Abstract
Lung adenocarcinoma is one of the most frequent causes of malignant pleural effusions (MPE). The presence of MPE bears a poor prognosis. Although epigenetic changes are commonly related to human neoplasia, scarce date is available on patients with MPE. We aimed to estimate the prognostic value of DNA methylation of tumor suppressor genes from pleural fluid. Thirty patients with MPE due to lung adenocarcinoma were prospectively included. Methylation-specific (MS) PCR was used to study the methylation status of the promoter region of tumor suppressor genes p16/INK4a, MGMT, BRCA1 and RARβ in pleural fluid. Clinical data and survival were collected. Survival analysis was performed using Kaplan-Meier plots and Cox regression. Hypermethylation in at least one gene was detected in 25 patients (83.3%). On multivariate analysis factors significantly associated with shorter survival were the lack of hypermethylation in any of the studied genes (hazard ratio = 9.3; p = 0.001), Charlson index ≥ 3 (hazard ratio = 9.6, p = 0.002) and no oncological treatment (hazard ratio = 11.1; p < 0.001). Analysis of aberrant promoter hypermethylation of tumor suppressor genes may be useful in predicting prognosis, but further studies are needed to validate our findings.
Keywords: lung adenocarcinoma, malignant pleural effusion, DNA hypermethylation, prognosis, survival, treatment, comorbidity
Introduction
Malignant pleural effusion (MPE) is a common complication in neoplastic disease1-3 and can be caused by almost any tumor type. One of the most frequent underlying causes is broncogenic carcinoma, especially lung adenocarcinoma.1-3 The diagnosis of MPE is associated with advanced and disseminated disease and reduced life expectancy depending on the type and stage.1-3 The current TNM classification of lung cancer has re-classified patients with metastasis, discriminating those having only pleural involvement from those with distant metastasis, as prognosis and survival differences exist between these two groups.4 Biochemical characteristics of pleural fluid5-7 and comorbidity8 are also related to survival. However, studies are scarce and therefore clinical guidelines are inconclusive1-3 to establish which factors influence prognosis.
Patients with pleural effusion (PE) secondary to adenocarcinoma may show differences in their evolution, therefore it would be of great utility the use of molecular markers that, in combination to clinical variables, could predict patient’s evolution and guide clinical decision making.
Epigenetic alterations are known to contribute in the development, invasion and metastases of many human tumors. DNA methylation is one of the most common epigenetic mechanisms studied, including genome-wide hypomethylation and site-specific CpG island hypermethylation.9-11 Methylation profiles at specific CpG islands of tumor suppressor genes have been described for various cancers and are considered promising markers for early detection, tumor classification and chemotherapy response.9-12 The transcriptional inactivation caused by promoter hypermethylation affects genes involved in important cellular pathways, and has been suggested as a diagnostic tool for PE.13-16 However, there is practically no evidence of its relation with prognosis in patients with MPE.17
Therefore we aim to analyze the hypermethylation status of the promoter region of tumor suppressor genes p16/INK4a, MGMT (O6- methylguanine-DNA-methyltransferase), BRCA1 (breast-cancer susceptibility gene 1) and RARβ (retinoic acid receptor β) in pleural fluid, and its association with survival and other common clinical parameters in patients with MPE. The four genes studied have known roles in the pathogenesis of NSCLC and are presumably and potentially important for growth and cell cycle, DNA repair and carcinogenesis suppression.11,13-16
Results
Patient characteristics
Thirty patients with lung adenocarcinoma were included: 20 (66.7%) men with a median age of 63 y (IQR: 53.2–80.0 y). Twenty-one patients (70%) had history of smoking (10 current smokers and 11 former smokers), and six patients (20%) had other malignancies (one with laryngeal squamous cell carcinoma, two prostate carcinoma, one basal cell carcinoma, one prostate carcinoma and lung adenocarcinoma, and one basal cell carcinoma and laringeal carcinoma).
The size of PE was greater than 2/3 according to X-ray in 13 patients (43.3%) and less than 1/3 in five (16.7%) cases. At present six patients (20%) are alive, with a minimum follow-up of seven months. Median survival was 275.5 d (IQR: 65–457).
Methylation status and survival
Promoter hypermethylation of p16/INK4a, BRCA1, RARβ and MGMT in MPE of lung adenocarcinoma was detected in 6 (20.0%), 16 (53.3%), 12 (40.0%) and 12 (40.0%) patients, respectively. Table 1 summarizes the methylation status of each gene and survival data. According to the total number of methylated genes, 5 patients (16.7%) showed lack of methylation in any of the four genes studied while the remaining 25 (83.3%) presented hypermethylation in at least one gene. One methylated gene was registered in 8 patients (26.7%), 2 methylated genes in 13 patients (43.3%), and 3 methylated genes were found in 4 patients (13.3%). Regarding survival time, statistically significant differences (Kruskal-Wallis; p = 0.049) were detected in relation to the number of methylated genes, suggesting that the number of methylated genes may be related to different survival times. The U Mann Whitney test for the pair-wise comparisons revealed statistically significant differences in survival for patients bearing 0 or 1 methylated gene (p = 0.013) and 1 or 3 methylated genes (p = 0.042).
Table 1. Methylation status of the genes studied and survival for the 30 patients studied.
| Case | p16/INKa | BRCA1 | RARβ | MGMT | Hypermethylation in any gene | Survival (days) |
|---|---|---|---|---|---|---|
|
1 |
|
X |
X |
|
X |
410 |
|
2 |
|
|
X |
X |
X |
322 |
|
3 |
|
X |
|
|
X |
200 |
|
4 |
|
|
|
|
|
55 |
|
5 |
|
X |
|
|
X |
174 |
|
6 |
X |
|
|
|
X |
412 |
|
7 |
|
X |
|
|
X |
374 |
|
8 |
|
|
X |
|
X |
777 |
|
9 |
|
|
|
|
|
386 |
|
10 |
|
|
X |
X |
X |
526 |
|
11 |
|
X |
|
|
X |
446 |
|
12 |
X |
|
|
|
X |
509 |
|
13 |
|
X |
X |
|
X |
36 |
|
14 |
X |
X |
|
X |
X |
230 |
|
15 |
|
X |
|
X |
X |
249 |
|
16 |
|
X |
|
X |
X |
89 |
|
17 |
X |
X |
|
X |
X |
59 |
|
18 |
|
X |
|
X |
X |
9 |
|
19 |
|
|
|
|
|
15 |
|
20 |
|
|
|
|
|
14 |
|
21 |
|
X |
X |
X |
X |
102 |
|
22 |
X |
|
X |
|
X |
85 |
|
23 |
|
X |
X |
X |
X |
302 |
|
24 |
|
|
|
X |
X |
1022 |
|
25 |
|
X |
X |
|
X |
419 |
|
26 |
|
X |
X |
|
X |
560 |
|
27 |
|
|
|
|
|
67 |
|
28 |
X |
|
X |
|
X |
3 |
|
29 |
|
|
X |
X |
X |
751 |
| 30 | X | X | X | 491 |
The boxes marked with a “X” indicate methylation, whereas white boxes correspond to unmethylation,
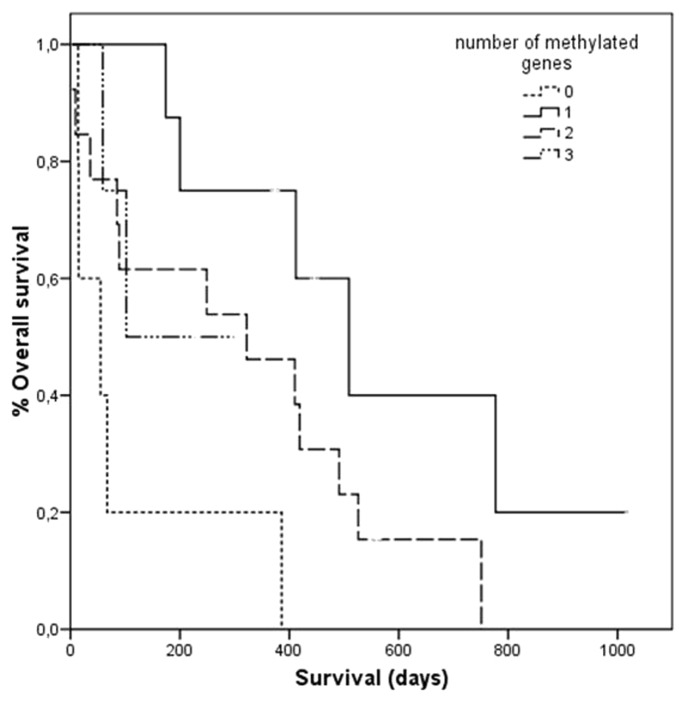
Table 2 shows the median survival for each potential prognostic factor obtained from Kaplan-Meier analyses. No differences in survival were observed for methylation of p16/INK4a, BRCA1, RARβ or MGMT. However, the lack of hypermethylation in any of the genes studied was surprisingly related to a shorter survival (median survival of 55 d vs. 322 d; log-rank p = 0.003). Regarding the number of methylated genes, statistically significant differences were observed with 0, 1, 2 or 3 methylated genes and survival (log-rank p = 0.021). As shown in Figure 1, patients with no methylation had shorter survival compared with patients with 1, 2 or 3 methylated genes. Conversely, survival was gradually shortened by the presence of 1, 2 or 3 hypermethylated genes. Cox multivariate analysis corroborated that the lack of hypermethylation in any of the genes significantly correlated with survival (p = 0.001; hazard ratio: 9.3, 95% CI 2.4–35.2), suggesting that hypermethylation in at least one gene is related to a better prognosis (Table 3).
Table 2. Univariate analysis of the survival predictive variables in patients with malignant pleural effusion of lung adenocarcinoma.
| |
Survival (days)* |
|
|
|---|---|---|---|
| Variable | YES | NO | p |
|
Sex (men) Age (> 67 y) p16/INK4a HPM |
215 (61 - 439) 102 (0 - 237) 157 (45 - 436) |
348 (80 - 582) 412 (359 - 465) 312 (72 - 479) |
0.5+ 0.1^ 0.5^ |
|
BRCA1 HPM |
239 (92 - 416) |
354 (45 - 582) |
0.7^ |
|
RARβ HPM |
366 (89 - 551) |
215 (58 - 420) |
0.3^ |
|
MGMT HPM Lack of HPM in any gene HPM in at least 2 genes HPM in at least 3 genes |
275 (92 - 517) 55 (14 - 226) 322 (60 - 508) 166 (69 - 284) |
287 (50 - 425) 322 (95 - 500) 230 (63 - 429) 348 (64 - 495) |
0.4^ 0.003+ 0.7^ 0.9^ |
|
pH (≤ 7.28) |
202 (54 - 347) |
374 (85 - 509) |
0.5^ |
|
Glucose (≤ 60 mg/dL) |
78 (28 - 194) |
380 (89 - 504) |
0.1^ |
|
LDH (≥ 1000 UI/L) |
302 (89 -486) |
275 (56 - 437) |
0.5^ |
|
Karnofsky index (≤ 70) |
85 (6 - 468) |
302 (78 - 468) |
0.1+ |
|
Charlson index (≥ 3) |
142 (28 - 236) |
348 (65 - 495) |
0.04 |
|
No oncological treatment |
15 (9 - 102) |
374 (174 - 509) |
< 0.001+ |
| Distant metastasis | 211 (40 - 473) | 348 (97 - 466) | 0.2^ |
HPM, hypermethylation of the promoter region; LDH, lactate dehydrogenase. *, Expressed as median (interquartile range). ^Breslow or +Log-Rank
Figure 1.
Overall survival of patients with malignant pleural effusion according to the number of hypermethylated genes.
Table 3. Multivariate analysis of the survival predictive variables in patients with malignant pleural effusion of lung adenocarcinoma.
| Variable | HR | CI 95% | p |
|---|---|---|---|
|
Lack of hypermethylation in any gene |
9.3 |
2.4 – 35.2 |
0.001 |
|
Charlson index (≥ 3) |
9.6 |
2.3 – 39.9 |
0.002 |
| No oncological treatment | 11.1 | 3.3 – 37.2 | < 0.001 |
HR, hazard ratio
Clinical characteristics and survival
Biochemical characteristics of the pleural fluid including pH ≤ 7.28, glucose ≤ 60 mg/dL and elevated LDH showed no correlation with median survival (Table 2), and therefore were not associated with prognosis. We found no relationship between a Karnofsky index ≤ 70 and survival, whereas a Charlson index ≥ 3 correlated significantly with a worse prognosis (p = 0.04).
Regarding oncological treatment, seven patients (23.3%) did not receive any treatment. On the other hand, the presence of distant metastasis (other than pleural) was not a significant prognosis factor. Cox multivariate model including Charlson’s index and oncological treatment showed significant hazard ratios greater than 9 for both variables (Table 3).
Discussion
Lung adenocarcinoma is one of the most common metastatic tumors to the pleura.1-3 The diagnosis of MPE is generally related to advanced disease and poor prognosis. Consequently, clinical guidelines1-3 state that treatment is essentially palliative, and emphasize that general health condition together with tumor histology are factors affecting prognosis in these patients.1 A biomarker detectable in pleural fluid capable of predicting patient outcome and guiding treatment would be of great utility.
Methylation of the promoter region of certain tumor suppressor genes has been proposed as potential markers for various types of cancer,9-11 including lung cancer.12 In our study a panel of four tumor suppressor genes related to various cell functions were analyzed to confirm their importance and relation with malignant pleural effusion due to lung adenocarcinoma. In general, hypermethylation studies involving tissue specimens from patients with lung cancer have reported contrasting and inconclusive results regarding patient outcome. The authors20 reported that hypermethylation of p16/INK4a was associated with shorter survival in patients with lung adeno and squamous cell carcinoma, while other studies in Korean21,22 and American23 patients found no correlation with survival. These latter studies also found no differences in survival with MGMT methylation, although Hayashi et al.24 found that smokers with methylated MGMT had worse outcome. Regarding RARβ, both studies performed in Korean patients reported methylation of this gene as a risk factor related to poor survival.21,25
Certain genes such as DAPK, RASSF1A, p15, p16/INK4a, RARβ, MGMT and APC, are known to be methylated in many cancers, and have been studied in patients with MPE.13-16 In these studies, methylation analyses (combined or not with cytology) improved diagnosis of MPE13-16 avoiding the need of other invasive diagnostic tests. However, the application of methylation as a prognostic tool is still poorly explored in patients with MPE.17
In our study, methylation of p16/INK4a, BRCA1, RARβ and MGMT in pleural fluid was not significantly related to median survival. However, patients with hypermethylation of p16/INK4a and BRCA1 showed shorter survival. In contrast, hypermethylation of RARβ was related to longer survival time. In the case of MGMT, survival was barely the same for the methylated or unmethylated status.
According to the number of methylated genes, statistically significant differences in survival were detected in patients that lacked hypermethylation in any gene. In these patients, the transcriptional inactivation caused by promoter hypermethylation in at least one gene resulted in a much better prognosis. This was corroborated in the multivariate analysis, showing a hazard ratio of 9.3 (95% CI 2.4 – 35.2). Considering that the transcriptional inactivation caused by promoter hypermethylation results in loss of expression, in the case of tumor suppressor genes, it seems contradictory to find longer survival times in this group of patients. However, methylation should be considered a continuous and gradual process were incomplete suppression is possible and can result in active gene expression. In this regard, Safar et al.23 reported that in certain tumor suppressor genes, the presence of hypermethylation seems to indicate a protective effect, in accordance to our findings concerning RARβ and the variable at least one hypermethylated gene. It should be noted that besides methylation, other mechanisms may lead to gene silencing like mutations or transcription modulators, among others.
When we analyzed the methylation status of the four genes studied codified as 0, 1, 2 or 3 hypermethylated genes, two tendencies are found: first, the absence of hypermethylation seems to be related to a very poor prognosis, while the presence of hypermethylated genes suggests favorable clinical outcome. Second, the progressive increase in the number of hypermethylated genes from 1 to 3 correlated with shorted survival times. The former observation may seem contradictory, however at first glance this could partially be explained by the fact that 40% of the patients with absence of hypermethylation did not receive oncological treatment compared with only 20% of the patients with at least one hypermetthylated gene. To determine its relation to treatment methylation frequencies were compared in patients with or without oncological treatment, finding no differences (data not shown). Therefore the first tendency observed seems more concerned with the gradual methylation process. On the other hand, the latter finding on shorter survival times with increasing methylation supports the concept that aberrant promoter methylation of tumor suppressor genes [p16/INK4a (cell-cycle regulation), MGMT and BRCA1 (DNA repair-related) and RARβ (thyroid-steroid hormone receptor)] is associated with a loss of gene function that can lead to a selective growth advantages to neoplastic cells.
As previously stated, to date only one study investigated the utility of methylation for predicting prognosis in MPE patients with lung adenocarcinoma. This study was conducted in 34 Japanese lung cancer patients and found, in contrast to us, similar survival in patients with methylation in at least one gene (MGMT, p16/INK4a, RASSF1A, DAPK or RARβ) and patients without methylation in a univariate analysis.17 Since this survival difference in our cohort resulted statistically significant in both uni- and multivariate analyses, discrepancies observed between our study and Katayama’s17 may be related to ethnic differences instead of experimental variations given that we used the same primer designs and therefore analyzed the same CpG sites contained in the promoter region of p16/INK4a, RARβ and MGMT.
Regarding hypermethylation of BRCA1, no studies on pleural fluid have so far included this gene. BRCA1 plays an important role in the repair of DNA double-strand breaks and epigenetic silencing of this gene promoter occurs frequently in many human cancers resulting in loss of heterozygosity and chromosome instability.25 Supporting evidence exists for the involvement of BRCA1 in non-small cell lung cancer tumorigenesis as a tumor suppressor promoter and hypermethylation as the predominant mechanism for deregulation, though BRCA1 was rarely hypermethylated in tumor specimens from patients with non-small cell lung cancer.12
Epigenetic alterations are also modified by diet, stress, aging, environmental exposures or hormones.26 Therefore, environmental factors, besides promoting tumor development, are thought to be related not only to the phenotype of the disease, but also to potential biomarkers27 which may be responsible for the differences between our results and others. However, determination of the methylation status (methylated or unmethylated) used in this study may be improved with the use of a methylation quantification strategy through quantitative real-time methylation-specific PCR. This could allow more accurate results and the estimation of a methylation percentage cut-off for each gene and their combination.
In relation to the biochemical characteristics of pleural fluid, none of the parameters studied showed statistically significant correlation with survival. Low pH, low glucose or elevated LDH are considered by many authors to correlate with poor prognosis,1,3,5,6 although survival is much more strongly related to tumor histology.7 In a systematic review Heffner et al.2 affirms a greater relationship between low pH and pleurodesis failure compared with low pH and shorter survival time. Regarding the variable Karnofsky index, and in contrast to other authors,7,8 we found no relation with survival using a cut-off of 70. On the other hand, comorbidity based on the Charlson index strongly predicted prognosis.
As expected, survival in patients with MPE due to lung cancer was correlated strongly with treatment. The indication of treatment was based on the oncologist’s criterion together with the patient’s willingness, independently of our study. Patients receiving oncological treatment showed significantly longer survival compared with untreated patients, independently of the methylation status of the four genes studied and other biochemical/clinical characteristics.
The current TNM classification, based on prognosis, separates patients with broncogenic carcinoma and metastatic disease limited to pleura (M1a) from those with distant metastasis (M1b).4 However, we did not find differences in survival between patients with distant vs. pleural metastasis.
Our study is the first to use a multivariate analysis to estimate prognosis combining methylation status, biochemical and clinical factors influencing survival in patients with MPE of lung adenocarcinoma. The hazard ratios observed for the factors lack of hypermethylation in any gene, Charlson index and no oncological treatment are considerably high and therefore are robustly related to survival in patients with MPE related to lung adenocarcinoma. Inclusion of patients from a single hospital in addition to the exclusion of patients receiving oncological treatment at the time of pleural effusion diagnosis largely explains the reduced number of cases in our study; moreover, patients with MPE related solely to lung adenocarcinoma were included. To confirm our results, further studies with larger populations of Caucasian patients and perhaps more candidate genes for methylation analysis using a quantitative approach are needed.
Material and Methods
Study subjects
Consecutive, unselected patients with MPE secondary to lung adenocarcinoma were prospectively enrolled. All patients were evaluated in the Bronchopleural Unit of Complexo Hospitalario Universitario de Vigo (CHUVI) over a three-year period.
Only patients with a confirmed diagnosis of MPE (presence of malignant cells in pleural fluid or in pleural transparietal biopsy or thoracoscopy)1-3 were included in the study. Patients receiving chemotherapy at the time of diagnosis or with a history of intrapleural instillation with symphyseal or chemotherapeutic agents were excluded.
Clinical and epidemiological characteristics, biochemical analysis of pleural fluid [pH, glucose and lactate dehydrogenase (LDH)], presence of other metastatic sites at the time of diagnosis of MPE and oncological treatment after diagnosis (at least one chemotherapy cycle and/or treatment for at least one month with epidermal growth factor receptor tyrosine kinase inhibitors) were recorded. Patient’s health status and comorbidity were estimated through the Karnofsky and Charlson indexes,18,19 respectively.
Clinical-radiological follow-up was accomplished for all patients until death or submission of this manuscript.
The local Ethical Committee approved this study. All patients provided informed consent.
Collection of samples and methylation-specific-PCR
During thoracocentesis or pleural biopsy 10 mL of pleural fluid were obtained for molecular analysis. The pleural fluid was centrifuged at 1,600 g for 15 min and stored in 1 mL aliquots at -20°C until testing.
DNA methylation status of the promoter regions of p16/INK4a, MGMT, BRCA1 and RARβ was assessed with methylation-specific (MS) PCR. Cell-free DNA from pleural fluid was extracted using QIAamp® DNA Blood Mini Kit (Qiagen; Valencia, CA, USA) according to manufacturer’s instructions. Sodium bisulfite modification and purification was done using EZ DNA Methylation-Direct kit (Zymo Research; Orange, CA, USA). Briefly, 20 μL of DNA were bisulfite-treated according to manufacturer’s instructions, and finally resuspended and eluted in 20 μL of elution buffer. Modified DNA was stored at -80°C until used. A positive methylated control treated with CpG methyltransferase (M.SssI; New England Biolabs, Ipswich, MA, USA) and a positive unmethylated control (untreated DNA) were included in each bisulfite treatment and PCR.
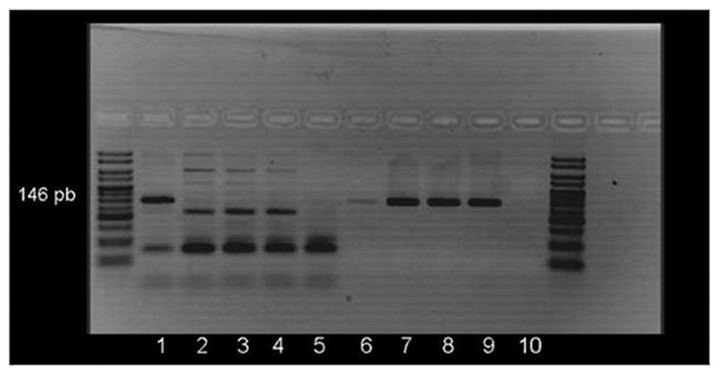
PCR was performed with primers specifically designed for methylated and unmethylated promoter sequences of p16/INK4a, MGMT, BRCA1 and RARβ as previously described20,21 and summarized in Table 4. To confirm bisulfite-treated DNA integrity both methylated and unmethylated specific PCRs were accomplished in parallel for all samples. Figure 2 shows a representative example of MS-PCR detection. PCR products were visualized in 3% agarose gels containing ethidium bromide.
Table 4. Primer sequences and characteristics.
| Gene | Methylated primer | Unmethylated primer | Annealing temp/ pb |
|---|---|---|---|
|
p16/INK4a |
F: TATTAGAGGGTGGG GCGGATCGC R: GACCCCGAACCGCGA CCGTAA |
F:TTATTAGAGGGTGGG GTGGATTGT R:CAACCCCAAACCACA ACCATAA |
65°C met (150 pb) 65°C unmet (151 pb) |
|
BRCA1 |
F:TCGTGGTAACGGAAA AGCGC R:AAATCTCAACGAACTC ACGCCG |
F: TTGGTTTTTGTGGTA ATGGAAAAGTGT R: CAAAAAATCTCAACA AACTCACACCA |
62°C met (75 pb) 62°C unmet (86 pb) |
|
RARβ |
F: CGAGAACGCGAGCG ATTCG R: GACCAATCCAACCGA AACGA |
F:TTGAGAATGTGAGTG ATTTGA R: AACCAATCCAACCAA AACAA |
59°C met (146 pb) 59°C unmet (146 pb) |
| MGMT | F:TTTCGACGTTCGTAG GTTTTCGC R: GCACTCTTCCGAAAA CGAAACG |
F: TTGTGTTTTGATGT TTGTAGGTTTTTGT R: AACTCCACACTCTTC CAAAAACAAAACA |
66°C met (81 pb) 66°C unmet (93 pb) |
Figure 2. Methylated and unmethylated RARβ PCR amplification of two patients with malignant pleural effusion. Lanes 1–5 correspond to the methylated-specific PCR and lanes 6–10 to the unmethylated-specific PCR. Lanes 1 and 6: methylated control; 2 and 7: unmethylated control; 3 and 4: two samples (no 146 pb band appears, indicating no methylation); 8 and 9: the same samples (a 146 pb appears indicating no methylation); 5 and 10: negative control.
The methylation status of each sample and gene was verified twice, in at least two independent PCRs using different aliquots of the pleural fluid sample (DNA extraction and bisulfite modification for each PCR). PCRs were performed in a blind fashion.
Statistical analysis
Statistical analyses were performed with the SPSS package (v16.0); tests were two-sided and p-values < 0.05 were considered significant. Results for qualitative variables are expressed as percentage or absolute frequency, and for numeric variables as median and interquartile range (IQR). For the evaluation of survival, variables that could influence survival were dichotomized as in previous studies.5,8,18,19
Survival curves were calculated by the Kaplan-Meier method and the significance of differences was estimated by the log-rank or breslow test. Multivariate analysis using Cox model was used to estimate hazard ratios.
Acknowledgments
This study was supported by grant PI081100 (FIS-FEDER), Research Intensification Activity in the National Health System from Fondos de Investigación Sanitaria (FIS-ISCIII), and grant PS08/18 from Xunta de Galicia - Consellería de Sanidade.
Glossary
Abbreviations:
- MPE
malignant pleural effusion
- PE
pleural effusion
- MGMT
O6- methylguanine-DNA-methyltransferase
- BRCA1
breast-cancer susceptibility gene 1
- RARβ
retinoic acid receptor β
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/22004
References
- 1.Antony VB, Loddenkemper R, Astoul P, Boutin C, Goldstraw P, Hott J, et al. ERS/ATS Statement: Management of malignant pleural effusion. Eur Respir J. 2001;18:402–19. doi: 10.1183/09031936.01.00225601. [DOI] [PubMed] [Google Scholar]
- 2.Heffner JE, Klein JS. Recent advances in the diagnosis and management of malignant pleural effusions. Mayo Clin Proc. 2008;83:235–50. doi: 10.4065/83.2.235. [DOI] [PubMed] [Google Scholar]
- 3.Roberts ME, Neville E, Berrisford RG, Antunes G, Ali NJ, BTS Pleural Disease Guideline Group Management of a malignant pleural effusion: British Thoracic Society pleural disease guideline 2010. Thorax. 2010;65(Suppl 2):ii32–40. doi: 10.1136/thx.2010.136994. [DOI] [PubMed] [Google Scholar]
- 4.Postmus PE, Brambilla E, Chansky K, Crowley J, Goldstraw P, Patz EF, Jr., et al. International Association for the Study of Lung Cancer International Staging Committee. Cancer Research and Biostatistics. Observers to the Committee. Participating Institutions The IASLC Lung Cancer Staging Project: proposals for revision of the M descriptors in the forthcoming (seventh) edition of the TNM classification of lung cancer. J Thorac Oncol. 2007;2:686–93. doi: 10.1097/JTO.0b013e31811f4703. [DOI] [PubMed] [Google Scholar]
- 5.Bielsa S, Salud A, Martínez M, Esquerda A, Martín A, Rodríguez-Panadero F, et al. Prognostic significance of pleural fluid data in patients with malignant effusion. Eur J Intern Med. 2008;19:334–9. doi: 10.1016/j.ejim.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Heffner JE, Heffner JN, Brown LK. Multilevel and continuous pleural fluid pH likelihood ratios for evaluating malignant pleural effusions. Chest. 2003;123:1887–94. doi: 10.1378/chest.123.6.1887. [DOI] [PubMed] [Google Scholar]
- 7.Ozyurtkan MO, Balci AE, Cakmak M. Predictors of mortality within three months in the patients with malignant pleural effusion. Eur J Intern Med. 2010;21:30–4. doi: 10.1016/j.ejim.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Burrows CM, Mathews WC, Colt HG. Predicting survival in patients with recurrent symptomatic malignant pleural effusions: an assessment of the prognostic values of physiologic, morphologic, and quality of life measures of extent of disease. Chest. 2000;117:73–8. doi: 10.1378/chest.117.1.73. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 10.Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–11. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303:21–8. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Ibáñez de Cáceres I, Cairns P. Methylated DNA sequences for early cancer detection, molecular classification and chemotherapy response prediction. Clin Transl Oncol. 2007;9:429–37. doi: 10.1007/s12094-007-0081-9. [DOI] [PubMed] [Google Scholar]
- 13.Benlloch S, Galbis-Caravajal JM, Martín C, Sanchez-Paya J, Rodríguez-Paniagua JM, Romero S, et al. Potential diagnostic value of methylation profile in pleural fluid and serum from cancer patients with pleural effusion. Cancer. 2006;107:1859–65. doi: 10.1002/cncr.22190. [DOI] [PubMed] [Google Scholar]
- 14.Brock MV, Hooker CM, Yung R, Guo M, Han Y, Ames SE, et al. Can we improve the cytologic examination of malignant pleural effusions using molecular analysis? Ann Thorac Surg. 2005;80:1241–7. doi: 10.1016/j.athoracsur.2005.05.088. [DOI] [PubMed] [Google Scholar]
- 15.Chen ML, Chang JH, Yeh KT, Chang YS, Chang JG. Epigenetic changes in tumor suppressor genes, P15, P16, APC-3 and E-cadherin in body fluid. Kaohsiung J Med Sci. 2007;23:498–503. doi: 10.1016/S1607-551X(08)70007-X. [DOI] [PubMed] [Google Scholar]
- 16.Katayama H, Hiraki A, Aoe K, Fujiwara K, Matsuo K, Maeda T, et al. Aberrant promoter methylation in pleural fluid DNA for diagnosis of malignant pleural effusion. Int J Cancer. 2007;120:2191–5. doi: 10.1002/ijc.22576. [DOI] [PubMed] [Google Scholar]
- 17.Katayama H, Hiraki A, Fujiwara K, Matsuo K, Maeda T, Chikamori K, et al. Aberrant promoter methylation profile in pleural fluid DNA and clinicopathological factors in patients with non-small cell lung cancer. Asian Pac J Cancer Prev. 2007;8:221–4. [PubMed] [Google Scholar]
- 18.Karnofsky DA, Abelman WH, Craver LF, Burchenal JH. The use of nitrogen mustards in the palliative treatment of cancer. Cancer. 1948;1:634–56. doi: 10.1002/1097-0142(194811)1:4<634::AID-CNCR2820010410>3.0.CO;2-L. [DOI] [Google Scholar]
- 19.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–83. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 20.Gu J, Berman D, Lu C, Wistuba II, Roth JA, Frazier M, et al. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006;12:7329–38. doi: 10.1158/1078-0432.CCR-06-0894. [DOI] [PubMed] [Google Scholar]
- 21.Kim YT, Lee SH, Sung SW, Kim JH. Can aberrant promoter hypermethylation of CpG islands predict the clinical outcome of non-small cell lung cancer after curative resection? Ann Thorac Surg. 2005;79:1180–8, discussion 1180-8. doi: 10.1016/j.athoracsur.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 22.Kim YT, Park SJ, Lee SH, Kang HJ, Hahn S, Kang CH, et al. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg. 2005;130:1378–84. doi: 10.1016/j.jtcvs.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Safar AM, Spencer H, 3rd, Su X, Coffey M, Cooney CA, Ratnasinghe LD, et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin Cancer Res. 2005;11:4400–5. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi H, Yazawa T, Okudela K, Nagai J, Ito T, Kanisawa M, et al. Inactivation of O6-methylguanine-DNA methyltransferase in human lung adenocarcinoma relates to high-grade histology and worse prognosis among smokers. Jpn J Cancer Res. 2002;93:184–9. doi: 10.1111/j.1349-7006.2002.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Horiuchi A, Imai T, Ohira S, Itoh K, Nikaido T, et al. Expression of BRCA1 protein in benign, borderline, and malignant epithelial ovarian neoplasms and its relationship to methylation and allelic loss of the BRCA1 gene. J Pathol. 2004;202:215–23. doi: 10.1002/path.1507. [DOI] [PubMed] [Google Scholar]
- 26.Bell CG, Beck S. The epigenomic interface between genome and environment in common complex diseases. Brief Funct Genomics. 2010;9:477–85. doi: 10.1093/bfgp/elq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herceg Z, Vaissière T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics. 2011;6:804–19. doi: 10.4161/epi.6.7.16262. [DOI] [PubMed] [Google Scholar]