Abstract
The filamentous cyanobacterial genus Moorea gen. nov., described here under the provisions of the International Code of Botanical Nomenclature, is a cosmopolitan pan-tropical group abundant in the marine benthos. Members of the genus Moorea are photosynthetic (containing phycocyanin, phycoerythrin, allophycocyanin and chlorophyll a), but non-diazotrophic (lack heterocysts and nitrogenase reductase genes). The cells (discoid and 25–80 µm wide) are arranged in long filaments (<10 cm in length) and often form extensive mats or blooms in shallow water. The cells are surrounded by thick polysaccharide sheaths covered by a rich diversity of heterotrophic micro-organisms. A distinctive character of this genus is its extraordinarily rich production of bioactive secondary metabolites. This is matched by genomes rich in polyketide synthase and non-ribosomal peptide synthetase biosynthetic genes which are dedicated to secondary metabolism. The encoded natural products are sometimes responsible for harmful algae blooms and, due to morphological resemblance to the genus Lyngbya, this group has often been incorrectly cited in the literature. We here describe two species of the genus Moorea: Moorea producens sp. nov. (type species of the genus) with 3LT as the nomenclature type, and Moorea bouillonii comb. nov. with PNG5-198R as the nomenclature type.
Benthic filamentous marine cyanobacteria from the tropics have been of increasing biomedical interest due to their extraordinary richness in bioactive secondary metabolites (Tidgewell et al., 2010). Many of these natural product molecules are potent toxins responsible for harmful algal blooms and thus are hazardous to humans as well as near-shore environments. At the same time, some of these cyanobacterial toxins and other natural products have properties of potential benefit to human health as pharmaceutical leads (Golubic et al., 2010). Surprisingly, the majority of these unique natural products have been ascribed as being produced by members of a single genus, Lyngbya, and a preponderance of these come from a single species, Lyngbya majuscula (Liu & Rein, 2010). However, an unfortunate consequence of using traditional morphology-based taxonomic systems in these identifications has been that cyanobacteria of many recently explored biological frontiers (e.g. tropical marine environments) have been forced into existing morphological groupings and, thus, the true biodiversity of this group has been greatly underestimated (Casamatta et al., 2005; Engene et al., 2011).
The proposed cyanobacterial genus Moorea gen. nov. is a cosmopolitan, pan-tropical group abundant in the marine benthos. Strains of Moorea gen. nov. have often been incorrectly classified as the cyanobacterial genus Lyngbya due to morphological similarities between the two groups (Engene et al., 2011). This misidentification of Moorea as Lyngbya has been a source of considerable taxonomic confusion as well as the major reason for the perceived chemical richness of the genus Lyngbya (Engene et al., 2011). Herein, we firmly differentiate between these two phylogenetically distinct groups and describe Moorea as a novel generic entity (gen. nov.). This description and naming of Moorea gen. nov. was performed under the provisions of the International Code of Botanical Nomenclature.
A total of 51 geographically distributed populations of the genus Moorea were included in this taxonomic revision (geographical data for Moorea specimens are available in Table S1 in IJSEM Online). Field collections of cyanobacteria were carefully rinsed with autoclaved SWBG-11 medium (Castenholz, 1988) and visible macro-organisms were mechanically removed with sterile tweezers under an Olympus VMZ dissecting microscope. Clonal, non-axenic strains were derived from phototactically isolated single filaments on 0.5 % agar plates with SWBG-11 and cultured in SWBG-11 medium at 28 °C with 33 g Instant Ocean salt l−1 (Aquarium Systems). The cultures were kept at a light intensity of 7 µmol photons s−1 m−2 (16 h light/8 h dark). Two isolated strains, Moorea producens 3LT and Moorea bouillonii PNG5-198R, were deposited in the Canadian Phycological Culture Centre (CPCC) and the national marine phytoplankton collection (CCMP) as reference strains. Additionally, the Lyngbya reference strain PCC 7419R was obtained from the Pasteur Culture Collection (PCC) for biological comparison. Light microscopy was performed using an Olympus IX51 epifluorescent microscope (1000×) equipped with an Olympus U-CMAD3 camera. Samples for scanning electron microscopy were placed on indium-tin-oxide glass slides that had been coated with 0.1 % polyethylenimine to facilitate adhesion. The samples were then fixed in 2.5 % glutaraldehyde buffered in 1× PBS for 30 min and a secondary fix of 2 % aqueous osmium tetroxide for 15 min. Dehydration was achieved with a graded ethanol series. The samples were then critical-point-dried and sputter-coated with gold palladium. A Hitachi SU6600 scanning electron microscope was used to view the samples. Samples for transmission electron microscopy were prepared using high pressure freezing and subsequent freeze substitution. The filaments were cut into pieces of <0.5 mm and placed into specimen holders with a drop of cryoprotectant hexadecane. The samples were frozen using a Bal-Tec HPM 010 high pressure freezing machine. Freeze substitution was done using a Leica EM AFS machine. Samples were transferred to pre-cooled 1 % glutaraldehyde with 0.2 % tannic acid in anhydrous acetone and left at −90 °C for 36 h, then washed with acetone three times for 15 min each and subsequently transferred to 1 % osmium tetroxide with 0.1 % uranyl acetate in acetone and held for 24 h. The temperature was raised to −60 °C, –30 °C and 0 °C, being held for 24 h between each step. At 0 °C, the samples were washed with acetone three times for 15 min each and transferred into 50 % Durcopan in acetone and held for 12 h. Once samples warmed to room temperature, they were embedded in Durcopan and left to polymerize for 48 h. Thin sections (70 nm) were obtained using a Reichart Ultracut E and then placed on Formvar-coated 75 and 200 mesh copper grids. The grids were subsequently stained with uranyl acetate and Sato lead. A JEOL 1200FX transmission electron microscope was used to view the samples. Photosynthetic pigments were characterized as described previously (Tandeau de Marsac & Houmard, 1988).
Genomic DNA was extracted using the Wizard Genomic DNA Purification kit (Promega) following the manufacturer’s specifications. DNA concentration and purity were measured on a DU 800 spectrophotometer (Beckman Coulter). The PCR volumes were 25 µl, containing 0.5 µl (~50 ng) of DNA, 2.5 µl of 10× PfuUltra IV reaction buffer, 0.5 µl (25 mM) dNTP mix, 0.5 µl of each primer (10 µM), 0.5 µl PfuUltra IV fusion HS DNA polymerase and 20.5 µl distilled H2O. PCRs were performed in an Eppendorf Mastercycler gradient as follows: initial denaturation for 2 min at 95 °C; 25 amplification cycles of 20 s at 95 °C, 20 s at 50 °C and 1.5 min at 72 °C; and final elongation for 3 min at 72 °C. PCR products were purified using a MinElute PCR Purification kit (Qiagen) before subcloning into the Zero Blunt TOPO PCR Cloning kit (Invitrogen). Plasmid DNA was isolated using the QIAprep Spin Miniprep kit (Qiagen) and sequenced bidirectionally with M13 vector-primers as well as internal primers. The gene sequences are available in DDBJ/EMBL/GenBank (see Table S1, available in IJSEM Online). The 16S (SSU) rRNA genes of all 51 Moorea specimens were included in the analysis. Representative reference strains were selected from Bergey’s Manual (Castenholz, 2001). The unicellular Gloeobacter violaceus PCC 7421T (GenBank accession no. NC_005125) was included as an evolutionarily distant outgroup. All gene sequences were aligned using the L-INS-I algorithm in MAFFT 6.717 (Katoh & Toh, 2008). The alignment was visually compared and refined using the SSU secondary structures model of Escherichia coli J01695 (Cannone et al., 2002) without data exclusion. The multiple sequence alignments are available in the TreeBASE database (http://www.treebase.org) under the submission ID 11599. Pair-wise sequence divergences were calculated in paup* 4.0b10. Appropriate nucleotide substitution models were compared and selected using uncorrected/corrected Akaike Information Criterion (AIC/AICc), Bayesian Information Criterion (BIC) and the Decision-theoretic (DT) in jModeltest 0.1.1 (Posada, 2008). The maximum-likelihood inference was performed using GARLI 1.0 (Zwickl, 2006). The analysis was run using the GTR+I+G model assuming a heterogeneous substitution rate and gamma substitution of variable sites [proportion of invariable sites (pINV) = 0.450, shape parameter (α) = 0.449, number of rate categories = 4]. Bootstrap resampling was performed on 1000 replicates. Bayesian analysis was conducted using MrBayes 3.1 (Ronquist & Huelsenbeck, 2003). Four Metropolis-coupled MCMC chains (one cold and three heated) were run for 10 000 000 generations. MCMC convergence was determined using AWTY; the first 1 000 000 generations (10 %) were discarded as burn-in and the following datasets were sampled with a frequency of every 1000 generations. The maximum-parsimony analysis was performed in paup* 4.0b10 using a heuristic search through the branch-swapping tree-bisection-reconnection algorithm with the addition of 10 000 random replicates to find the most parsimonious tree. Bootstrap support was obtained from 1000 replicates.
Draft genomes from M. producens strain 3LT (GenBank accession no. AEPQ01000000) and M. bouillonii strain PNG5-198R have been obtained recently and were used for phylogenomic and functional genomics comparison. Phylogenomic inference was performed bioinformatically on the basis of the DNA-G, FRR, rpsB, NusA, PGK, PyrG, rpoB, rpsC, rpl2, rpl3, rpl4 and TSF genes. These gene sequences were downloaded from all 59 publicly available cyanobacterial genomes and concatenated for phylogenetic inference with the two Moorea genomes. Maximum-likelihood (RaxML) inference was performed on the WAG+I+G model assuming heterogeneous substitution rates and gamma substitution of variable sites [proportion of invariable sites (pINV) = 0.265, shape parameter (α) = 0868, number of rate categories = 4] with a bootstrap resampling of 500 replicates.
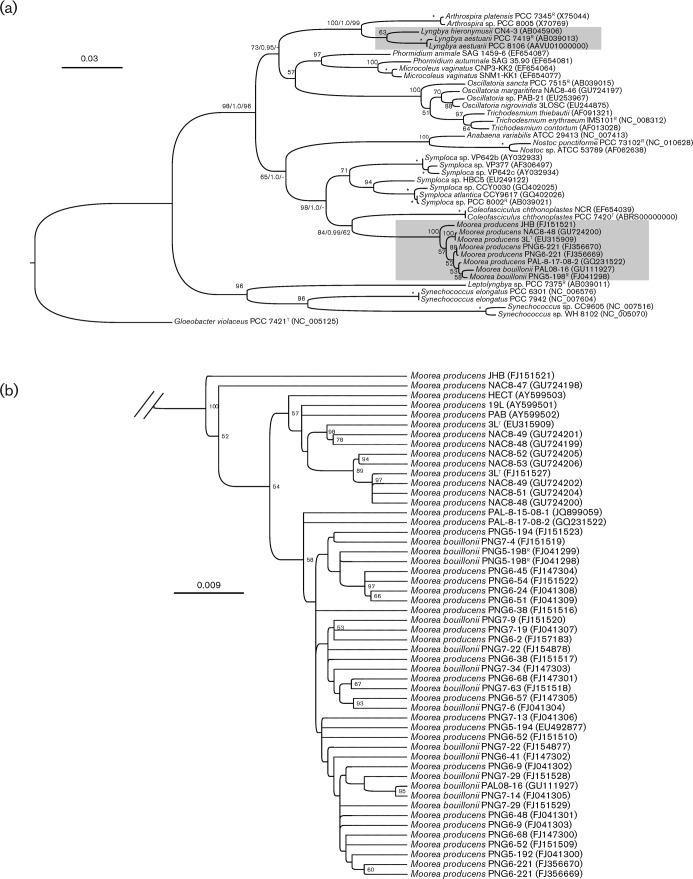
Phylogenetic inference based on the 16S rRNA gene revealed that the Moorea lineage was evolutionarily distinct and distant from the Lyngbya sensu stricto (reference strain = PCC 7419T; p-distance = 9.2 %) (Fig. 1a). The Moorea lineage was nested between members of the closest related genera Symploca (reference strain = PCC 8002R; p-distance = 6.1 %) and Coleofasciculus (reference strain = PCC 7420T; p-distance = 6.9 %). The phylogenetic positions of Moorea and evolutionary distances from Lyngbya were corroborated by analysis of the RNA polymerase gamma subunit (rpoC1) gene (an evolutionary tree of the rpoC1 gene is available as Fig. S1 in IJSEM Online). An additional 12 other protein-coding genes (DNA-G, FRR, rpsB, NusA, PGK, PyrG, rpoB, rpsC, rpl2, rpl3, rpl4 and TSF) were selected from the Moorea genome drafts and the evolutionary histories of these genes were individually constructed and compared with all available sequenced genomes of cyanobacteria. As a result, each protein-coding gene showed an evolutionary history that was relatively congruent with that of the 16S rRNA gene phylogram. All 12 phylogenetically informative genes were concatenated for a more robust phylogenomic inference. The combined genes supported the phylogenetic distance between Lyngbya (i.e. PCC 8106) and Moorea (i.e. 3LT and PNG5-198R) as well as the overall evolutionary history of the phylum (a phylogenomic inference is available as Fig. S2 in IJSEM Online).
Fig. 1.
(a) Phylogenetic inferences (GARLI) of Lyngbya and Moorea diversification based on SSU (16S) rRNA nucleotide sequences (these two genera are highlighted with shaded boxes). Species and strain numbers of the specimens are given with accession numbers in parentheses. Reference strain (R) and type strain (T) numbers were obtained from Bergey’s Manual (Castenholz, 2001). Support values are indicated as bootstrap and posterior probability for the maximum-likelihood/Bayesian inference/maximum-parsimony methods (* = bootstrap of 100% and a posterior probability of 1.0). Bar, 0.03 estimated nucleotide substitutions per site using the GTR+I+G substitution model. (b) A close-up of the Moorea lineage. Bar, 0.009 expected substitutions per site.
On a subgeneric level, the Moorea specimens formed a tight clade with low interior sequence divergence (p-distance: mean = 0.5 %; max. = 1.4 %) (Fig. 1b). This high DNA bar-coding gap of the Moorea clade of more than 12 times further supports the exclusivity of this clade and the need to distinguish it from neighbouring genera (Fig. 1a). However, the genomes of Moorea specimens contain multiple and variable copies of their 16S rRNA genes (Engene et al., 2010) and this relatively high level of intra-genomic gene heterogeneity in combination with the low subgeneric sequence divergence makes the 16S rRNA gene inadequate for speciation. The lack of phylogenetic resolution for species delineation was further indicated by low statistical node support at the terminal nodes and incongruence using different phylogenetic methods (Fig. 1b). The less conserved internal transcribed spacer (ITS) region linking the 16S and 23S ribosomal genes has been proposed to be taxonomically more informative on a subgeneric level and has often been used for species delineation in cyanobacteria (Otsuka et al., 1999; Boyer et al., 2001; Gugger et al., 2005). Primer-sites on the adjacent 16S and 23S rRNA genes were used to PCR-amplify the 16S–23S ITS regions of 41 Moorea specimens. However, the 16S–23S ITS regions were, in accordance with the 16S rRNA genes, present in multiple and variable gene copies and, thus, this gene region was also not able to definitively distinguish between Moorea specimens (Fig. S3). In addition, the intra-genomic sequence heterogeneity of the 16S–23S ITS region was found to influence structurally informative domains, such as the D1-D1′ helix and the Box-B, secondary structures which are frequently used for taxonomic delineation (Boyer et al., 2001). In the case of Moorea, we argue that the 16S–23S ITS regions are not able to further resolve species delineation.
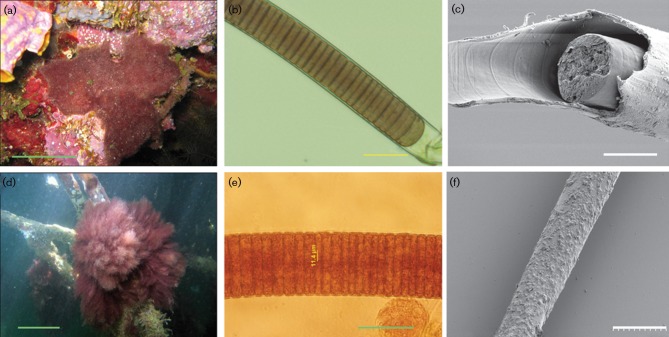
Morphologically, the Moorea specimens were composed of long isopolar filaments enclosed in thick exopolysaccharide sheaths with discoid cells arranged in trichomes (Fig. 2). The exteriors of the sheaths were consistently covered by a rich fauna of heterotrophic bacteria and other micro-organisms (Fig. 2). The two Moorea species M. producens and M. bouillonii had distinctively different colony morphologies. Environmental specimens of M. bouillonii always formed characteristic reddish cobweb-like mats firmly attached to surrounding substrate and each colony was also always found with an associated snapping shrimp (Alpheus frontalis) (Fig. 2).
Fig. 2.
Morphological characterization of Moorea gen. nov. (a–c) M. bouillonii PNG5-198R and (d–e) M. producens 3LT. (a) Underwater pictures of M. bouillonii at 10 m depth forming a characteristic cobweb mat firmly attached to the surrounding corals. (d) Tuft colony morphology of M. producens growing on shallow-water mangrove roots. Microphotographs of cyanobacterial filaments of (b) M. bouillonii and (e) M. producens and scanning electron micrographs of (c) M. bouillonii and (f) M. producens. Bars: a, 10 cm; b, 29.5 µm; c, ca. 20 µm; d, 10 cm; e, 50 µm; f, 50 µm.
M. producens has often been reported in the literature as either Lyngbya majuscula or L. sordida. The primary reason for combining tropical marine L. majuscula and L. sordida into a single species, Moorea producens, was variability in the morphological characters of these two former morphotypes. M. bouillonii was, in contrast to L. majuscula and L. sordida, initially described from tropical marine environments and will consequently keep its species nomenclature in order to preserve taxonomic stability (Hoffman & Demoulin, 1991).
The ultrastructure of Moorea cells contained a high degree of compartmentalization and cells were rich in intrathylakoidal spaces (widened thylakoids) (Fig. 3). The thylakoid membranes were arranged parallel to the cell walls. The filaments were surrounded by thick (2–3 µm) firm and laminated sheaths.
Fig. 3.
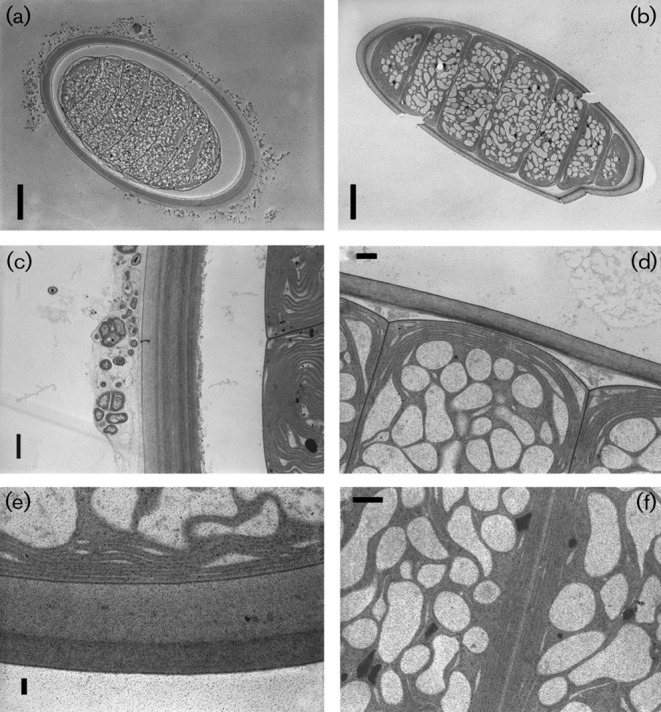
Microphotographs of cyanobacterial filaments obtained by transmission electron microscopy. Filament transections of Moorea producens 3LT (a) and M. producens JHB (b); polysaccharide sheaths and thylakoid arrangements in M. producens 3LT with heterotrophic bacteria on the exterior (c), M. producens JHB (d), polysaccharide sheath of M. producens 3LT (e) and thylakoid arrangements in adjacent cells in M. producens JHB (f). Bars: a, 10 µm; b, 10 µm; c, 1 µm; d, 2 µm; e, 0.5 µm; f, 2 µm.
Geographically, Moorea is a widely distributed group that is abundant in tropical marine regions (see Table S1, available in IJSEM Online). The latitudinal distribution of this group, according to current sampling and records, ranges approximately between the Tropic of Cancer and the Tropic of Capricorn. The most northern reported collection of Moorea is Florida (26° 04′ N) just north of the Tropic of Cancer (Sharp et al., 2009). The habitats of Moorea include diverse shallow-water marine environments such as coral reefs, sandy beaches and mangroves. While M. producens is a cosmopolitan species and has been found pantropically in shallow marine waters, M. bouillonii has only been reported from tropical Pacific locations (see Table S1, available in IJSEM Online).
Biochemically, extracts from the three Moorea strains 3LT, PNG5-198R, and JHB showed UV absorption at 565, 620, 650 and 665 nm, corresponding to the photosynthetic pigments phycocyanin, phycoerythrin, allophycocyanin and chlorophyll a, respectively (Table 1). In addition to these basic cyanobacterial photosynthetic pigments, all three Moorea strains contained at least two structurally unique bioactive secondary metabolites, as characterized by LC-MS and NMR (Table 1).
Table 1. Genomic and biochemical characteristics of Moorea.
Strains: 1, Moorea producens 3LT; 2, Moorea bouillonii PNG5-198R. Both strains had PKS/NRPS secondary metabolite genes, possessed chlorophyll a, and had the phycobiliproteins phycocyanin, phycoerythrin and allophycocyanin. Neither strain had nitrogen-fixing genes. nd, Not determined.
| Characteristic | 1 | 2 |
| Genome size | 8.5 Mbp | nd |
| DNA G+C content (mol%) | 41 | 42 |
| Protein-coding genes | 7415 | nd |
| Secondary metabolites* | cur/car/bar | apr/lbn |
cur, Curacins; car, carmabins; bar, barbamide; apr, apratoxins; lbn, lyngbyabellins.
The DNA G+C contents of M. producens 3LT and M. bouillonii PNG5-198R were 41.0 mol% and 42.3 mol%, respectively (Table 1), which were comparable to other filamentous cyanobacteria (mean DNA G+C content = 41.2 mol%). The genome size of M. producens 3LT (8.5 Mbp) was larger than the mean genome of filamentous cyanobacteria (6.1 Mbp) and the second largest after the evolutionarily related Coleofasciculus PCC 7420T (genome size = 8.7 Mbp). The relatively large genome of M. producens 3LT was reflected in a high abundance of protein-coding genes (7415 compared with the mean copy number of protein-coding genes in filamentous cyanobacteria of 5468 copies). A potential reason for the large genome is the relatively large number of genes involved in the biosynthesis of bioactive secondary metabolites. For example, genome analysis of strain 3LT has revealed that approximately 3 % of its genome contains polyketide synthase (PKS), non-ribosomal protein synthetase (NRPS), or other biosynthetic genes dedicated to secondary metabolism (Jones & Monroe et al., 2011). The partial genome of M. bouillonii PNG05-198T also contained multiple copies of PKS and NRPS genes with high identity to biosynthetic genes involved in secondary metabolite production. The genome of M. producens 3LT has been shown to lack genes involved in nitrogen fixation (Jones & Monroe et al., 2011). This was further supported by a blast search (E-value = 1e-5) of the M. bouillonii PNG5-198R genome, which also lacked the nifHDK genes necessary for nitrogen fixation. The presence of nif genes in the closely related genera Coleofasciculus and Symploca suggests a loss of these vertically inherited genes in Moorea as a relatively recent evolutionary event, which further supports the delineation of Moorea as an exclusive genus.
Latin diagnosis of Moorea gen. nov.
Filamenta solitaria vel in fasciculis irregularis, ad macroscopica, ad <10 cm longa, 25–65 (82) µm lata, non divaricata nec ramosa. Trichoma cylindrical, ad dissepimenta non vel paucim constrincta, aeruginosa, olivacea vel rubentes. Vaginae firmae, plus minusve tenues vel paucim dilatatae, externe saepe mucosae, sine colore, paucim lamellosae. Cellulae semper distincte brevior quam latae [20–55 (70) × (2) 3–10 µm], discoidae; cellula apicalis late rotundata. Reproductio hormogoniis necridiis separatur. Heterocytae akinetaeque carentes.
Typus generis: Moorea producens spec. nova.
Etymologia: ad honorem in memoriam Professor Richard E. Moore nominate.
Description of Moorea gen. nov.
Moorea gen. nov. (Mo.o.re′a. N.L. fem. n. Moorea in memory of Professor Richard E. Moore).
Large filamentous cyanobacteria common in tropical marine oceans, abundant on coral reefs, rocks or mangroves at depths ranging between 0.3–30 m. Filaments are unbranched, <10 cm in length, with wide diameters [25–65 (82) µm]. Trichomes are cylindrical, not attenuated towards ends, constricted or not constricted on crosswalls, surrounded by thick (3–5 µm) and distinct polysaccharide sheaths. The sheaths are typically covered by a rich diversity of mucus (often containing heterotrophic bacteria and other micro-organisms). The cells are discoid, always shorter than they are wide [20–55 (70) µm wide and (2) 3–10 µm long]. The trichomes contain necridic cells separating the trichomes into hormogonia. The terminal cells of the filaments and those of the hormogonia are rounded. Non-diazotrophic and the filaments lack heterocysts or other specialized cells. Members of the genus are photosynthetic and contain phycobiliproteins (phycocyanin, phycoerythrin, and allophycocyanin) and chlorophyll a. Strains are often rich in bioactive secondary metabolites typically biosynthesized by PKS, NRPS or mixed PKS/NRPS pathways.
Type species: Moorea producens sp. nov.
Latin diagnosis of Moorea producens sp. nov.
Thalus caespitosus vel prostratus, coloratus, rubescens ad viride-fuscus. Filamenta 30–67 (82) µm crassa. Vaginae sine colore, plus minusve tenues, 1–2 (12) µm latae, paucim lamellosae. Trichomata rubra vel praecipue olivaceae, cylindrica, apicem non attenuata, ad dissepimenta constricta (25) 30–65 (70) µm lata. Cellulae 3–7 µm longae, cellula apicalis rotundata, calyptra nulla.
DNA G+C contentus = 41.2 mol%.
Holotypus: cultura 3L, in CPCC et CCMP deposita; exemplum conservatum in herbario Musei Moravici Brno (BRNM/HY 2364) depositum; icona typical Fig. 2.
Habitatio: ad radices arborum mangrovis, ad oras Antillarum Hollandicum, in profunditate 2–3 m.
Etymologia: contentuu multis producti chemicis.
Description of Moorea producens sp. nov.
Moorea producens (pro.du′cens. L. part. adj. producens making, producing, referring to the fact that the species is rich in metabolic products).
The thallus morphology ranges from tuft to extensive mats. The coloration is highly varied, ranging from dark red to greenish-brown. Filaments 30–67 (82) µm width. Sheaths are colourless, thin (1–2 µm, but can be 12 µm wide in extreme situations), slightly lamellose. Trichomes are cylindrical, attenuated on the end, constricted on the cell walls, cells (25) 30–65 (70) µm wide and 3–7 µm long. Apical cells are rounded, without calyptra.
The type strain, 3LT, was isolated from coral rubble and rocks at 2–3 m depth in Curaçao, Netherlands Antilles. The genomic DNA G+C content of the type strain is 41.2 mol%.
Holotype: strain 3L, deposited in the CPCC and CCMP collections; dried material deposited at the herbarium of the Moravian Museum Brno (BRNM/HY 2364); typical morphology is shown in Fig. 2.
This species has often been incorrectly cited in the literature as L. majuscula or L. sordida.
Description of Moorea bouillonii comb. nov.
Basionym: Lyngbya bouillonii (Hoffmann et Demoulin Belg J Bot 124: 85, 1991).
For a basic description see Hoffmann & Demoulin (1991).
The colony morphology is mat-like and tenaciously attached to surrounding substrate. The colonies are found in association with the snapping shrimp (Alpheus frontalis). Coloration fluctuates between brownish-red and dark red depending on depth.
The reference strain, PNG5-198R, was isolated from coral reefs at a depth of 10 m in New Ireland, Papua New Guinea. The DNA G+C content of the reference strain is 42.3 mol%.
Acknowledgements
We thank the governments of Papua New Guinea, Curaçao, Panama and Jamaica and the Palmyra Atoll Research Consortium for permission to collect Moorea specimens. We are also grateful to J. P. Euzéby for help with nomenclature. We also acknowledge the generous funding from the Sea Grant Program R/NMP-103EPD and the Ministry of Education of the Czech Republic MSM6007665801 and AVOZ60050516. Some of the work reported here was carried out at the National Center for Microscopy and Imaging Research, which is supported by NIH grant P41-RR004050 to M. H. G. This manuscript and the naming of Moorea are dedicated to the late R. E. Moore (U. Hawaii) for his landmark contributions to natural products discovery from tropical marine cyanobacteria.
Abbreviations:
- ITS
internal transcribed spacer
- NRPS
non-ribosomal peptide synthetase
- PKS
polyketide synthase
Footnotes
Three supplementary figures and a supplementary table are available with the online version of this paper.
References
- Boyer S. L., Flechtner V. R., Johansen J. R. (2001). Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol 18, 1057–1069 10.1093/oxfordjournals.molbev.a003877 [DOI] [PubMed] [Google Scholar]
- Cannone J. J., Subramanian S., Schnare M. N., Collett J. R., D’Souza L. M., Du Y., Feng B., Lin N., Madabusi L. V. & other authors (2002). The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamatta D. A., Johansen J. R., Vis M. L., Broadwater S. T. (2005). Molecular and morphological characterization of ten polar and near-polar strains within the Oscillatoriales (Cyanobacteria). J Phycol 41, 421–438 10.1111/j.1529-8817.2005.04062.x [DOI] [Google Scholar]
- Castenholz R. W. (1988). Culturing of cyanobacteria. Methods Enzymol 167, 68–93 [Google Scholar]
- Castenholz R. W. (2001). Phylum BX. Cyanobacteria oxygenic photosynthetic bacteria. In Bergey’s Manual of Systematic Bacteriology, pp. 473–599 Edited by Boone D. R., Castenholz R. W., Garrity G. M. New York: Springer; 10.1007/978-0-387-21609-6_27 [DOI] [Google Scholar]
- Engene N., Coates R. C., Gerwick W. H. (2010). 16S rRNA gene heterogeneity in the filamentous marine cyanobacterial genus Lyngbya. J Phycol 46, 591–601 10.1111/j.1529-8817.2010.00840.x [DOI] [Google Scholar]
- Engene N., Choi H., Esquenazi E., Rottacker E. C., Ellisman M. H., Dorrestein P. C., Gerwick W. H. (2011). Underestimated biodiversity as a major explanation for the perceived rich secondary metabolite capacity of the cyanobacterial genus Lyngbya. Environ Microbiol 13, 1601–1610 10.1111/j.1462-2920.2011.02472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubic S., Abed R. M. M., Palińska K., Pauillac S., Chinain M., Laurent D. (2010). Marine toxic cyanobacteria: diversity, environmental responses and hazards. Toxicon 56, 836–841 10.1016/j.toxicon.2009.07.023 [DOI] [PubMed] [Google Scholar]
- Gugger M., Molica R., Le Berre B., Dufour P., Bernard C., Humbert J.-F. (2005). Genetic diversity of Cylindrospermopsis strains (cyanobacteria) isolated from four continents. Appl Environ Microbiol 71, 1097–1100 10.1128/AEM.71.2.1097-1100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L., Demoulin V. (1991). Marine cyanophyceae of Papua New Guinea. II. Lyngbya bouillonii sp. nov., a remarkable tropical reef-inhabiting blue-green alga. Belg J Bot 124, 82–88 [Google Scholar]
- Jones A. C., Monroe E. A., Podell S., Hess W. R., Klages S., Esquenazi E., Niessen S., Hoover H., Rothmann M. & other authors (2011). Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proc Natl Acad Sci U S A 108, 8815–8820 10.1073/pnas.1101137108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9, 286–298 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- Liu L., Rein K. S. (2010). New peptides isolated from Lyngbya species: a review. Mar Drugs 8, 1817–1837 10.3390/md8061817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S., Suda S., Li R., Watanabe M., Oyaizu H., Matsumoto S., Watanabe M. M. (1999). Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol Lett 172, 15–21 10.1111/j.1574-6968.1999.tb13443.x [DOI] [PubMed] [Google Scholar]
- Posada D. (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253–1256 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sharp K., Arthur K. E., Gu L., Ross C., Harrison G., Gunasekera S. P., Meickle T., Matthew S., Luesch H. & other authors (2009). Phylogenetic and chemical diversity of three chemotypes of bloom-forming Lyngbya species (Cyanobacteria: Oscillatoriales) from reefs of southeastern Florida. Appl Environ Microbiol 75, 2879–2888 10.1128/AEM.02656-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandeau de Marsac N., Houmard J. (1988). Complementary chromatic adaption: physiological conditions and action spectra. Methods Enzymol 167, 318–328 10.1016/0076-6879(88)67037-6 [DOI] [Google Scholar]
- Tidgewell K., Clark B. R., Gerwick W. H. (2010). The natural products chemistry of cyanobacteria. In Comprehensive Natural Products II Chemistry and Biology, vol. 2, pp. 141–188 Edited by Mander L., Lui H.-W. Oxford: Elsevier; 10.1016/B978-008045382-8.00041-1 [DOI] [Google Scholar]
- Zwickl D. J. (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD Thesis, The University of Texas at Austin. [Google Scholar]