell culture supernatants were collected at 12 h intervals until 60 h post-inoculation. The virus titre was determined by titration on MDCK cells.
Virus rescue from cloned cDNA.
SD-PJEC, 293T and MDCK cells were grown to 100 % confluence in a T75 flask and then trypsinized with trypsin-EDTA (Invitrogen) and resuspended in 10 ml Opti-MEM I (Invitrogen). Cells were counted and seeded into each well of a six-well tissue culture plate (3 ml per well with 1×106 cells). For co-culture of 293T with SD-PJEC or MDCK cells, cells were seeded at a 3 : 1 ratio for 293T/MDCK and 3 : 1 ratio for 293T/SD-PJEC. Cells were incubated at 37 °C for 16–18 h, and transfected with 1 µg each plasmid DNA using the Fugene HD reagent (Promega) following the manufacturer’s instructions. Six hours post-transfection, the DNA-transfection mixture was replaced by Opti-MEM I. At 30 h post-transfection, 1 ml Opti-MEM I containing TPCK-treated trypsin (1 µg ml−1 for MDCK and 0.1 µg ml−1 for SD-PJEC) was added to the cells. At 24, 36, 48 and 60 h post-transfection, 200 µl of culture supernatant was collected at each time point. The amount of viruses present in the supernatant was determined by titration on MDCK cells and virus titres were calculated as TCID50 ml−1.
Statistical analysis.
Statistical analysis was performed using GraphPad InStat version 3.06 (GraphPad Software). Comparison was performed by one-way analysis of variance with Tukey’s multiple comparison tests to determine the mean significance. Differences between treatment groups were considered statistically significant at P<0.05.
Acknowledgements
We thank Bruce Schultz (Kansas State University) for providing IPEC-J2 cells and Pam Steen (South Dakota State University) for providing swine influenza viruses. Special thanks to Yanhua Li and Steven Lawson (South Dakota State University) for excellent technical assistance. This work was supported by National Institutes of Health (to Y. F. and V. C. H. grant # R15AI090582-01), and South Dakota Agricultural Experiment Station (to R. S. K. grant # SDOOH326-09).
References
- Bateman A. C., Busch M. G., Karasin A. I., Bovin N., Olsen C. W. (2008). Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for α2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J Virol 82, 8204–8209 10.1128/JVI.00718-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser J. A., Blixt O., Chen L. M., Pappas C., Maines T. R., Van Hoeven N., Donis R., Busch J., McBride R. & other authors (2008). Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci U S A 105, 7558–7563 10.1073/pnas.0801259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berschneider H. M. (1989). Development of normal cultured small intestinal epithelial cell lines which transport Na and Cl [Abstract]. Gastroenterology 96, A41 [Google Scholar]
- Buonagurio D. A., Nakada S., Fitch W. M., Palese P. (1986). Epidemiology of influenza C virus in man: multiple evolutionary lineages and low rate of change. Virology 153, 12–21 10.1016/0042-6822(86)90003-6 [DOI] [PubMed] [Google Scholar]
- Chakrabarti A. K., Vipat V. C., Mukherjee S., Singh R., Pawar S. D., Mishra A. C. (2010). Host gene expression profiling in influenza A virus-infected lung epithelial (A549) cells: a comparative analysis between highly pathogenic and modified H5N1 viruses. Virol J 7, 219 10.1186/1743-422X-7-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutinimitkul S., Herfst S., Steel J., Lowen A. C., Ye J., van Riel D., Schrauwen E. J., Bestebroer T. M., Koel B. & other authors (2010). Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol 84, 11802–11813 10.1128/JVI.01136-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedson D. S. (2008). NEW technologies for meeting the global demand for pandemic influenza vaccines. Biologicals 36, 346–349 10.1016/j.biologicals.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Govorkova E. A., Kodihalli S., Alymova I. V., Fanget B., Webster R. G. (1999a). Growth and immunogenicity of influenza viruses cultivated in Vero or MDCK cells and in embryonated chicken eggs. Dev Biol Stand 98, 39–51, discussion 73–74 [PubMed] [Google Scholar]
- Govorkova E. A., Matrosovich M. N., Tuzikov A. B., Bovin N. V., Gerdil C., Fanget B., Webster R. G. (1999b). Selection of receptor-binding variants of human influenza A and B viruses in baby hamster kidney cells. Virology 262, 31–38 10.1006/viro.1999.9892 [DOI] [PubMed] [Google Scholar]
- Gregersen J. P., Schmitt H. J., Trusheim H., Bröker M. (2011). Safety of MDCK cell culture-based influenza vaccines. Future Microbiol 6, 143–152 10.2217/fmb.10.161 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Sakai-Tagawa Y., Kiso M., Goto H., Kawakami C., Mitamura K., Sugaya N., Suzuki Y., Kawaoka Y. (2005). Enhanced expression of an α2,6-linked sialic acid on MDCK cells improves isolation of human influenza viruses and evaluation of their sensitivity to a neuraminidase inhibitor. J Clin Microbiol 43, 4139–4146 10.1128/JCM.43.8.4139-4146.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heynisch B., Frensing T., Heinze K., Seitz C., Genzel Y., Reichl U. (2010). Differential activation of host cell signalling pathways through infection with two variants of influenza A/Puerto Rico/8/34 (H1N1) in MDCK cells. Vaccine 28, 8210–8218 10.1016/j.vaccine.2010.07.076 [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Webster R. G. (2000). Unidirectional RNA polymerase I-polymerase II transcription system for the generation of influenza A virus from eight plasmids. J Gen Virol 81, 2843–2847 [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Hobom G., Webster R. G., Kawaoka Y. (2000). “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology 267, 310–317 10.1006/viro.1999.0140 [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Krauss S., Perez D., Webby R., Webster R. G. (2002). Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20, 3165–3170 10.1016/S0264-410X(02)00268-2 [DOI] [PubMed] [Google Scholar]
- Hussain A. I., Cordeiro M., Sevilla E., Liu J. (2010). Comparison of egg and high yielding MDCK cell-derived live attenuated influenza virus for commercial production of trivalent influenza vaccine: in vitro cell susceptibility and influenza virus replication kinetics in permissive and semi-permissive cells. Vaccine 28, 3848–3855 10.1016/j.vaccine.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Couceiro J. N. S. S., Kelm S., Baum L. G., Krauss S., Castrucci M. R., Donatelli I., Kida H., Paulson J. C. & other authors (1998). Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 72, 7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeffer B., Bottreau E., Velge P., Pardon P. (1993). Epithelioid and fibroblastic cell lines derived from the ileum of an adult histocompatible miniature boar (d/d haplotype) and immortalized by SV40 plasmid. Eur J Cell Biol 62, 152–162 [PubMed] [Google Scholar]
- Kaushik R. S., Begg A. A., Wilson H. L., Aich P., Abrahamsen M. S., Potter A., Babiuk L. A., Griebel P. (2008). Establishment of fetal bovine intestinal epithelial cell cultures susceptible to bovine rotavirus infection. J Virol Methods 148, 182–196 10.1016/j.jviromet.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiabanian H., Holmes A. B., Kelly B. J., Gururaj M., Hripcsak G., Rabadan R. (2010). Signs of the 2009 influenza pandemic in the New York-Presbyterian Hospital electronic health records. PLoS ONE 5, e12658 10.1371/journal.pone.0012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Hu A. Y. (2012). A cell-based backup to speed up pandemic influenza vaccine production. Trends Microbiol 20, 103–105 10.1016/j.tim.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Maines T. R., Chen L. M., Van Hoeven N., Tumpey T. M., Blixt O., Belser J. A., Gustin K. M., Pearce M. B., Pappas C. & other authors (2011). Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology 413, 139–147 10.1016/j.virol.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhov M. P., Aschenbrenner L. M., Smee D. F., Wandersee M. K., Sidwell R. W., Gubareva L. V., Mishin V. P., Hayden F. G., Kim D. H. & other authors (2006). Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob Agents Chemother 50, 1470–1479 10.1128/AAC.50.4.1470-1479.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M., Tuzikov A., Bovin N., Gambaryan A., Klimov A., Castrucci M. R., Donatelli I., Kawaoka Y. (2000). Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74, 8502–8512 10.1128/JVI.74.18.8502-8512.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A., Saito T., Iverson A. R. (2004). Multiple genotypes of influenza B virus circulated between 1979 and 2003. J Virol 78, 12817–12828 10.1128/JVI.78.23.12817-12828.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroz D., Yoon S. W., Ducatez M. F., Fabrizio T. P., Webby R. J., Hertz T., Ben-Tal N. (2011). Putative amino acid determinants of the emergence of the 2009 influenza A (H1N1) virus in the human population. Proc Natl Acad Sci U S A 108, 13522–13527 10.1073/pnas.1014854108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J. M., Chan R. W., Russell R. J., Air G. M., Peiris J. S. (2008). Evolving complexities of influenza virus and its receptors. Trends Microbiol 16, 149–157 10.1016/j.tim.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Osterhaus A. D., Rimmelzwaan G. F., Martina B. E., Bestebroer T. M., Fouchier R. A. (2000). Influenza B virus in seals. Science 288, 1051–1053 10.1126/science.288.5468.1051 [DOI] [PubMed] [Google Scholar]
- Ozawa M., Basnet S., Burley L. M., Neumann G., Hatta M., Kawaoka Y. (2011). Impact of amino acid mutations in PB2, PB1-F2, and NS1 on the replication and pathogenicity of pandemic (H1N1) 2009 influenza viruses. J Virol 85, 4596–4601 10.1128/JVI.00029-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads J. M., Chen W., Chu P., Berschneider H. M., Argenzio R. A., Paradiso A. M. (1994). L-glutamine and L-asparagine stimulate Na+ -H+ exchange in porcine jejunal enterocytes. Am J Physiol 266, G828–G838 [DOI] [PubMed] [Google Scholar]
- Rimmelzwaan G. F., Boon A. C. M., Geelhoed-Mieras M. M., Voeten J. T. M., Fouchier R. A. M., Osterhaus A. D. M. E. (2004). Human airway epithelial cells present antigen to influenza virus-specific CD8+ CTL inefficiently after incubation with viral protein together with ISCOMATRIX. Vaccine 22, 2769–2775 10.1016/j.vaccine.2004.01.052 [DOI] [PubMed] [Google Scholar]
- Rogers G. N., Daniels R. S., Skehel J. J., Wiley D. C., Wang X. F., Higa H. H., Paulson J. C. (1985). Host-mediated selection of influenza virus receptor variants. Sialic acid-α2,6Gal-specific clones of A/duck/Ukraine/1/63 revert to sialic acid-α2,3Gal-specific wild type in ovo. J Biol Chem 260, 7362–7367 [PubMed] [Google Scholar]
- Roth B., Mohr H., Enders M., Garten W., Gregersen J. P. (2012). Isolation of influenza viruses in MDCK 33016PF cells and clearance of contaminating respiratory viruses. Vaccine 30, 517–522 10.1016/j.vaccine.2011.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusu D., Loret S., Peulen O., Mainil J., Dandrifosse G. (2005). Immunochemical, biomolecular and biochemical characterization of bovine epithelial intestinal primocultures. BMC Cell Biol 6, 42 10.1186/1471-2121-6-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C., Bürger H., Bachmann P. A., Hannoun C. (1983). Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 129, 521–523 10.1016/0042-6822(83)90194-0 [DOI] [PubMed] [Google Scholar]
- Shinde V., Bridges C. B., Uyeki T. M., Shu B., Balish A., Xu X., Lindstrom S., Gubareva L. V., Deyde V. & other authors (2009). Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N Engl J Med 360, 2616–2625 10.1056/NEJMoa0903812 [DOI] [PubMed] [Google Scholar]
- Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. (2006). Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436 10.1038/440435a [DOI] [PubMed] [Google Scholar]
- Sidorenko Y., Reichl U. (2004). Structured model of influenza virus replication in MDCK cells. Biotechnol Bioeng 88, 1–14 10.1002/bit.20096 [DOI] [PubMed] [Google Scholar]
- Smith G. J. D., Vijaykrishna D., Bahl J., Lycett S. J., Worobey M., Pybus O. G., Ma S. K., Cheung C. L., Raghwani J. & other authors (2009). Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- Solórzano A., Webby R. J., Lager K. M., Janke B. H., García-Sastre A., Richt J. A. (2005). Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol 79, 7535–7543 10.1128/JVI.79.12.7535-7543.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Ito T., Suzuki T., Holland R. E., Jr, Chambers T. M., Kiso M., Ishida H., Kawaoka Y. (2000). Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 74, 11825–11831 10.1128/JVI.74.24.11825-11831.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N., Ruttanapumma R., Parchariyanon S., Yoneyama S., Hayashi T., Hiramatsu H., Sriwilaijaroen N., Uchida Y., Kondo S. & other authors (2010). Alterations in receptor-binding properties of swine influenza viruses of the H1 subtype after isolation in embryonated chicken eggs. J Gen Virol 91, 938–948 10.1099/vir.0.016691-0 [DOI] [PubMed] [Google Scholar]
- Trebbien R., Larsen L. E., Viuff B. M. (2011). Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J 8, 434 10.1186/1743-422X-8-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanitchang A., Kramyu J., Jongkaewwattana A. (2010). Enhancement of reverse genetics-derived swine-origin H1N1 influenza virus seed vaccine growth by inclusion of indigenous polymerase PB1 protein. Virus Res 147, 145–148 10.1016/j.virusres.2009.10.010 [DOI] [PubMed] [Google Scholar]
- Webby R. J., Webster R. G. (2003). Are we ready for pandemic influenza? Science 302, 1519–1522 10.1126/science.1090350 [DOI] [PubMed] [Google Scholar]
- Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. (1992). Evolution and ecology of influenza A viruses. Microbiol Rev 56, 152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]