Abstract
The cariogenic bacterium Streptococcus mutans has two paralogues of the YidC/Oxa1/Alb3 family of membrane protein insertases/chaperones. Disruption of yidC2 results in loss of genetic competence, decreased membrane-associated ATPase activity and stress sensitivity (acid, osmotic and oxidative). Elimination of yidC1 has less severe effects, with little observable effect on growth or stress sensitivity. To examine the respective roles of YidC1 and YidC2, a conditional expression system was developed allowing simultaneous elimination of both endogenous YidCs. The function of the YidC C-terminal tails was also investigated and a chimeric YidC1 protein appended with the C terminus of YidC2 enabled YidC1 to complement a ΔyidC2 mutant for stress tolerance, ATP hydrolysis activity and extracellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity. Elimination of yidC1 or yidC2 affected levels of extracellular proteins, including GtfB, GtfC and adhesin P1 (AgI/II, PAc), which were increased without YidC1 but decreased in the absence of YidC2. Both yidC1 and yidC2 were shown to contribute to S. mutans biofilm formation and to cariogenicity in a rat model. Collectively, these results provide evidence that YidC1 and YidC2 contribute to cell surface biogenesis and protein secretion in S. mutans and that differences in stress sensitivity between the ΔyidC1 and ΔyidC2 mutants stem from a functional difference in the C-termini of these two proteins.
Introduction
Acidogenic and acidophilic bacteria within the supragingival plaque on teeth are responsible for dental caries. Streptococcus mutans is highly acidogenic and mounts an acid tolerance response to cope with the acid produced, thus enabling it to outcompete other bacteria in the oral cavity (Hamilton & Buckley, 1991). An avid biofilm former, S. mutans adheres to the tooth surface through sucrose-dependent [glucosyltransferases (GTFs) and fructosyltransferases (FTFs)] and sucrose-independent (antigen I/II, aka P1 or PAc) mechanisms (Gibbons et al., 1986; Jenkinson & Lamont, 1997). During a search for S. mutans genes that contribute to acid tolerance, a mutant disrupted in ffh expression was isolated (Gutierrez et al., 1996). The ffh gene encodes an integral component of the signal recognition particle (SRP) co-translational targeting pathway, which was previously believed to be essential for viability in all cells, including bacteria (Honda et al., 1993; Phillips & Silhavy, 1992). Subsequent analysis of the S. mutans genome revealed genes encoding two paralogues of the YidC/Oxa1/Alb3 family, YidC1 and YidC2, that appear to supplement the SRP pathway in this organism (Funes et al., 2009). This family of proteins is universally conserved in all three domains of life (Zhang et al., 2009): Oxa1 is in the inner mitochondrial membrane; YidC is in the bacterial cytoplasmic membrane; and Alb3 is in the thylakoid membrane of chloroplasts. These proteins serve as membrane-integral chaperones and insertases with conserved functions in the insertion of respiratory chain complexes, F1F0 ATP synthase components and light-harvesting chlorophyll-binding proteins (Bonnefoy et al., 2009; Kiefer & Kuhn, 2007; van der Laan et al., 2003). In mitochondria, chloroplasts and most Gram-positive bacteria, there are often two or more paralogues, whereas Escherichia coli and other Gram-negative bacteria have only one.
Elimination of yidC2 in S. mutans results in a stress-sensitive phenotype similar to S. mutans SRP pathway mutants, including growth impairment under acid, osmotic and oxidative stress conditions, decreased membrane-associated ATPase activity, decreased genetic competence and impaired biofilm formation (Hasona et al., 2005). Disruption of yidC1 has a much less severe effect, with no obvious growth defects or stress sensitivity. YidC2, but not YidC1, can complement a yeast Oxa1 mutant for growth on a non-fermentable carbon source (Funes et al., 2009). This ability requires the C terminus of YidC2. In contrast, either YidC1 or YidC2 can complement an E. coli YidC depletion strain for growth and restore functional activity to the F1F0 ATP synthase (Dong et al., 2008). Previous attempts to isolate double mutants in the SRP pathway and yidC2 have been unsuccessful. It has also not been possible to simultaneously eliminate yidC1 and yidC2, suggesting overlapping functions in the SRP and YidC2 pathways, as well as between YidC1 and YidC2.
The work presented here highlights the complex nature of protein translocation and maturation in S. mutans and supports the hypothesis that pathway redundancies allow the organism to thrive under environmentally stressful conditions, establish biofilms and cause disease. We developed a conditional expression system for yidC2 to enable the simultaneous elimination of endogenous YidC1 and YidC2. In addition, C-terminal-domain substitutions between YidC1 and YidC2 indicated that the C terminus of YidC2 is critical for the ability of S. mutans to tolerate environmental stress. Our studies show further that both YidC1 and YidC2 contribute to S. mutans protein secretion, cell surface biogenesis, biofilm formation and cariogenicity, albeit in different ways and to different extents.
Methods
Bacterial strains, plasmids and growth conditions.
Table 1 lists strains and plasmids, and Table S1 (available with the online version of this paper) lists primers used in this study. A schematic representation of the strains used in this study is shown in Fig. S1. S. mutans strains were routinely grown at 37 °C in Todd–Hewitt broth (BBL, Becton Dickinson) supplemented with 0.3 % yeast extract (THYE) or in FMC (Terleckyj et al., 1975), modified as described in the Supplementary Methods [Terleckyj’s defined medium (TDM)], with 0.5 % (w/v) sugar when indicated. Spectinomycin (1 mg ml−1), kanamycin (500 µg ml−1) or erythromycin (5 µg ml−1) were added where appropriate. E. coli strain Top10 (Invitrogen), used for intermediate cloning steps, was grown aerobically at 37 °C in Luria–Bertani (LB) broth or agar with kanamycin (50 µg ml−1), spectinomycin (100 µg ml−1) or erythromycin (150 µg ml−1) where appropriate.
Table 1. Bacterial strains and plasmids used in this study.
em, Erythromycin; km, kanamycin; ap, ampicillin; sp, spectinomycin; tc, tetracycline; MCS, multiple cloning site.
| Strain/plasmid | Relevant characteristic(s) | Source |
| S. mutans | ||
| UA159 | Wild-type | R. A. Burne |
| NG8 | Wild-type | Knox et al. (1986) |
| PC3370 | NG8 ΔspaP : : tc | Crowley et al. (1999) |
| AH398 | NG8 ΔyidC2 : : em | Hasona et al. (2005) |
| AH378 | NG8 ΔyidC2 : : km | A. Hasona |
| AH374 | NG8 ΔyidC1 : : em | Hasona et al. (2005) |
| AH412 | NG8 yidC2ΔC255–310 : : em | Funes et al. (2009) |
| SP10 | AH398 gtfA : : PcelB–yidC2–km | This study |
| SP13 | AH378 gtfA:PgtfA : : yidC11–249–yidC2246–310–em | This study |
| SP14 | AH378 gtfA:PgtfA : : yidC21–247–yidC1228–271–em | This study |
| SP16 | AH412 ΔyidC1 : : sp | This study |
| SP17 | AH378 gtfA : : PgtfA–yidC2–em | This study |
| SP20 | SP10 ΔyidC1 : : sp | This study |
| SP22 | NG8 gtfA : : em | This study |
| Plasmids | ||
| pDL289 | E. coli–Streptococcus shuttle vector; Kmr(aphA) | Buckley et al. (1995) |
| pDL278 | E. coli–Streptococcus shuttle vector, Spr (aad9) | LeBlanc et al. (1992) |
| pBGK2 | Streptococcus integration vector at the gtfA locus (same as pBGK with bla deleted) | Wen & Burne (2001) |
| pBGE | Streptococcus integration vector, genes are expressed from the gtfA promoter in the chromosome | Zeng & Burne (2009) |
| pSP10 | pBGK2 with PcelA–yidC2 cloned into SmaI site | This study |
| pSP11 | pBGK2 with PcelB–yidC2 cloned into SmaI site | This study |
| pCR2.1 | T/A cloning vector, LacZα MCS, Apr, Kmr | Invitrogen |
| pCR2.1-ΔyidC1SpR | pCR2.1 with ΔyidC1 : : sp for allelic replacement of yidC1 | This study |
Strain construction
YidC2 conditional expression strains.
UA159 genomic DNA was used as the template in PCRs unless otherwise indicated. The celB promoter (PcelB; −1 to −263) was amplified by PCR using primers SP13F and SP13R. The yidC2 gene was amplified by PCR using primers SP14F and SP05R. Each PCR product was ligated to pCR2.1, and a 1 kb fragment from a properly oriented yidC2-containing-plasmid, cut with BamHI and NdeI, was ligated to a 5.0 kb fragment containing PcelB to create the promoter fusion PcelB–yidC2. The PcelB–yidC2 fragment was excised with SmaI and ligated to SmaI-digested pBGK2. The resulting plasmid, pSP10, was used to transform a S. mutans ΔyidC2 mutant strain, AH398, resulting in strain SP10 in which PcelB-yidC2 had integrated into the chromosome at the gtfA locus (illustrated in Fig. S1). Strain SP10 was confirmed by PCR using primers SP17F and SP16R. Primers SP17F-RC and SP18R were used to confirm orientation of the insert in the gtfA locus. Strain SP10 was used to create strain SP20, where yidC1 was deleted using pCR2.1 ΔyidC1 : : SpecR (see below).
YidC1 deletion strain.
yidC1 was eliminated by allelic replacement with a spectinomycin-resistance (spec) gene cassette constructed as follows: splice overlap extension (SOE) PCR (Heckman & Pease, 2007) was used to combine PCR-amplified fragments upstream and downstream of yidC1 with an intervening spec gene amplified from pDL278. Primer pairs used were SP29F and SP24RSOE, SP25FSOE and SP25RSOE, and SP26FSOE and AH25R, respectively. Primers SP29F and SP25RSOE were used to combine the yidC1 upstream and spec fragments by SOE PCR. This product was combined with the yidC1 downstream fragment by SOE PCR using primers SP29F and AH25R to generate a 2087 nt product that was subsequently ligated to pCR2.1, generating plasmid pCR2.1ΔyidC1SpR. This plasmid was linearized with HindIII and used to naturally transform S. mutans resulting in replacement of the yidC1 gene with the spectinomycin-resistance gene via double-crossover recombination.
Construction of genes encoding chimeric YidC1–YidC2 proteins.
SOE PCR was used to create a chimeric gene encoding a protein composed of amino acids 1–229 of YidC1 and the C-terminal tail of YidC2 (amino acids 247–310). The yidC1 fragment (−131 to +687) was amplified by PCR using primers SP21F and SP22RSOE. The yidC2 C-terminal gene fragment (+741 to +1009) was amplified by PCR using primers SP22FSOE and SP21R. Each fragment was gel-purified and combined via SOE PCR, using primers SP21F and SP21R. The product was ligated to pCR2.1, excised with XbaI and BsrGI and ligated to integration vector pBGE that had been similarly digested. Plasmid pBGE–yidC1–C2–Erm was used to transform AH378 to generate strain SP13 (see Figs S1 and S2).
A chimeric protein comprising amino acids 1–247 of YidC2 and C-terminal amino acids 227–271 of YidC1 was constructed in the same manner as described above. The yidC2 gene fragment (−43 to +741) was amplified by PCR with primers SP27F2 and SP27RSOE. The yidC1 C-terminal fragment (+685 to +875) was amplified by PCR using primers SP28FSOE and SP28R. Fragments were used as templates in an SOE PCR with primers SP27F2 and SP28R. The amplified product was ligated to pCR2.1, excised and ligated to pBGE, and the XbaI site in pCR2.1 was used to restrict the fragment from the plasmid for subcloning. The resultant strain was named SP14. The control strain SP17, where unaltered yidC2 was amplified by PCR using primers SP27F2 and SP21R and placed under control of the gtfA promoter in strain NG8, was generated for experiments with SP13 and SP14. The control strain SP22 was created by transforming strain NG8 with pBGE. Proper integration of chimeric and control genes was verified by PCR using the PgtfA-based forward primer SP37F and reverse primer AH31R for strains SP13 and SP17, or SP21R for SP14, followed by DNA sequencing.
YidC2 depletion conditions.
To determine the conditions necessary to deplete YidC2 in strains SP20 and SP10, cells were grown for 16 h in TDM broth with 0.5 % cellobiose then diluted 1 : 20 into fresh TDM cellobiose (inducing sugar) or TDM-glucose, -fructose or -mannose (repressing sugars). Cultures were grown to OD600 0.9 and diluted in an equal volume of fresh TDM containing the same sugar with continued incubation. This was repeated three times or until growth slowed, indicating depletion of YidC2. Growth in TDM-mannose was the only condition that resulted in reduced growth and depletion of YidC2 occurred after an additional 8 h of growth, as determined by Western blot analysis of cell lysates.
For evaluation of growth on agar, cells were grown 16 h in TDM-cellobiose and diluted 1 : 10 into fresh TDM-cellobiose and grown to mid-exponential phase. They were then streaked on TDM agar containing 0.5 % cellobiose or 0.5 % mannose and incubated at 37 °C in a 5 % CO2 incubator for 48 h.
Western blot analysis.
To determine depletion conditions of YidC1 and YidC2 in SP20 and SP10, cells from a mid-exponential-phase culture (OD600 0.5) were pelleted and washed once in 25 mM Tris/HCl, pH 7.5 and then resuspended in 500 µl of the same buffer. Cells were lysed with glass beads (0.1 mm, 0.5 g, BioSpec Products) in a Mini Bead Beater (BioSpec Products), for two 1 min cycles with cooling on ice in between. Proteins were separated by SDS-PAGE through a 10 % gel followed by transfer to an Immobilon PVDF membrane (GE Healthcare). Western blots were performed using the ECL kit (GE Healthcare) according to the manufacturer’s instructions. Membranes were reacted with affinity-purified YidC1 C-terminal or YidC2 C-terminal antibodies (Dong et al., 2008) at 1 : 4000 and 1 : 8000 dilutions, respectively.
For detection of extracellular Gtf B and C, proteins in 500 µl filter-sterilized supernatant (0.2 µm filter) from 16 h THYE cultures were precipitated with an equal volume of 20 % (v/v) TCA on ice for 20 min. Pellets were washed twice with 300 µl acetone, resuspended in 100 µl 50 mM Tris–HCl (pH 7.5) containing 10 mM EDTA. Protein samples were separated by SDS-PAGE through a 4–15 % gradient mini gel (Bio-Rad). Western blot analysis was performed as described above using a polyclonal rabbit antiserum (1 : 500), kindly provided by Dr William Bowen, University of Rochester, that recognizes both GtfB and GtfC (Wunder & Bowen, 2000).
Evaluation of bacterial growth.
Cultures of S. mutans strains grown for 16 h were diluted 1 : 20 in THYE, pH 7.0, without antibiotics and grown to OD600 0.4–0.45. Strains then were diluted 1 : 10 into THYE buffered to pH 7.0, pH 5.0 or pH 7.0 with 3 % (w/v) NaCl and 300 µl was loaded in triplicate wells on a 100-well Bioscreen C plate and overlaid with mineral oil. Growth was monitored in the Bioscreen C Machine (Growth Curves USA) at 37 °C for 16 h with OD600 recorded every 15 min. Doubling times were calculated as described by Khalichi et al. (2004). Statistical significance was determined by one-way ANOVA.
ATP hydrolysis activity assay.
Permeabilized cells were made essentially as described by Belli & Marquis (1991) using a 25 ml culture of each strain grown to mid-exponential phase (OD600 approximately 0.5) in THYE. For assay, 100 µl permeablized cells was added to 3 ml assay buffer [50 mM Bistris–HCl (pH 6.0), 10 mM MgCl2], and the assay was started by adding 3 mM ATP. Reactions were stopped at time points 0, 5 and 10 min by removing an aliquot to iced stop buffer [1.3 parts H2O, 0.6 parts HCl/molybdate (2.5 % w/v NH4Mo4O2.4H2O, 4.0 M HCl), 0.4 parts 10 %, w/v, SDS]. Inorganic phosphate was measured by addition of a 1 : 10 dilution of Eikonogen (1 M NaHSO3, 0.1 M Na2SO3, 0.01 M 4-amino-3-hydroxyl-1-naphthalenesulfonic acid) followed by a 30 min incubation at room temperature with absorbance read at OD700. Assays were performed in quadruplicate. Whole-cell protein concentrations were determined by BCA assay with BSA standards. Enzyme activity was calculated as nmol Pi min−1 (mg protein)−1. Statistical differences were determined using Student’s t-test.
GAPDH assay of whole cells.
Whole cell GAPDH activity was determined using cells grown for 16 h in THYE as described previously (Seifert et al., 2003) with the following modifications. Cells were harvested by centrifugation and suspended to OD600 1.0, in 25 mM Tris–HCl (pH 7.5), 5 mM EDTA. For assay, 1 ml cell suspension was pelleted and resuspended in 900 µl 40 mM triethanolamine, 50 mM Na2HPO4, 5 mM EDTA, pH 8.6, and OD600 measured again. For each assay, 7 µl glyceraldyhyde-3-phosphate (50 mg ml−1, Sigma) and 100 µl 10 mM NAD+ were added, and the mixtures were incubated at 37 °C for 30 min. Cells were removed by centrifugation and A340 of the supernatants was read and standardized by dividing this value by the measured OD600. Assays were performed in triplicate for each strain. Statistical differences were determined using Student’s t- test.
Comparison of cell-surface-localized P1 by whole cell dot-blot.
Wild-type and YidC mutant strains were compared for their reactivity with a panel of extensively characterized monoclonal antibodies (McArthur et al., 2007) directed against the surface-localized protein adhesin P1 by whole-cell dot-blot as described previously (Seifert et al., 2004) using anti-P1 mAbs 4-10A, 1-6F, 5-5D, 6-11A, 3-10E ascites fluids at a 1 : 500 dilution.
Evaluation of S. mutans adherence by surface plasmon resonance.
Adherence of S. mutans wild-type and YidC mutant strains to a salivary agglutinin-coated chip surface was evaluated by surface plasmon resonance. Studies were performed on a BIAcoreR 3000 using a Pioneer F1 sensor chip as described previously (Oli et al., 2006).
Biofilm formation and analysis.
Formation of wild-type and YidC mutant strain biofilms was assessed as described previously (Hasona et al., 2007). Statistical significance relative to the wild-type value at each time point and growth condition was determined using Student’s t-test.
Assessment of S. mutans colonization and cariogenicity in gnotobiotic rats.
In vivo colonization and cariogenicity was assessed in gnotobiotic rats using a previously described method (Crowley et al., 1999). Statistical significance in mean caries scores, c.f.u. per mandible and body weights between groups of rats infected with the wild-type strain compared with those infected with the YidC mutant strains was determined by one-way ANOVA with the Tukey–Kramer multiple comparison test using the InStat program (Graphpad Software). Differences between groups were considered significant at a P-value <0.05.
Results
Double elimination of yidC1 and yidC2 is lethal in S. mutans
To enable simultaneous elimination of endogenous YidC1 and YidC2 in S. mutans, a conditional expression system was developed for yidC2. The yidC2 gene was fused to the celB promoter and placed into the gtfA locus of the ΔyidC2 mutant (AH398) chromosome to create strain SP10, where expression of yidC2 is under the control of the carbon-catabolite-repressible celB promoter (Zeng & Burne, 2009). Strain SP20 was produced by replacing yidC1 in strain SP10 with a spectinomycin marker (see Fig. S1 for schematic). Following growth in different carbohydrates, YidC2 production was evaluated by Western blot using YidC2 and YidC1 C-terminal antibodies (Fig. 1a). Growth in TDM-cellobiose (inducing sugar) or THYE (rich media) resulted in a high level of YidC2 production (Fig. 1a, lanes 4 and 8, respectively). Growth in TDM-fructose did not repress expression of yidC2 (Fig. 1a, lane 5); however, growth in TDM-mannose resulted in a substantial decrease of YidC2 after 4 h (lane 6), with near complete repression after 8 h (lane 7). Suppression of YidC2 occurred in the presence of both glucose and mannose in strain SP10 where YidC1 was present (Fig. 1a, lanes 9 and 10). Growth of SP10, SP20 and appropriate control strains was assessed on TDM agar with 0.5 % cellobiose or 0.5 % mannose (Fig. 1b). Both SP10 and SP20 grew on agar containing cellobiose (PcelB–yidC2-inducing conditions), while only SP10 grew on agar with mannose (PcelB–yidC2-repressing conditions). This confirms that double elimination of YidC1 and YidC2 is lethal in S. mutans.
Fig. 1.
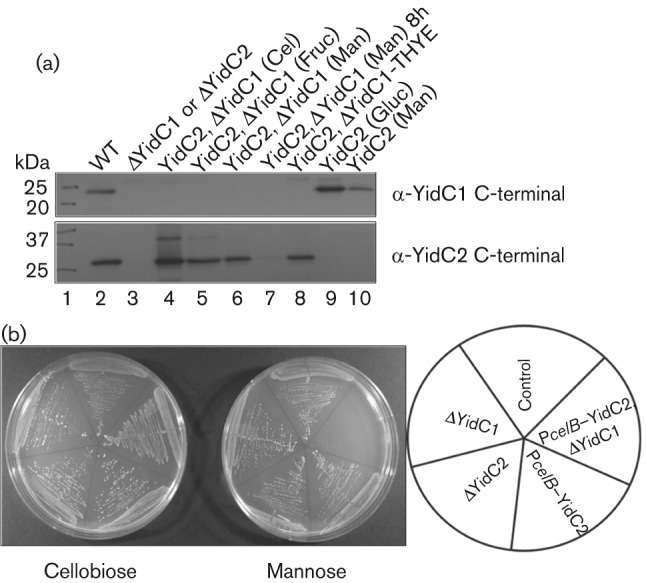
Western blot of whole-cell lysates of yidC2 depletion strains (PcelB–yidC2) SP20 and SP10 grown in various sugars. (a) Lysates were prepared from cultures grown in TDM supplemented with 0.5 % of the indicated sugar, reacted with anti-YidC1 C-terminal antibody (top panel) or with anit-YidC2 C-terminal antibody (bottom panel). The wild-type NG8 strain (WT) and AH374 (ΔyidC1) or AH398 (ΔyidC2) mutant strains were used as positive and negative controls, respectively. Cel, cellobiose; Fruc, fructose; Man, mannose; Gluc, glucose. (b) Growth of various strains on TDM 0.5 % mannose or 0.5 % cellobiose agar plates. The key is shown to the right. Strains used: control (SP22) NG8 gtfA : : em, ΔYidC1 (AH374), ΔYidC2 (AH398), PcelB–YidC2 (SP10), PcelB–YidC2, ΔYidC1 (SP20).
The C terminus of YidC2 is important for stress tolerance
YidC1 and YidC2 have approximately 30 % sequence identity and approximately 75 % similarity. The major differences are in their C-terminal tails with YidC2′s tail being longer and more basic (illustrated in Fig. S2). To investigate the functional difference between the tails, genes encoding chimeric proteins YidC1–C2 and YidC2–C1 were engineered and placed into the gtfA locus in the chromosome of a ΔyidC2 mutant to create strains SP13 and SP14 (illustrated in Figs S1 and S2), respectively. Strain SP17 also was generated in which full-length yidC2 was placed in the gtfA locus for use as a control; proper expression was confirmed by Western blot with C-terminal-specific antibodies against YidC1 and YidC2, as well as a non-C-terminal anti-YidC2 antibody (not shown). All strains were tested for the ability to tolerate acid and osmotic stress by growth in THYE broth at pH 5.0 or with 3 % NaCl (pH 7.0) compared with growth under non-stress (pH 7.0) conditions. Mean generation times are shown in Table 2. YidC1–C2 (SP13) was able to restore acid tolerance to the ΔyidC2 mutant to the same level as the positive control SP17 strain. Salt tolerance was also partially restored and similar in strains SP13 and SP17. In contrast with the complementation observed in strain SP13, the growth defect of strain SP14 (expressing YidC2–C1) was significantly worse under non-stress conditions than that of the ΔyidC2 mutant. This shows that substitution of the C terminus of YidC2 with that of YidC1 interferes with normal YidC2 function, resulting perhaps in a non-functional sink for YidC2 substrates. The critical nature of the YidC2 C terminus is also supported by the fact that placing it onto YidC1 (SP13) confers on YidC1–C2 the ability to restore acid tolerance to the ΔyidC2 mutant. The yidC2 C-terminal deletion strain AH412 showed similar acid and salt sensitivity to the complete yidC2 deletion strain AH378. Deletion of yidC1 in the yidC2ΔC background (strain SP16) further exacerbated the growth defects of this mutant.
Table 2. Mean generation times of S. mutans yidC mutants and complemented strains grown in THYE broth under non-stress, acid-stress and osmotic-stress conditions.
Mean generation times (min)±sd were calculated based on growth curves completed in triplicate in a Bioscreen C 100-well microtitre plate; OD600 was monitored in the Bioscreen C. NG, No growth. Statistical differences compared with: SP22, *P<0.001, **P<0.05; AH378 †P<0.001, ††P<0.01; AH412 ‡P<0.01.
| Strain | Relevant characteristics | pH 7.0 | pH 5.0 | 3 % NaCl |
| SP22 | NG8 gtfA : : em | 73±4.7 | 167±6.6† | 240±17.6 |
| SP17 | ΔyidC2 gtfA : : yidC2 | 77±1.5† | 285±4.2*†† | 334±27.5 |
| AH378 | NG8 ΔyidC2 : : km | 114±1.7* | 346±19.4* | 401±24.0* |
| AH374 | NG8 ΔyidC1 : : em | 77±11.9† | 186±5.6† | 266±27.8†† |
| SP13 | AH378 gtfA : : yidC1C2 | 80±1.5† | 282±19.0*†† | 346±2.7** |
| SP14 | AH378 gtfA : : yidC2C1 | 171±5.3*† | 390±41.7* | NG |
| AH412 | NG8 yidC2ΔC : : em | 83±10.0† | 367±0.0* | 399±40.0* |
| SP16 | AH412 ΔyidC1 : : sp | 143±3.2*† | 431±17.8*†‡ | NG |
ATPase activity is restored by chimeric YidC1–C2
In S. mutans the F1F0 ATPase plays a large role in acid tolerance by pumping excess protons out of the cytoplasm. The mechanisms of insertion of the F1F0 ATPase in S. mutans membranes are not known; however, mutations of ffh or yidC2 decrease acid tolerance and membrane-associated ATPase activity (Crowley et al., 2004; Hasona et al., 2005). In previous work, a deletion of yidC1 had no apparent effect on acid tolerance or membrane-associated ATPase activity (Hasona et al., 2005), leading to the hypothesis that, in S. mutans, YidC2 and the SRP pathways are involved in the assembly of the F-ATPase, while YidC1 is not. In the current study, ATP hydrolysis activity was evaluated using permeablized whole cells, as opposed to membranes, prepared by the method of Vadeboncoeur et al. (1991) as described previously. As shown in Fig. 2(a), there was a significant decrease in whole-cell ATPase activity in both the ΔyidC1 (AH374) and ΔyidC2 (AH378) mutants compared with the wild-type, albeit more pronounced in the ΔyidC2 mutant. ATPase activity was significantly increased, although not to wild-type levels, by expression of yidC2 (SP17) or chimeric yidC1–C2 (SP13) from the gtfA promoter in the ΔyidC2 mutant background. ATPase activity was not complemented in the ΔyidC2 mutant strain expressing the chimeric yidC2–C1 gene (SP14).
Fig. 2.
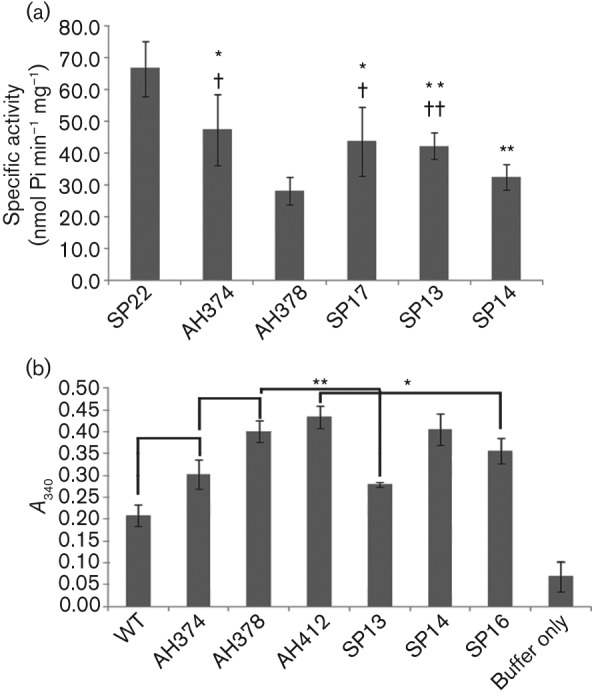
ATPase and extracellular GAPDH activity of wild-type S. mutans and yidC mutants. (a) ATPase activity of permeabilized whole cells. Average specific activity is expressed in nmol Pi min−1 mg−1 whole-cell protein. Bars represent mean±sd for triplicate assays. Statistically significant differences are indicated, *P≤0.05 and **P≤0.01 compared with SP22, or †P≤0.05 and ††P≤0.01 compared with AH378. (b) Extracellular GAPDH activity. Results are expressed as A340, standardized for cell number. Statistically significant differences are indicated, *P≤0.02, **P≤0.001. Descriptions of strains are as follows: WT (wild-type NG8), SP22 (gtfA : : em), AH374 (ΔyidC1), AH378 (ΔyidC2), AH412 (yidC2ΔC), SP13 (ΔyidC2, gtfA : : yidC1–C2), SP14 (ΔyidC2, gtfA : : yidC2–C1) and SP16 (yidC2ΔC, ΔyidC1).
Extracellular GAPDH activity is increased in yidC mutants
GAPDH, as well as a number of other cytoplasmic proteins, has been found on the surface of S. mutans and other streptococci (Henderson & Martin, 2011; Ling et al., 2004; Severin et al., 2007). An investigation by Biswas & Biswas (2005) found that deletion of the HtrA extracellular protease/chaperone resulted in increased levels of extracellular GAPDH in culture media. It is not known how GAPDH or other cytoplasmic proteins are transported, as they do not contain signal peptides. To analyse a potential role of YidC1 or YidC2 in GAPDH transport, we tested for differences in cell-surface-localized extracellular GAPDH activity using whole cells. There was a significant increase in extracellular GAPDH activity in the ΔyidC2 mutant compared with the ΔyidC1 mutant or wild-type NG8 (Fig. 2b). The yidC2ΔC (AH412) strain also demonstrated significantly increased surface-localized GAPDH activity, at a level similar to ΔyidC2 mutant. Expression of yidC1–C2 (SP13) restored GAPDH activity toward wild-type levels in the ΔyidC2 mutant. Expression of yidC2–C1 (SP14) did not result in complementation. When yidC1 was deleted in the yidC2ΔC background (SP16), GAPDH activity was significantly decreased compared with the yidC2ΔC strain (AH412). Thus, whatever the cause for increased GAPDH activity that occurs in the absence of YidC2’s C terminus, the effect can be alleviated to some extent by additionally deleting yidC1.
Effects of YidC1 and YidC2 on secreted proteins
The ability of S. mutans to form biofilms is dependent on the secretion of extracellular GTFs and FTFs (Banas & Vickerman, 2003) and the cell surface-localized-protein adhesin P1 (antigen I/II, PAc) (Jenkinson & Demuth, 1997). The ΔyidC2 mutant is impaired in sucrose-dependent biofilm formation and shows decreased clumping in defined media containing 0.5 % sucrose compared with wild-type NG8 (Fig. 3a). The yidC2ΔC mutant also shows diminished clumping under these conditions. Conversely, the ΔyidC1 mutant displays a hyper-clumping phenotype. The amount of extracellular secreted proteins in culture supernatants of wild-type and mutant strains was evaluated following standardization of bacterial numbers of respective cultures by optical density and is shown in Fig. 3(b) (protein stain). The presence of GtfB and GtfC in the culture supernatants was investigated by Western blot using a polyclonal rabbit antiserum that recognizes both GtfB (162 kDa) and GtfC (160 kDa). A decreased level of GtfB was observed in the ΔyidC2 as well as in the yidC2ΔC mutant culture supernatants (Fig. 3b). The level of GtfB was restored in the ΔyidC2 mutant background when yidC2 was expressed from the gtfA promoter (ΔYidC2+YidC2). Complementation of the ΔyidC2 mutant by chimeric yidC1–C2 (ΔYidC2+YidC1–C2) was minimal compared with complementation with the unaltered yidC2. When chimeric yidC2–C1 was expressed in the ΔyidC2 mutant (ΔYidC2+YidC2–C1), a more severe dominant-negative phenotype was observed compared with simple deletion of yidC2. There was a pronounced decrease in the overall level of secreted proteins detected in the culture supernatant of this strain (Fig. 3b, protein stain), including GtfB and GtfC (Fig. 3b, α-GtfB/C). On the other hand, the ΔyidC1 mutant displayed an overall increase in the level of secreted proteins, particularly GtfB and GtfC, in the culture supernatant compared with the wild-type (protein stain, Fig. 3b). This is consistent with the hyper-clumping seen with this mutant.
Fig. 3.
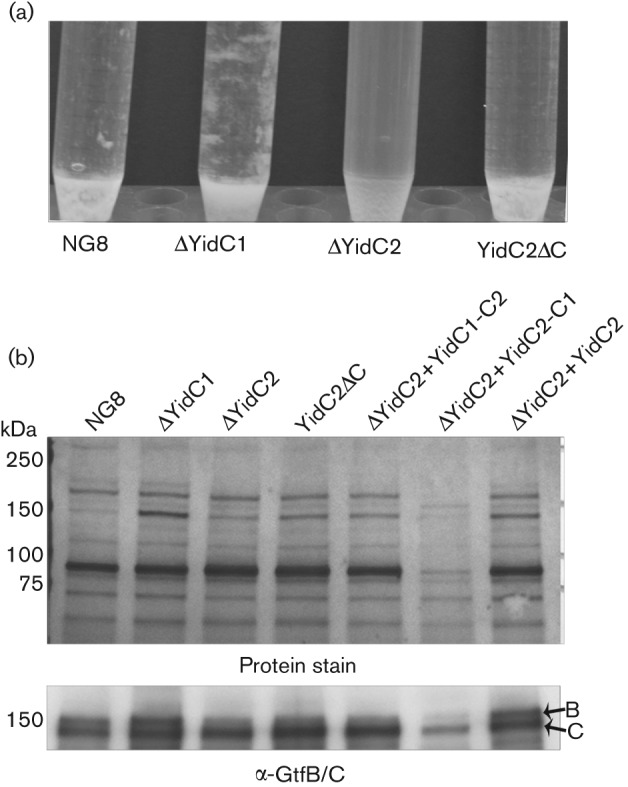
Cell clumping of stationary-phase cultures and detection of secreted GtfB/GtfC in culture supernatants from S. mutans wild-type and yidC mutants. (a) Cells were grown in TDM supplemented with 0.5 % sucrose for 16 h. (b) Culture supernatants from S. mutans cells grown 16 h in THYE were TCA-precipitated and proteins were separated on a 4–15 % Bio-Rad TGX gradient gel. The top panel shows the colloidal gold protein stain. The bottom panel shows the Western blot using polyclonal anti-GtfB/C serum.
When the ΔyidC1, ΔyidC2 and yidC2ΔC mutant strains were tested for the presence of surface-localized adhesin P1, results were consistent with those observed with the GTFs, with the exception that eliminating the C terminus of YidC2 had no effect on P1. There was notably more reactivity of a panel of anti-P1 monoclonal antibodies with S. mutans whole cells lacking YidC1, but less reactivity with the mutant lacking YidC2, compared with the parent NG8 strain (Fig. 4a). P1 has a highly unusual tertiary structure (Larson et al., 2010) and the binding sites of the mAbs, several of which recognize complex conformational epitopes (McArthur et al., 2007), have now been mapped on the 3D model (Robinette et al., 2011). While there was an overall decrease in immunoreactivity of all anti-P1 antibodies tested in the absence of YidC2, the binding of some antibodies was affected more than others. mAbs 1-6F and 3-10E were most affected by the deletion, while 4-10A was least affected. This was a consistent finding in multiple experiments. This suggests that the presence of YidC2 affects both the amount and conformation of P1 localized on the cell surface. Results of the immunoassays were mirrored when the adherence of S. mutans to P1’s physiological binding partner, human salivary agglutinin (SAG), was evaluated by surface plasmon resonance (Fig. 4b). The ΔyidC1 mutant was hyper-adherent, the ΔyidC2 mutant was impaired in P1-mediated adherence and the yidC2ΔC mutant was unaffected in its interaction with immobilized SAG. Strain PC3370, which lacks the spaP gene encoding P1 (Crowley et al., 1999), was non-adherent. The fact that the ΔyidC2 mutant was so severely impaired in P1-mediated adherence to SAG, despite detection of the adhesin on the cell surface, is consistent with an effect on its conformation and indicates that the P1 present is not functionally active as an adhesin. In contrast, P1-mediated aggregation in the presence of fluid-phase SAG was far less affected and only slightly diminished in the ΔyidC1, ΔyidC2 and yidC2ΔC mutant strains (data not shown). The binding of P1 to immobilized versus fluid-phase SAG represents two types of interaction that can be inhibited by different subsets of anti-P1 mAbs (Brady et al., 1992). Therefore, the preservation of S. mutans aggregation by fluid-phase SAG supports the conclusion that the functional form of P1 in addition to its total amount was influenced by the elimination of YidC2.
Fig. 4.
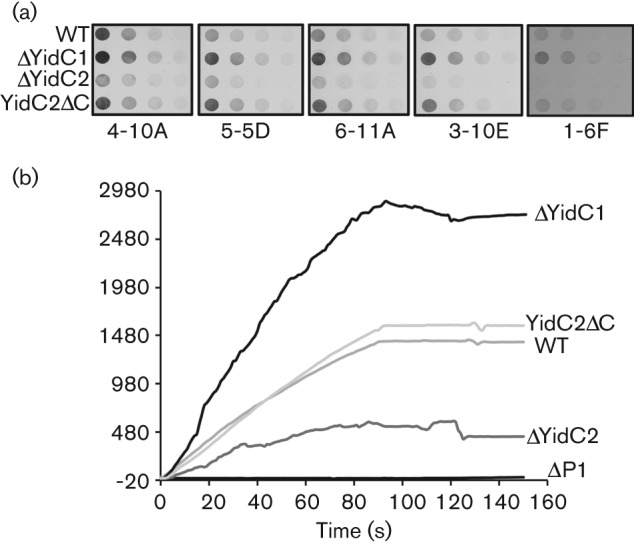
Anti-P1 mAb reactivity with wild-type (WT) and yidC mutant whole cells and comparison of bacterial binding to an agglutinin-coated chip surface. (a) Whole cell dot-blot. Whole bacterial cells serially diluted twofold beginning at 5×106 c.f.u. were applied to nitrocellulose membranes and reacted with the anti-P1 mAb indicated below each blot. (b) Surface plasmon resonance (BIAcore) was used to measure adherence of wild-type (WT) and ΔYidC and ΔP1 mutant cells to a salivary agglutinin-coated chip surface. The sensogram shows the change in resonance units following injection of a suspension of the indicated S. mutans background.
Effect of YidC1 and YidC2 on biofilm formation
The contribution of YidC1 and YidC2 to biofilm formation by S. mutans was evaluated using hexidium-iodide staining and confocal microscopy (Fig. 5). Biofilm formation under non-stress (pH 7.0) and acid-stress (pH 5.0) conditions was tested using growth in a semi-synthetic medium supplemented with 0.5 % sucrose on a glass surface. While growth of the ΔyidC1 mutant is not sensitive to acid (Hasona et al., 2005), biofilm formation by both the ΔyidC1 and ΔyidC2 mutants was notably altered at pH 5.0. The biofilms that did form under this acid-stress condition were too patchy for accurate thicknesses to be measured. Although both were impaired, the biofilm phenotypes of ΔyidC1 and ΔyidC2 mutants differed under non-stress conditions. Elimination of YidC2 resulted in a significantly thinner biofilm at 48 h compared with the wild-type, while elimination of YidC1 did not. Interestingly, biofilm thickness at the earlier 24 h time point was significantly greater in the ΔyidC1 mutant. This is consistent with the increased secretion and cell surface localization of the GtfB/C and P1 proteins that contribute to bacterial adhesion, an early event in biofilm formation.
Fig. 5.
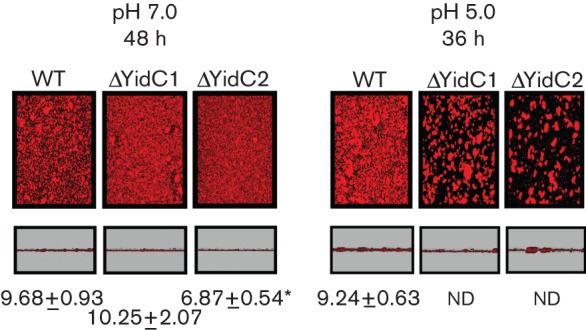
Biofilm formation by wild-type (WT) and yidC mutant strains. Biofilms were allowed to develop for up to 48 h (pH 7.0) or 36 h (pH 5.0) in THYE 0.5 % sucrose using hexidium iodide-labelled mid-exponential-phase cultures at the pH conditions indicated. Two areas from each biofilm that formed on the cover glass were visualized and the thicknesses of three sections through each area were measured. Representative images of overhead and side views of biofilms made by each strain are shown. The measured thickness values of biofilm (in µm; mean±sd) of three cross-sections from each of two fields per well performed for duplicate wells are given on the figure. For WT, ΔYidC1 and ΔYidC2 at pH 7.0 after development for 24 h the values were 7.33±0.50, 8.75±0.89* and 6.14±0.71*, respectively (significance given below). Each experiment was performed twice. Statistically significant differences compared with the wild-type are indicated. *P≤0.001. ND, not determined, growth was too patchy to accurately measure thickness.
Effect of YidC1 and YidC2 on S. mutans colonization and cariogenicity in rats
A previously described gnotobiotic rat model (Michalek et al., 1975) was used to test the effect of elimination of the YidC proteins of S. mutans on bacterial colonization and cariogenicity in vivo (see Table 3). Statistically significant differences were observed in the sulcal and proximal surface caries scores of animals infected with the ΔyidC1, ΔyidC2 and yidC2ΔC mutants compared with the NG8 parent strain. In addition, there were significantly fewer buccal caries in animals infected with the ΔyidC2 mutant. Animals from all of the experimental groups were colonized with S. mutans at the time they were killed for caries evaluation, 6 weeks post-infection, indicating that the differences in caries scores stemmed from a difference in virulence of the test strains, rather than a simple impairment in their ability to colonize. Interestingly, animals in the ΔyidC1 mutant group, in which there were significantly decreased caries scores, also had a significantly higher degree of colonization at the 6 week time point compared with the other groups.
Table 3. Colonization and cariogenicity of S. mutans wild-type and YidC deletion mutants in gnotobiotic rats.
n = 5 per group. Weights (g): Group A, 162±11; Group B, 149±11; Group C, 152±8; Group D, 145±11.4. There were no significant differences between the weight of any of the groups. Carious lesion location and severity: E, enamel; Ds, slight dentinal; Dm, moderate dentinal; Dx, extensive dentinal. Significant difference in mean caries score compared with the wild-type strain at each location: *P<0.01, **P<0.05, ***P<0.001.
| Group (strain) | Mean caries score | 10−6×c.f.u. per mandible | |||||||||||
| Buccal | Sulcal | Proximal | |||||||||||
| E | Ds | Dm | Dx | E | Ds | Dm | Dx | E | Ds | Dm | Dx | ||
| A (wild-type) | 13.8±0.4 | 11.4±0.4 | 6.2±0.4 | 3.8±0.6 | 19.8±0.7 | 15.8±0.7 | 7.8±0.9 | 2.6±0.7 | 7.2±0.5 | 4.4±0.4 | 0.0±0.0 | 0.0±0.0 | 11.8±1.5 |
| B (ΔYidC1) | 12.8±0.9 | 9.4±0.8 | 6.4±0.7 | 5.4±0.7 | 17.0±0.4*** | 12.0±0.9** | 6.0±0.4 | 2.0±0.8 | 4.0±0.0** | 2.2±0.9** | 0.2±0.2 | 0.0±0.0 | 23.1±2.7*** |
| C (ΔYidC2) | 9.8±1.0* | 6.8±0.9* | 4.6±0.7 | 3.4±0.7 | 16.2±0.2*** | 9.8±0.7*** | 5.4±0.8 | 1.4±0.5 | 4.8±0.8 | 0.4±0.4*** | 0.0±0.0 | 0.0±0.0 | 13.8±0.7 |
| D (YidC2ΔC) | 10.8±0.4 | 7.6±0.4** | 6.0±0.3 | 3.8±0.6 | 15.6±0.5** | 10.4±0.7* | 5.6±0.9 | 1.2±0.5 | 4.8±0.5 | 0.0±0.0*** | 0.0±0.0 | 0.0±0.0 | 8.4±1.3 |
Discussion
Streptococci have several redundant pathways for protein translocation and secretion, making it difficult to evaluate functions of individual proteins. There are two YidC homologues, including one (YidC2) that may overlap functionally with the SRP to facilitate co-translational translocation (Funes et al., 2009). In this study, a conditional expression system to allow controlled expression of S. mutans yidC2 upon deletion of yidC1 revealed that double deletion of YidC1 and YidC2 is lethal in this organism.
A number of strains were constructed to investigate functional differences between the two S. mutans YidC proteins and evaluated for growth under no stress, acid-stress and osmotic-stress conditions, as well as for whole-cell ATPase activity, a determinant for acid tolerance (Belli & Marquis, 1991). Results indicated a clear role of the C terminus of YidC2 in stress tolerance. When the C-terminal tail of YidC2 was deleted, bacteria exposed to acid or osmotic stress were similarly impaired for growth and ATPase activity (Funes et al., 2009) compared with those with a complete deletion of yidC2. On the other hand, when transformation efficiency was evaluated, deletion of its C terminus had less of an effect compared with elimination of YidC2 in its entirety. Complementation experiments with the yeast mitochondrial homologues Oxa1 and Cox18 have suggested that S. mutans YidC2 functions in at least two pathways, and that there are distinct functions of YidC2 that do and do not depend on an intact C terminus (Funes et al., 2009). An intermediate growth phenotype was seen in the yidC2ΔC partial mutant under non-stress conditions. Chimeric YidC1–C2 restored stress tolerance and ATPase activity to the ΔyidC2 mutant, while YidC2–C1 slowed growth even further compared with the un-complemented ΔyidC2 mutant. YidC2–C1 was unable to complement ATPase activity in the ΔyidC2 mutant, and this strain displayed a more pronounced defect in secretion with very little protein found in the culture supernatants. Hence, it is likely that the chimeric YidC2–C1 is non-functional and acts as a sink for YidC2 and possibly YidC1 substrates. Taken together, our results indicate that the C terminus is integral to each YidC protein’s individual function. Also of note is that in strain SP17, where yidC2 was expressed from the gtfA promoter, growth to wild-type levels under stress conditions and ATPase activity were not fully restored. This suggests that the yidC2 promoter and consequent level of yidC2 expression also contributes to stress tolerance. In studies of the LiaSR two-component system, yidC2 expression was upregulated in response to membrane stress through the LiaR response regulator (Suntharalingam et al., 2009). Despite the ability of chimeric YidC1–C2 to complement the ΔyidC2 mutant strain, knockout of the yidC1 gene in this background was not possible. It appears therefore that the overall level of YidC expression is important and use of the yidC2 promoter to express YidC1–C2 may have resulted in full complementation.
A study by Biswas & Biswas (2005) investigated the effects of deletion of the HtrA extracellular protease/chaperone and found that levels of extracellular GAPDH in culture media increased when HtrA was absent. Extracellular GAPDH activity was increased in both ΔyidC1 and ΔyidC2 mutants compared with the parental strain, with a greater effect seen with ΔyidC2. The yidC2ΔC mutant displayed GAPDH activity similar to the ΔyidC2 mutant. The YidC1–C2 protein was able to partially restore GAPDH activity toward wild-type levels in the ΔyidC2 mutant, indicating that the C-terminal tail is important for this YidC function but that this is not the whole story. It is possible that YidC1 and YidC2 are involved in the localization of HtrA. In that case, one would expect their disruption to have a similar effect on extracellular GAPDH to deletion of HtrA.
Cell surface localization of the adhesin P1 was negatively affected by deletion of yidC2, as shown by decreased immunoreactivity of anti-P1 antibodies with the ΔyidC2 strain. Conversely, elimination of yidC1 resulted in increased detection of P1 on the cell surface. This implies a balanced surface biogenesis mechanism that involves both YidC1 and YidC2, although it is unclear whether the increased immunodetection of P1 on the surface of the ΔyidC1 mutant reflects an increased abundance of P1 or a decrease in another surface component that allows improved exposure to the antibodies. The respective changes in anti-P1 antibody reactivity in the ΔyidC1 and ΔyidC2 mutant strains were associated with corresponding functional differences in P1-mediated adherence determined using surface plasmon resonance. There were similar effects on the levels of extracellular GtfB and GtfC detected in culture supernatants of the yidC mutants compared with the wild-type, with a decrease seen in the ΔyidC2 mutant and an increase in the ΔyidC1 mutant. The increased secretion of GtfB and GtfC in the ΔyidC1 mutant suggests that the increased immunodetection of P1 on the cell surface of this strain stems from a real increase in the amount of cell-surface-localized protein secreted rather than an effect on antibody accessibility.
The increase in colonization of rats with the ΔyidC1 strain compared with the wild-type is consistent with an increase in secretion of the GTFs and the P1 adhesin. The more rapid biofilm formation of the ΔyidC1 mutant under non-stress conditions is not surprising in light of these findings. Virulence of both the ΔyidC1 and ΔyidC2 mutants was negatively affected in these strains as evidenced by decreases in caries scores in rats and aberrant biofilm formation of the ΔyidC2 mutant under all conditions tested, and the ΔyidC1 mutant under acid stress at pH 5.0. While the ΔyidC2 mutant displays a growth defect under both non-stress and acid-stress conditions, the ΔyidC1 mutant does not (Hasona et al., 2005). Thus the pH 5.0 biofilm phenotype does not appear to stem from poor growth or acid sensitivity, but from another mechanism.
The findings that S. mutans YidC1 and YidC2 both contribute to pathways mediating cell surface biogenesis and secretion of extracellular proteins as well as the cellular response to stress furthers our understanding of the complex series of events involved in S. mutans pathogenesis, beginning with bacterial adhesion and colonization, and ending with the generation of acid end products and resilience in the face of environmental stress. Taken together, these results suggest that while YidC2 appears to play a more critical role in stress tolerance of S. mutans, YidC1 also contributes to its biology. YidC1 and YidC2 display unique functions and cooperate in an integrated balanced manner in protein secretion and cell wall biogenesis, thus providing an explanation for the inability to simultaneously eliminate both paralogues in this species.
Acknowledgements
We would like to thank Drs Lin Zeng and R. A. Burne (University of Florida) for advice in the development of a conditional expression system in S. mutans and for the gift of plasmids pBGK2 and pBGE. We would also like to acknowledge Dr Nathan Lewis (University of Florida) for his critical reading of the manuscript. This work was supported by the National Institute of Dental and Craniofacial Research grants F31 DE019605, R01 DE08007 and R01 DE09081
Abbreviations:
- SOE
splice overlap extension
- SRP
signal recognition particle
Footnotes
Three supplementary tables, two supplementary figures and a set of supplementary methods are available with the online version of this paper.
References
- Banas J. A., Vickerman M. M. (2003). Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med 14, 89–99. 10.1177/154411130301400203 [DOI] [PubMed] [Google Scholar]
- Belli W. A., Marquis R. E. (1991). Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol 57, 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Biswas I. (2005). Role of HtrA in surface protein expression and biofilm formation by Streptococcus mutans. Infect Immun 73, 6923–6934. 10.1128/IAI.73.10.6923-6934.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy N., Fiumera H. L., Dujardin G., Fox T. D. (2009). Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta 1793, 60–70. 10.1016/j.bbamcr.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady L. J., Piacentini D. A., Crowley P. J., Oyston P. C., Bleiweis A. S. (1992). Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun 60, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley N. D., Lee L. N., LeBlanc D. J. (1995). Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J Bacteriol 177, 5028–5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley P. J., Brady L. J., Michalek S. M., Bleiweis A. S. (1999). Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun 67, 1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley P. J., Svensäter G., Snoep J. L., Bleiweis A. S., Brady L. J. (2004). An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol Lett 234, 315–324. 10.1111/j.1574-6968.2004.tb09550.x [DOI] [PubMed] [Google Scholar]
- Dong Y., Palmer S. R., Hasona A., Nagamori S., Kaback H. R., Dalbey R. E., Brady L. J. (2008). Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J Bacteriol 190, 2458–2469. 10.1128/JB.01366-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S., Hasona A., Bauerschmitt H., Grubbauer C., Kauff F., Collins R., Crowley P. J., Palmer S. R., Brady L. J., Herrmann J. M. (2009). Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci U S A 106, 6656–6661. 10.1073/pnas.0809951106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons R. J., Cohen L., Hay D. I. (1986). Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors. Infect Immun 52, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez J. A., Crowley P. J., Brown D. P., Hillman J. D., Youngman P., Bleiweis A. S. (1996). Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol 178, 4166–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton I. R., Buckley N. D. (1991). Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol Immunol 6, 65–71. 10.1111/j.1399-302X.1991.tb00453.x [DOI] [PubMed] [Google Scholar]
- Hasona A., Crowley P. J., Levesque C. M., Mair R. W., Cvitkovitch D. G., Bleiweis A. S., Brady L. J. (2005). Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc Natl Acad Sci U S A 102, 17466–17471. 10.1073/pnas.0508778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasona A., Zuobi-Hasona K., Crowley P. J., Abranches J., Ruelf M. A., Bleiweis A. S., Brady L. J. (2007). Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J Bacteriol 189, 1219–1230. 10.1128/JB.01146-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman K. L., Pease L. R. (2007). Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2, 924–932. 10.1038/nprot.2007.132 [DOI] [PubMed] [Google Scholar]
- Henderson B., Martin A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79, 3476–3491. 10.1128/IAI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Nakamura K., Nishiguchi M., Yamane K. (1993). Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J Bacteriol 175, 4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H. F., Demuth D. R. (1997). Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol 23, 183–190. 10.1046/j.1365-2958.1997.2021577.x [DOI] [PubMed] [Google Scholar]
- Jenkinson H. F., Lamont R. J. (1997). Streptococcal adhesion and colonization. Crit Rev Oral Biol Med 8, 175–200. 10.1177/10454411970080020601 [DOI] [PubMed] [Google Scholar]
- Khalichi P., Cvitkovitch D. G., Santerre J. P. (2004). Effect of composite resin biodegradation products on oral streptococcal growth. Biomaterials 25, 5467–5472. 10.1016/j.biomaterials.2003.12.056 [DOI] [PubMed] [Google Scholar]
- Kiefer D., Kuhn A. (2007). YidC as an essential and multifunctional component in membrane protein assembly. Int Rev Cytol 259, 113–138. 10.1016/S0074-7696(06)59003-5 [DOI] [PubMed] [Google Scholar]
- Knox K. W., Hardy L. N., Wicken A. J. (1986). Comparative studies on the protein profiles and hydrophobicity of strains of Streptococcus mutans serotype c. J Gen Microbiol 132, 2541–2548. [DOI] [PubMed] [Google Scholar]
- Larson M. R., Rajashankar K. R., Patel M. H., Robinette R. A., Crowley P. J., Michalek S., Brady L. J., Deivanayagam C. (2010). Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of α- and PPII-helices. Proc Natl Acad Sci U S A 107, 5983–5988. 10.1073/pnas.0912293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Lee L. N., Abu-Al-Jaibat A. (1992). Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28, 130–145. 10.1016/0147-619X(92)90044-B [DOI] [PubMed] [Google Scholar]
- Ling E., Feldman G., Portnoi M., Dagan R., Overweg K., Mulholland F., Chalifa-Caspi V., Wells J., Mizrachi-Nebenzahl Y. (2004). Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin Exp Immunol 138, 290–298. 10.1111/j.1365-2249.2004.02628.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur W. P., Rhodin N. R., Seifert T. B., Oli M. W., Robinette R. A., Demuth D. R., Brady L. J. (2007). Characterization of epitopes recognized by anti-Streptococcus mutans P1 monoclonal antibodies. FEMS Immunol Med Microbiol 50, 342–353. 10.1111/j.1574-695X.2007.00260.x [DOI] [PubMed] [Google Scholar]
- Michalek S. M., McGhee J. R., Navia J. M. (1975). Virulence of Streptococcus mutans: a sensitive method for evaluating cariogenicity in young gnotobiotic rats. Infect Immun 12, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oli M. W., McArthur W. P., Brady L. J. (2006). A whole cell BIAcore assay to evaluate P1-mediated adherence of Streptococcus mutans to human salivary agglutinin and inhibition by specific antibodies. J Microbiol Methods 65, 503–511. 10.1016/j.mimet.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Phillips G. J., Silhavy T. J. (1992). The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359, 744–746. 10.1038/359744a0 [DOI] [PubMed] [Google Scholar]
- Robinette R. A., Oli M. W., McArthur W. P., Brady L. J. (2011). A therapeutic anti-Streptococcus mutans monoclonal antibody used in human passive protection trials influences the adaptive immune response. Vaccine 29, 6292–6300. 10.1016/j.vaccine.2011.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K. N., McArthur W. P., Bleiweis A. S., Brady L. J. (2003). Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein–protein interactions. Can J Microbiol 49, 350–356. 10.1139/w03-042 [DOI] [PubMed] [Google Scholar]
- Seifert T. B., Bleiweis A. S., Brady L. J. (2004). Contribution of the alanine-rich region of Streptococcus mutans P1 to antigenicity, surface expression, and interaction with the proline-rich repeat domain. Infect Immun 72, 4699–4706. 10.1128/IAI.72.8.4699-4706.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin A., Nickbarg E., Wooters J., Quazi S. A., Matsuka Y. V., Murphy E., Moutsatsos I. K., Zagursky R. J., Olmsted S. B. (2007). Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J Bacteriol 189, 1514–1522. 10.1128/JB.01132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam P., Senadheera M. D., Mair R. W., Lévesque C. M., Cvitkovitch D. G. (2009). The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol 191, 2973–2984. 10.1128/JB.01563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B., Willett N. P., Shockman G. D. (1975). Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadeboncoeur C., Brochu D., Reizer J. (1991). Quantitative determination of the intracellular concentration of the various forms of HPr, a phosphocarrier protein of the phosphoenolpyruvate: sugar phosphotransferase system in growing cells of oral streptococci. Anal Biochem 196, 24–30. 10.1016/0003-2697(91)90112-7 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Urbanus M. L., Ten Hagen-Jongman C. M., Nouwen N., Oudega B., Harms N., Driessen A. J., Luirink J. (2003). A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc Natl Acad Sci U S A 100, 5801–5806. 10.1073/pnas.0636761100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2001). Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45, 31–36. 10.1006/plas.2000.1498 [DOI] [PubMed] [Google Scholar]
- Wunder D., Bowen W. H. (2000). Effects of antibodies to glucosyltransferase on soluble and insolubilized enzymes. Oral Dis 6, 289–296. 10.1111/j.1601-0825.2000.tb00141.x [DOI] [PubMed] [Google Scholar]
- Zeng L., Burne R. A. (2009). Transcriptional regulation of the cellobiose operon of Streptococcus mutans. J Bacteriol 191, 2153–2162. 10.1128/JB.01641-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J., Tian H. F., Wen J. F. (2009). The evolution of YidC/Oxa/Alb3 family in the three domains of life: a phylogenomic analysis. BMC Evol Biol 9, 137. 10.1186/1471-2148-9-137 [DOI] [PMC free article] [PubMed] [Google Scholar]


