Abstract
Human enterovirus 68 (EV-D68) is a historically rarely reported virus linked with respiratory disease. In the past 3 years, a large increase in respiratory disease associated with EV-D68 has been reported, with documented outbreaks in North America, Europe and Asia. In several outbreaks, genetic differences were identified among the circulating strains, indicating the presence of multiple clades. In this report, we analyse archived and novel EV-D68 strains from Africa and the USA, obtained from patients with respiratory illness. Phylogenetic analysis of all EV-D68 sequences indicates that, over the past two decades, multiple clades of the virus have emerged and spread rapidly worldwide. All clades appear to be currently circulating and contributing to respiratory disease.
Introduction
Enteroviruses (family Picornaviridae, genus Enterovirus) are non-enveloped, positive-sense ssRNA viruses. Enteroviruses can cause a wide range of clinical symptoms, ranging from mild febrile illness to fatal meningitis and encephalitis and are among the most common human pathogens. A typical enterovirus genome consists of an RNA strand of approximately 7500 nt and contains a single ORF encoding a polyprotein that is processed post-translationally to yield individual viral proteins. The polyprotein ORF is flanked by a UTR at each end, with the 5′ UTR containing the highly conserved internal ribosome entry site (IRES) (Racaniello, 2001). The VP1 gene, which encodes one of four capsid proteins, has traditionally been used to distinguish between enterovirus serotypes and, based on molecular and biological characteristics, four human enterovirus (HEV) species are currently recognized, designated HEV-A, -B, -C and -D (Oberste et al., 1999a, b).
Human enterovirus 68 (EV-D68) was first isolated from samples obtained in California in 1962 from four children with pneumonia and bronchiolitis (Schieble et al., 1967). Along with EV-D70, EV-D94 and EV-D-111, EV-D68 is one of four serotypes assigned to HEV-D. Unlike other enteroviruses, EV-D68 is acid-labile and biologically more similar to human rhinoviruses in being mainly associated with respiratory disease; however, until recently, reports of respiratory disease due to EV-D68 were rare (Oberste et al., 2004). Between 1970 and 2005 only 26 clinical isolates of EV-D68 were reported in the USA, representing 0.1 % of all clinical EV isolates (Khetsuriani et al., 2006). Over the past 3 years, however, outbreaks in Japan, the Philippines and the Netherlands, as well as several clusters in the USA, have implicated EV-D68 as an emerging respiratory pathogen (Hasegawa et al., 2011; Imamura et al., 2011; Kaida et al., 2011; Meijer et al., 2012; Rahamat-Langendoen et al., 2011; Tokarz et al., 2011; Ikeda et al., 2012; Jacobson et al., 2012). The clinical presentation of EV-D68 infections in these outbreaks has ranged from mild illness to complications requiring hospitalization and, in rare instances, death. In all reports, children represented the majority of symptomatic infections. In several clusters, novel genetic variants were described (Hasegawa et al., 2011; Imamura et al., 2011; Kaida et al., 2011; Meijer et al., 2012; Rahamat-Langendoen et al., 2011; Ikeda et al., 2012; Jacobson et al., 2012).
In the context of pursuing respiratory virus surveillance during the 2009 H1N1 pandemic, we detected a cluster of EV-D68 cases in New York City (NYC) in the autumn of 2009 (Tokarz et al., 2011). Thereafter we detected EV-D68 in respiratory disease samples collected from several other countries within the last decade. Here, we present the first analysis of EV-D68 from Africa, and compare it to the virus responsible for the NYC cluster, as well as currently circulating strains. Our analysis indicates that several recently emerged distinct clades are circulating globally.
Results
Sample origin
The EV-D68-positive samples originated from both children and adults (Table 1). All South African samples were collected from hospitalized children, while all US samples were originally collected in outpatient clinics. The US sample from Arizona came from a child with fever, cough and rhinitis, while 14 NYC samples came from patients between 14 and 47 years of age that were seen in outpatient clinics throughout the city from individuals presenting with respiratory illness. The initial survey of the samples by MassTag PCR identified EV-D68 as the lone viral pathogen present, with the exception of two South African samples that were co-infected with human respiratory syncytial virus A and one Gambian sample co-infected with human parainfluenza virus 1.
Table 1. Origin of EV-D68-positive samples.
| Location | No. of samples | Collection date | Age |
| The Gambia | 5 | June 2008 | <2 years |
| Senegal | 3 | February–March 2010 | 2, 23 and 32 years |
| South Africa | 8 | May 2000–May 2001 | 5–23 months |
| USA (NYC) | 16 | August–October 2009 | 14–47 years* |
| USA (Arizona) | 1 | October 2009 | 23 months |
Age unknown for two cases.
Prevalence of EV-D68 varied between sample sets. The Gambian and South African EV-D68-positive samples originated from large studies of paediatric respiratory disease and accounted for <1 % of all screened samples from each region. The eight South African cases occurred over a 2 year period between late April and July, which corresponds to the autumn–winter season in the southern hemisphere. All five samples from the Gambia were collected within a 1 month period, indicating a focal outbreak at the beginning of the rainy season. In contrast, the three EV-D68 samples from Senegal accounted for 7 % of the total number of respiratory disease samples collected during a 2 month interval during the dry season. In NYC, the EV-D68-positive samples accounted for approximately 2 % of all respiratory samples analysed over 13 months. However, EV-D68 was the second most frequently detected respiratory virus during a 4 week period in September and October 2009, when detection peaked (Tokarz et al., 2011).
Phylogenetic analysis
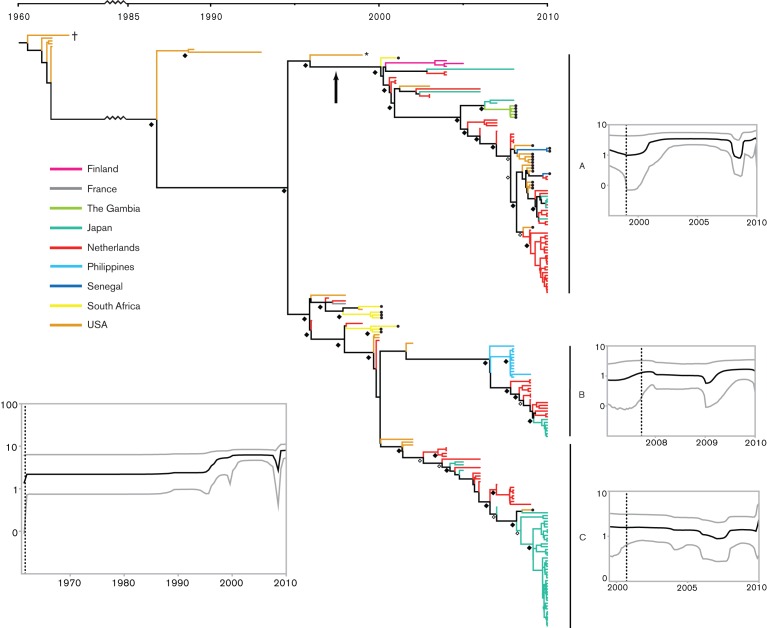
The Bayesian phylogenetic tree inferred using the VP1 gene of all available EV-D68 sequences revealed the presence of three primary clades (A, B and C) with strong nodal support [Bayesian posterior probability (BPP) = 1 in all cases, Fig. 1]. Notably, all three clades were distributed globally, although clade A contained a larger proportion of sequences from all four sampled continents (Asia, Europe, Africa and North America). All newly sequenced strains from NYC (2009), the Gambia (2008) and Senegal (2010), along with one South African sequence (2001), were found within clade A, while the remaining South African samples (2000–2001) formed a cluster with sequences from Europe and the USA ancestral to clades B and C. The sequence from Arizona (2009) belonged to clade C. We did not detect any EV-D68 that clustered within clade B in our sample set. Based on the data currently available, the USA appears to have been a source for much of the EV-D68 diversity that we have sampled. Sequences from the USA are present at the root of the tree, as well as positioned at the base of all major lineages. However, the EV-D68 phylogeny is characterized by the frequent presence of long branches throughout the tree, indicative of a general lack of global surveillance and detection of EV-D68, as well as the presence of in situ evolution in individual locations. It is therefore possible that retrospective or broader sampling may reveal additional (and deeper) diversity.
Fig. 1.
Evolutionary history of EV-D68 based on complete VP1 sequences. The maximum clade credibility tree is shown with BPP values of 0.7–0.8 and 0.9–1.0 indicated by ⧫ and ◊ at the nodes, respectively. Clades A, B and C are indicated and supported by BPP = 1.0. The Bayesian skyline plots (BSPs) are shown for the overall dataset (bottom left) and for clades A, B and C. The black line indicates the median relative genetic diversity (log10 scale) over time and the grey curves represent the upper and lower 95 % HPD intervals. • at the tips of the branches indicates novel sequences generated in this study. The vertical arrow indicates the branch along which the VP1 asparagine deletion occurred; † indicates the Fermon strain (1962), and * indicates the initial clade A strain from Maryland, USA (1999).
The high correlation between root-to-tip genetic distance and sampling time (R2 = 0.84; Fig. S1, available in JGV Online) indicates that EV-D68 evolution has been characterized by rapid, clock-like dynamics over the timescale of sampling. The combination of a relaxed molecular clock and the SRD06 model of nucleotide substitution resulted in the best fit of the model to the data (data not shown). Despite the strong clock-like dynamics suggested by our regression analysis, the values for the coefficient of variation of the relaxed molecular clock were always >0, indicating a significant deviation from strict clock-like evolution. The mean rate of nucleotide substitution for the VP1 gene for EV-D68 was 6.2×10−3 substitutions per site year−1 [95 % highest probability density (HPD) value, 5.4–7.1×10−3 substitutions per site year−1] with a corresponding time to most recent common ancestor (TMRCA) for the entire EV-D68 sample set of 1961 (1960–1962). This value corresponds closely to the TMRCA estimated under a strict clock, as well as by the regression analysis (1960).
The diversity within each of the three primary clades appears to have arisen only recently, with mean TMRCAs of 1997 and 1999 for clades A and C, respectively, and of 2007 for clade B (Table 2). However, the long branch leading to clade B indicates that the ancestral diversity of this group has probably not been sampled, and the age of this group may correspond more closely to that of the other clades. Analysing the population dynamics of EV-D68 revealed an overall trend towards increasing genetic diversity in the present, as well as within each of the three main clades (Fig. 1). However, based on the 95 % HPDs, no significant departure from a constant level of genetic diversity could be detected. Again, the long branches observed in the tree between the initial characterization of EV-D68 in 1962 and the recent burst of sampling that began around the year 2000 suggest strongly that a significant fraction of the evolutionary history of EV-D68 has not been sampled, making conclusions about the origin and spread of this virus difficult.
Table 2. Estimated mean TMRCA (years) and nucleotide substitution rates (nucleotide substitutions per site year−1) for EV-D68 based on all available complete VP1 gene sequences, as well as for clades A, B and C.
| Mean TMRCA (95 % HPD) | Mean substitution rate (95 % HPD) | |
| Complete dataset: relaxed clock | 1961 (1960–1962) | 6.2×10−3 (5.4–7.1×10−3) |
| Complete dataset: strict clock | 1960 (1959–1961) | 5.8×10−3 (5.2–6.4×10−3) |
| Clade A | 1997 (1995–1999) | 7.1×10−3 (5.9–8.3×10−3) |
| Clade B | 2007 (2006–2008) | 6.8×10−3 (5.6–8.1×10−3) |
| Clade C | 1999 (1998–2001) | 6.2×10−3 (5.1–7.2×10−3) |
Full genome comparison
In addition to the Fermon strain, there are only three other full-length EV-D68 sequences currently available, all from clade C. These consist of strain 37-99 isolated from France in 1999, and strains JPOC10-290 and JPOC10-378 from a 2009 Japanese outbreak. We sequenced the full genome of one of the clade A strains from NYC and compared it with the published genomes (Table 3). The overall length of the NYC strain was 7340 nt. The nucleotide identity compared with clade C strains ranged from 91.4 to 93.8 %, and was 88.5 % compared with the Fermon strain. The predicted polyprotein identity was 97.4–98 % compared with clade C and 95.3 % compared with the Fermon strain.
Table 3. Percentage nucleotide identity between clade A representative and EV-D68 strain Fermon as well as clade C strains 37-99, JPOC10-378 and JPOC10-290.
| Gene | Clade A NYC403 | Fermon | Clade C | ||
| 37-99 | JPOC10-378 | JPOC10-290 | |||
| VP4 | 100 | 93.2 | 95.7 | 94.2 | 93.7 |
| VP2 | 100 | 86.4 | 93.3 | 90.7 | 90.7 |
| VP3 | 100 | 85.8 | 93.5 | 89.9 | 90.1 |
| VP1 | 100 | 87.6 | 92.5 | 90.7 | 90.3 |
| 2A | 100 | 87.3 | 93.0 | 90.2 | 90.9 |
| 2B | 100 | 86.2 | 92.9 | 90.6 | 90.2 |
| 2C | 100 | 89.4 | 95.3 | 93.5 | 93.4 |
| 3A | 100 | 90.3 | 93.3 | 90.6 | 90.3 |
| 3B | 100 | 93.9 | 90.9 | 92.4 | 90.9 |
| 3C | 100 | 88.4 | 94.7 | 90.5 | 90.6 |
| 3D | 100 | 88.9 | 93.8 | 91.8 | 92.2 |
| ORF | 100 | 88.1 | 93.6 | 91.5 | 91.5 |
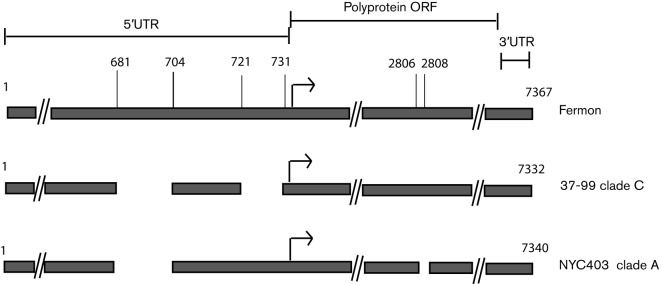
Among the major differences observed in the structure of the genome, all but one of the clade A sequences contained a 3 nt deletion in the VP1 gene at positions 2806–2808, resulting in a missing asparagine residue that is present in all sequences of clades B and C (Fig. 2). The lone clade A sequence without the deletion was a 1999 isolate from Maryland, USA, which is the earliest known representative of the clade (Fig. 1). In addition, we observed variation in the 5′ UTR spacer region between the end of the IRES and the beginning of the polyprotein ORF (Fig. 2). The UTR region between positions 162 and 623 was highly conserved and was >95 % identical across all EV-D68 strains. Within nt 624–732, both clades A and C had a 24 nt deletion at positions 681–704 relative to the Fermon strain, and clade C had an additional 11 nt deletion at positions 721–731 that was absent in clade A. 5′ UTR sequences from representatives of clade B were not available for analysis. To determine whether the pattern of deletions was conserved throughout clades A and C, we amplified and sequenced a fragment containing the 5′ UTR spacer region in all our remaining samples. The UTR pattern was observed in all clade A and C sequences, irrespective of year of collection and geographical area.
Fig. 2.
Major variations in the genomes of clades A and C in comparison with the EV-D68 Fermon strain. Dashed lines (//) indicate extended genomic regions without variation. Arrows represent the start of the polyprotein ORF.
Discussion
Reports identifying EV-D68 as the aetiological agent of respiratory disease have increased steadily over the last decade, most notably over the last 3 years. EV-D68 was identified in San Diego, CA, USA, in seven military recruits in 2004–2005 and in respiratory disease surveillance screens in France in 2008 and Italy in 2008–2009 (Petitjean-Lecherbonnier et al., 2011; Piralla et al., 2011; Wang et al., 2010). In the Philippines in late 2008 and early 2009, EV-D68 was detected in 21 patients, two of whom died (Imamura et al., 2011). In Japan, the virus has been detected each year since 2003, culminating in a report describing 14 cases in the autumn of 2010 (Kaida et al., 2011). In the USA, we documented the EV-D68 cluster in NYC in 2009, and the Centers for Disease Control and Prevention (CDC) reported EV-D68 clusters in several states in 2010 (CDC, 2011; Tokarz et al., 2011). In the Netherlands, 24 EV-D68 cases were recorded between August and November of 2010 (Rahamat-Langendoen et al., 2011). A more recent retrospective study of EV-D68 from the Netherlands and Finland described the presence of three distinct clades within the past 10 years (Meijer et al., 2012). Another recent report documented the circulation of these clades within the past decade in Japan (Ikeda et al., 2012). Our analysis of EV-D68 sequences from these reports, in addition to new sequences reported here, indicates that there are at least three major clades of EV-D68 currently in circulation worldwide, consistent with the data in these recent reports.
Between the early 1960s and mid-1990s, the EV-D68 genome underwent a rearrangement in the spacer region of the 5′ UTR between the end of the IRES and the polyprotein ORF. This resulted in a 24 nt deletion in all EV-D68 strains that we examined. The virus then underwent a large diversification in the mid-1990s that culminated in a division into clades A and C. Not long after this division, the genomes of clade C-type viruses underwent further rearrangements, resulting in an additional 11 nt deletion in the spacer region. This deletion, first reported by Kaida et al. (2011), may have a significant effect on the initiation of translation. Although it is established that variations within the IRES can have major effects on virulence (Gromeier et al., 1996; Li et al., 2011), little is known about the spacer region and its role in viral fitness. The length of the EV 5′ UTR spacer region is approximately 150 nt, and deletions such as those that have occurred in EV-D68 are not common. In rhinoviruses, the spacer is >50 nt and it may be noteworthy that EV-D68, which shares respiratory tropism with rhinoviruses, appears to be undergoing a reduction in the UTR length. Additionally, these deletions may affect the virulence of the virus by enhancing translational efficiency and may be correlated with the recent increase in EV-D68 cases worldwide.
Clade A lacks the secondary deletion observed in the spacer region of clade C viruses, but has a 3 nt deletion in the VP1 gene. According to our calculations, the deletion arose early after the split from clades C and B, sometime between 1997 and 2000. Meijer et al. (2012) showed that this deletion, as well as additional VP1 amino acid substitutions in clades A–C relative to the Fermon strain, were in the exposed portions of the VP1. Such changes were proposed to have a role in modulating the immune response to the virus and thus may aid in viral persistence. Following the divergence of the three clades, clades A and C appear to have expanded rapidly as both clades were present in North America, Europe and Africa by 2001. Clade B appears to have diverged from clade C around 2007, although the lack of sampling prior to the early 2000s makes this assumption difficult to corroborate.
It has been suggested that EV-D68 may have a shifted seasonality in comparison to other enteroviruses, which in temperate climates traditionally circulate in the summer and early autumn (Khetsuriani et al., 2006). The recently reported EV-D68 outbreaks appeared later than this time frame, with most cases reported throughout the autumn and into winter. EV-D68-positive samples in this report originating from the USA and South Africa were obtained between late summer and winter. Outside of an outbreak in the Philippines, there is little information on the seasonality of EV-D68 in tropical regions. We analysed EV-D68-positive samples from Senegal and the Gambia, where the climate consists of primarily dry and rainy seasons. Within the small sample set that we analysed, the circulation pattern was unclear, as EV-D68 cases in the Gambia occurred during the rainy season, while the cases from Senegal occurred during the dry season. A much larger sample set would be necessary to assess the seasonal circulation pattern of EV-D68 in non-temperate regions.
In previously documented clusters, approximately 80 % of reported EV-D68 cases occurred in children. One explanation for this finding is that most reports highlighted only paediatric cases. Our EV-D68 sequences from the Gambia, South Africa and Arizona were all obtained from children. However, a recent report from the Netherlands, which focused on screening for EV-D68 in archived respiratory samples without age bias, noted a preponderance of adult cases (Meijer et al., 2012). Additionally, NYC cases presented in our report were from individuals aged 14 years or older, which is the largest EV-D68 cluster of non-paediatric origin to date. We anticipate that, with increased screening, EV-D68 may ultimately be recognized as a respiratory pathogen in all age groups.
Although there appears to be a global upsurge of EV-D68-associated respiratory disease, we cannot ignore the possibility that this increase merely reflects improved case ascertainment. In previous studies of respiratory disease, EV-D68 may have been misidentified as a rhinovirus, which would lead to underestimation of its prevalence and importance (CDC, 2011). Continued efforts towards the improvement of diagnostic techniques and surveillance are necessary to further assess the epidemiology of EV-D68 and to assess its role in respiratory disease.
Methods
Sample collection and sequencing.
A total of 33 EV-D68-positive nasal-pharyngeal swab samples were identified from five different respiratory disease sample sets (Table 1). Samples were initially analysed by MassTag PCR for respiratory pathogens (Briese et al., 2005; Lamson et al., 2006) and EV-positive samples were typed based on amplification of the VP4/VP2 gene region (Coiras et al., 2004; Lamson et al., 2006). Sixteen EV-D68-positive samples from NYC were identified from August to October 2009. One sample collected in October 2009 came from Arizona in south-west USA. Sixteen samples originated from Africa: eight samples from South Africa were obtained between May 2000 and May 2001: two were collected in May, four in July of 2000 and one each in April and May of 2001. Five samples from the Gambia were collected in June 2008 and three EV-D68-positive samples were obtained from an outbreak of respiratory disease in Senegal in February and March 2010.
To acquire the sequence of the full-length VP1 gene from these samples, two sets of primer pairs were used. The first (5′fwd1, CCTTAATAGGGTTCATAGCAGC and 5′rev1, CTGGGCCGGTGGTYACTA) generated a 1009 bp fragment, while the second pair (5′fwd2, ATGAGAGAYAGYCCTGACATTG and 5′rev2, CATTGAGBGCATTTGGTGCT) amplified a 900 bp fragment. In instances where the first primer failed to generate the desired PCR product, the second primer pair was used. An additional primer pair (5′fwd3, GGTCAAGCACTTCTGTTTCCC and 5′rev3, TGGCAATGTTGGCATTYTC) was used to amplify a 606 bp fragment spanning most of the 5′ UTR and a portion of VP4. The full genome sequence of a single NYC representative of clade A was generated by overlapping consensus PCR using existing EV-D68 genomes as reference. All sequences were deposited in GenBank under accession numbers JX101786–JX101846. All base pair coordinates are provided relative to the original 1962 EV-D68 isolate, the Fermon strain (GenBank accession no. AY426531).
Phylogenetic analysis.
A nucleotide alignment incorporating all EV-D68 sequences available in GenBank was constructed manually (928 nt, n = 210) using Se-Al (v2.0a11 Carbon, http://tree.bio.ed.ac.uk/software/seal). To assess the molecular clock-like behaviour of the data, a maximum-likelihood (ML) phylogenetic tree was first constructed using PhyML (v3.0, Guindon & Gascuel, 2003) incorporating the Hasegawa, Kishino and Yano 1985 (HKY85) model of nucleotide substitution with an among-site rate heterogeneity parameter (gamma, G) and a heuristic SPR (subtree pruning and regrafting) branch-swapping search. The root-to-tip genetic distance inferred from the ML tree was then regressed against the time of sampling (in years) using the program Path-O-Gen (v1.2, http://tree.bio.ed.ac.uk/software/pathogen/). In this analysis, the correlation coefficient provides an estimate of the amount of variation in genetic distance that is explained by sampling time and the x-intercept serves as an estimator of the time to most recent common ancestor (TMRCA).
The evolutionary relationships of the viral sequences, along with the rate of nucleotide substitution per site year−1 and the TMRCA, were inferred using a Bayesian Markov chain Monte Carlo (MCMC) method implemented in the beast package (v1.7, Drummond & Rambaut, 2007). Both strict and relaxed (uncorrelated lognormal) molecular clocks were employed with a flexible Bayesian skyline plot (BSP) coalescent prior (10, 15 and 20 piece-wise constant groups) and both the HKY85+G and the SRD06 models of nucleotide substitution. The MCMC chains were run for 200 million iterations, with subsampling every 20 000 iterations. A 10 % burnin was removed and maximum clade credibility trees were summarized using TreeAnnotator (v.1.5.4), with Bayesian posterior probability (BPP) values providing a measure of statistical support at each node. Statistical confidence in the parameter estimates was represented by values for the 95 % highest probability density (HPD) intervals around the marginal posterior parameter means. The output of the BSP model was also used to infer the population dynamics of EV-D68 through time, both for the complete dataset and for each of three monophyletic clades we identified with BPP = 1.0.
Acknowledgements
We thank Niranjan Bhat and Eric Jaffe for their assistance. This work was supported by National Institutes of Health grant AI57158 (Northeast Biodefense Center-Lipkin), and US Department of Defense grant W911SR-11-C-0077.
Footnotes
A supplementary figure is available with the online version of this paper.
References
- Briese T., Palacios G., Kokoris M., Jabado O., Liu Z., Renwick N., Kapoor V., Casas I., Pozo F. & other authors (2005). Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis 11, 310–313 10.3201/eid1102.040492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2011). Clusters of acute respiratory illness associated with human enterovirus 68 – Asia, Europe, and United States, 2008–2010. MMWR Morb Mortal Wkly Rep 60, 1301–1304 [PubMed] [Google Scholar]
- Coiras M. T., Aguilar J. C., García M. L., Casas I., Pérez-Breña P. (2004). Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol 72, 484–495 10.1002/jmv.20008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromeier M., Alexander L., Wimmer E. (1996). Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci U S A 93, 2370–2375 10.1073/pnas.93.6.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704 [DOI] [PubMed] [Google Scholar]
- Hasegawa S., Hirano R., Okamoto-Nakagawa R., Ichiyama T., Shirabe K. (2011). Enterovirus 68 infection in children with asthma attacks: virus-induced asthma in Japanese children. Allergy 66, 1618–1620 10.1111/j.1398-9995.2011.02725.x [DOI] [PubMed] [Google Scholar]
- Ikeda T., Mizuta K., Abiko C., Aoki Y., Itagaki T., Katsushima F., Katsushima Y., Matsuzaki Y., Fuji N. & other authors (2012). Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol 56, 139–143 10.1111/j.1348-0421.2012.00411.x [DOI] [PubMed] [Google Scholar]
- Imamura T., Fuji N., Suzuki A., Tamaki R., Saito M., Aniceto R., Galang H., Sombrero L., Lupisan S., Oshitani H. (2011). Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis 17, 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson L. M., Redd J. T., Schneider E., Lu X., Chern S. W., Oberste M. S., Erdman D. D., Fischer G. E., Armstrong G. L. & other authors (2012). Outbreak of lower respiratory tract illness associated with human enterovirus 68 among American Indian children. Pediatr Infect Dis J 31, 309–312 10.1097/INF.0b013e3182443eaf [DOI] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Sekiguchi J., Kohdera U., Togawa M., Shiomi M., Nishigaki T., Iritani N. (2011). Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg Infect Dis 17, 1494–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetsuriani N., Lamonte-Fowlkes A., Oberst S., Pallansch M. A., Centers for Disease Control and Prevention (2006). Enterovirus surveillance – United States, 1970–2005. MMWR Surveill Summ 55, 1–20 [PubMed] [Google Scholar]
- Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J., Dean A., St George K., Briese T., Lipkin W. I. (2006). MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis 194, 1398–1402 10.1086/508551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zou Q., Chen L., Zhang H., Wang Y. (2011). Molecular analysis of virulent determinants of enterovirus 71. PLoS ONE 6, e26237 10.1371/journal.pone.0026237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A., van der Sanden S., Snijders B. E., Jaramillo-Gutierrez G., Bont L., van der Ent C. K., Overduin P., Jenny S. L., Jusic E. & other authors (2012). Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology 423, 49–57 10.1016/j.virol.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R., Flemister M. R., Brown B. A., Pallansch M. A. (1999a). Typing of human enteroviruses by partial sequencing of VP1. J Clin Microbiol 37, 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R., Pallansch M. A. (1999b). Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73, 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Schnurr D., Flemister M. R., Lovchik J. C., Peters H., Sessions W., Kirk C., Chatterjee N. & other authors (2004). Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol 85, 2577–2584 10.1099/vir.0.79925-0 [DOI] [PubMed] [Google Scholar]
- Petitjean-Lecherbonnier J., Dina J., Nguyen E., Gouarin S., Lebigot E., Vabret A. (2011). [Molecular diagnosis of respiratory enterovirus infections: use of PCR and molecular identification for a best approach of the main circulating strains during 2008]. Pathol Biol (Paris) 59, 113–121 (in French). 10.1016/j.patbio.2010.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piralla A., Baldanti F., Gerna G. (2011). Phylogenetic patterns of human respiratory picornavirus species, including the newly identified group C rhinoviruses, during a 1-year surveillance of a hospitalized patient population in Italy. J Clin Microbiol 49, 373–376 10.1128/JCM.01814-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello R. V. (2001). Picornaviridae: the viruses and their replication. In Fields Virology, 4th edn, vol. 1, pp. 685–722 Edited by Fields B. N., Knipe D. M., Howley. P. M. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Rahamat-Langendoen J., Riezebos-Brilman A., Borger R., van der Heide R., Brandenburg A., Schölvinck E., Niesters H. G. (2011). Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol 52, 103–106 10.1016/j.jcv.2011.06.019 [DOI] [PubMed] [Google Scholar]
- Schieble J. H., Fox V. L., Lennette E. H. (1967). A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85, 297–310 [DOI] [PubMed] [Google Scholar]
- Tokarz R., Kapoor V., Wu W., Lurio J., Jain K., Mostashari F., Briese T., Lipkin W. I. (2011). Longitudinal molecular microbial analysis of influenza-like illness in New York City, May 2009 through May 2010. Virol J 8, 288 10.1186/1743-422X-8-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Malanoski A. P., Lin B., Long N. C., Leski T. A., Blaney K. M., Hansen C. J., Brown J., Broderick M. & other authors (2010). Broad spectrum respiratory pathogen analysis of throat swabs from military recruits reveals interference between rhinoviruses and adenoviruses. Microb Ecol 59, 623–634 10.1007/s00248-010-9636-3 [DOI] [PubMed] [Google Scholar]