Abstract
Background
Cyanobacteria can form massive toxic blooms in fresh and brackish bodies of water and are frequently responsible for the poisoning of animals and pose a health risk for humans. Anabaena is a genus of filamentous diazotrophic cyanobacteria commonly implicated as a toxin producer in blooms in aquatic ecosystems throughout the world. The biology of bloom-forming cyanobacteria is poorly understood at the genome level.
Results
Here, we report the complete sequence and comprehensive annotation of the bloom-forming Anabaena sp. strain 90 genome. It comprises two circular chromosomes and three plasmids with a total size of 5.3 Mb, encoding a total of 4,738 genes. The genome is replete with mobile genetic elements. Detailed manual annotation demonstrated that almost 5% of the gene repertoire consists of pseudogenes. A further 5% of the genome is dedicated to the synthesis of small peptides that are the products of both ribosomal and nonribosomal biosynthetic pathways. Inactivation of the hassallidin (an antifungal cyclic peptide) biosynthetic gene cluster through a deletion event and a natural mutation of the buoyancy-permitting gvpG gas vesicle gene were documented. The genome contains a large number of genes encoding restriction-modification systems. Two novel excision elements were found in the nifH gene that is required for nitrogen fixation.
Conclusions
Genome analysis demonstrated that this strain invests heavily in the production of bioactive compounds and restriction-modification systems. This well-annotated genome provides a platform for future studies on the ecology and biology of these important bloom-forming cyanobacteria.
Keywords: Cyanobacteria, Anabaena, Mobile genetic elements, Insertion sequences, Biosynthetic gene clusters, Restriction-modification system, nifH excision element
Background
Cyanobacteria are evolutionarily important prokaryotic organisms that created the oxygenic atmosphere on Earth via oxygenic photosynthesis and were the progenitors of chloroplasts in eukaryotic algae and plants [1]. Cyanobacteria often dominate phytoplankton as surface scum in freshwater lakes and brackish water during the summer months [2]. A small number of cyanobacterial genera are typically involved in bloom formation [2]. Gas vesicles are common in planktonic cyanobacteria and allow the organisms to regulate their buoyancy [3]. Bloom-forming cyanobacteria produce an array of potent hepatotoxins and neurotoxins [4]. Microcystins are commonly reported hepatotoxic heptapeptides that inhibit eukaryotic protein phosphatases 1 and 2A [2]. Toxic blooms are responsible for the toxicoses of wild and domestic animals [5] and are a health risk for humans through the consumption or recreational use of water [6].
Anabaena is a genus of filamentous nitrogen-fixing cyanobacteria [7] that is especially common in aquatic environments, both in fresh and brackish waters worldwide [8,9]. Nitrogen fixation occurs in specialized cells called heterocysts that differentiate from the vegetative cells [10]. This property combined with photosynthesis makes Anabaena cyanobacteria autotrophic organisms that are able to live in a wide range of environments. Strains of the planktonic Anabaena genus are some of the most common cyanobacteria capable of forming blooms [4]. Blooms of Anabaena are a serious health risk, due to the production of a range of toxins such as microcystins, anatoxins and saxitoxins [4,11].
Cyanobacteria, including Anabaena, are prolific sources of natural products, many of which have biotechnological and biomedical importance [11]. In recent years, many new compounds and their biosynthetic pathways have been discovered [11,12]. The cyanobacterial hepatotoxins, microcystins and nodularins, are the end products of nonribosomal biosynthetic pathways [13]. Recently, it has been shown that cyanobacteria also use diverse ribosomally encoded pathways for the production of small linear and cyclic peptides [14,15]. To understand the role of these bioactive compounds in cyanobacteria, as well as the biotechnological exploitation of the biosynthetic machinery used to assemble them, more information on the regulation and association of these biosynthetic pathways with other metabolic processes is needed.
Only a small number of genomes for planktonic bloom-forming cyanobacteria are known, including Microcystis aeruginosa PCC 7806 and NIES-843 [16,17], Planktothrix rubescens NIVA CYA 98 [18], Cylindrospermopsis raciborskii CS-505 and Raphidiopsis brookii D9 [19], which produce microcystins, cylindrospermopsin or saxitoxin. Here, we present the complete genome of Anabaena sp. strain 90, a bloom-forming, microcystin-producing strain originating from a freshwater lake in Finland.
Results
Genome overview
The Anabaena sp. 90 genome was assembled with Sanger reads that were sequenced from libraries with different size inserts (2, 6 and 40 kb) and amounted to a 12.5 X depth of coverage. The remaining physical gaps that were derived from the unclonable regions were linked through combinatorial multiplex PCR screening of primers designed from the contig ends. The genome consists of five circular replicons, two chromosomes and three plasmids (Figure 1, Table 1). The total size of the genome amounted to 5,305,675 bp with an average G+C content of 38.1%. The quality of the genome sequence was very high and the estimated overall sequence error of the genome was 0.12 bp (Table 1). A total of 4,738 ORFs were annotated with putative functions assigned to 2,954 (62.35%) ORFs from manual annotation. The remaining 1,784 (37.65%) were assigned as hypothetical ORFs (see Additional file 1: Table S1). They were further subgrouped as 480 (10.13%) conserved hypothetical proteins that have more than 30 counterparts in other bacterial genomes, and 205 (4.33%) unique proteins that have no full-length counterparts (see Methods). In addition, there are 1099 (23.19%) hypothetical ORFs that lie in between, having few counterparts in other genomes. Five rRNA operons were identified and dispersed throughout chromosome I, two in the leading and three in the lagging strand (Table 1). They have nearly identical rRNA genes. Sequence variations in the spacer regions separate them into two groups. Two operons with consecutive tRNAs organized as 16S-trnI-trnA-23S-5S form one group. Members in another group have no tRNA genes. A total of 44 tRNAs were distributed over both chromosome I (40) and II (4).
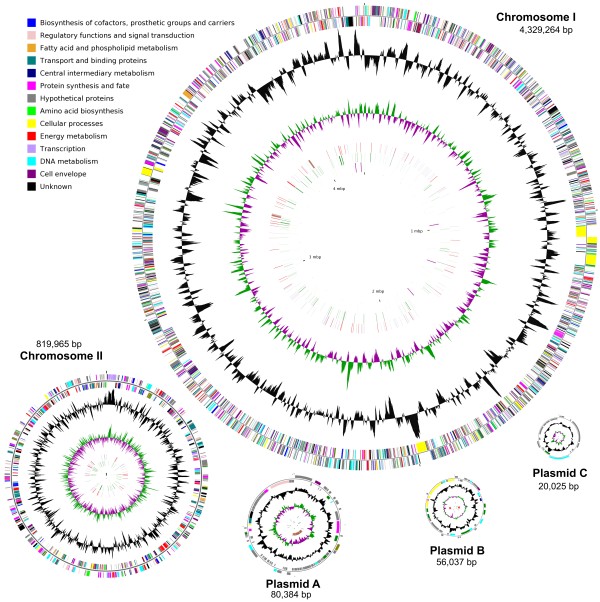
Figure 1.
Circular demonstration of the two chromosomes and three plasmids of the Anabaena sp. 90 genome. The outermost and second circles of all replicons indicate genes in forward and reverse orientation colour-coded by their role categories as shown. The third circles of all replicons show G+C content in black and the fourth circles show the G+C skew in green (+) and purple (−). The fifth circles show the insertion sequences in red, putative prophage remnants in brown and MITEs in blue. The sixth circles show the pseudogenes with disrupted ORFs in green. The innermost circles of chromosome I and II show the rRNA genes in purple and tRNA genes in orange. The circular sizes of the replicons are not proportional to the actual sizes
Table 1.
Summary of the Anabaena sp. 90 genome
| Length (bp) | * Errors/Mb | GC (%) |
# ORFs |
# RNAs |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Hypothetical | (%) | Pseudogene | (%) | rRNA | tRNA | ||||
| Chromosome I |
4,329,264 |
0.02 |
38.1 |
3,817 |
1,414 |
(37.04) |
169 |
(4.43) |
15 |
40 |
| Chromosome II |
819,965 |
0.03 |
38.2 |
755 |
256 |
(33.91) |
49 |
(6.49) |
0 |
4 |
| Plasmid A |
80,384 |
0.03 |
37.2 |
93 |
71 |
(76.34) |
5 |
(5.38) |
0 |
0 |
| Plasmid B |
56,037 |
0.01 |
37.3 |
51 |
23 |
(45.10) |
4 |
(7.84) |
0 |
0 |
| Plasmid C |
20,025 |
0 |
37.42 |
22 |
20 |
(90.91) |
0 |
(0) |
0 |
0 |
| Total | 5,305,675 | 0.022 | 38.1 | 4,738 | 1,784 | (37.65) | 227 | (4.79) | 15 | 44 |
Gene composition between the chromosomes and plasmids is highly biased. All enzymes of essential biological pathways, such as photosynthesis, are encoded in the two chromosomes. The three plasmids mostly encode integrases, recombinases, transposases, phage-related proteins and a high percentage of hypothetical proteins (Table 1).
Prophage remnants
Remnants of three putative prophages were found, due to the presence of phage genes with a highly compact organization (Figure 2). However, ORFs encoding phage terminases and capsid protein-encoding genes were absent. Two of the putative prophage remnants are 11 kb in size and display high sequence similarity (93.5%) but are located 500 kb apart in chromosome I. The third prophage remnant is located in plasmid A with a slightly larger size (13 kb). These three prophage remnants have similar gene organization and share homologous genes (Figure 2), suggesting a close relationship between them. This indicates that Anabaena sp. 90 may have been a lysogen at some point in the past. Prophages are frequently inserted into tRNA genes [20]. A truncated C-terminal tRNA (Thr) gene was found 2.2 kb downstream of the first prophage remant in chromosome I. In addition, there is a 14-kb gene cluster containing clustered regularly interspaced short palindromic repeats (CRISPRs) and associated cas genes found in this genome in plasmid B. It was classified as subtype I-D, according to recent studies [21,22]. This gene cluster may provide acquired phage resistance for the strain.
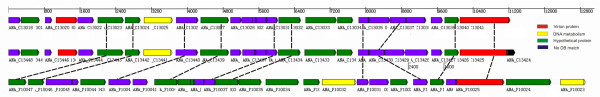
Figure 2.
Organization of genes in the regions of the three putative prophage remnants. The remnants of the three putative prophages were found in the presence of the phage genes and highly compact organization. Two of the putative prophage remnants (~11 kb) on chromosome I share high sequence similarity (93.5%) and are located 500 kb apart. The third region of the prophage remnants (~13 kb) was located in plasmid A. Genes identified with BLASTp as having similarity to known phage genes are marked in red, DNA metabolism genes in yellow, hypothetical genes in green, and the novel genes without database counterpart in purple. The homologue genes shared among the three prophages were indicated by broken lines
Pseudogenes and transposable elements
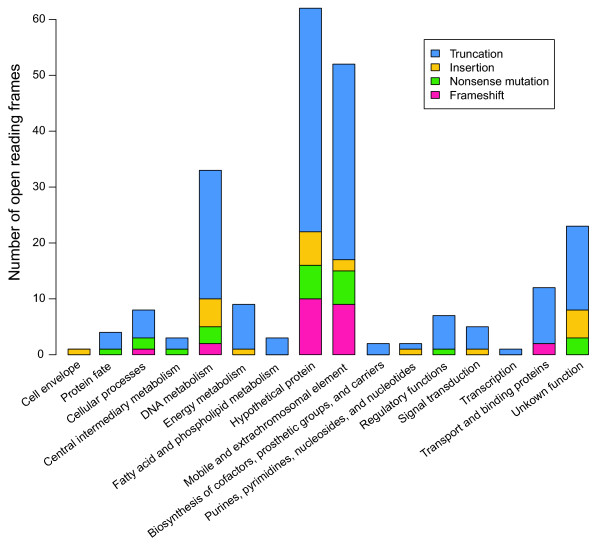
Surprisingly, a total of 227 pseudogenes were discovered across the genome through inspection of the disrupted ORFs (see Additional file 1: Table S2). More than half of the pseudogenes (114) were assigned to the ‘hypothetical protein’ and ‘mobile and extrachromosomal element’ categories, based on the Comprehensive Microbial Resource database [23] (Figure 3). There are 33 pseudogenes related to DNA metabolism, 13 of which were predicted with functions involved in DNA restriction and modification. Most of the pseudogenes (69.6%) were derived from truncation of the ORFs. Other types of pseudogenes that were found either contain frame shifts (10.6%), nonsense mutations (10.1%) or insertions (9.7%). The distribution of these pseudogenes overlaps that of the mobile genetic elements, especially insertion sequences (ISs) (Figure 1).
Figure 3.
Compositional distribution of 227 pseudogenes among different functional categories. Four types of pseudogenes were found and colour-coded as truncation in blue, insertion in orange, nonsense mutation (premature stop codon) in green and frameshift in pink. The functional category of the Comprehensive Microbial Resource database was applied [23]
In all, 81 complete and partial ISs, with a total size of 96.6 kb, were annotated from the Anabaena sp. 90 genome (see Additional file 1: Table S3). Sixty-five of these ISs were classified into 22 subfamilies, according to the ISfinder database [24], 18 of which occurred in two to seven copies (Table 2). Sixteen of the elements could not be assigned to a family in the ISfinder database. Pseudogenes with disrupted ORFs were often found adjacent to ISs (Figure 1), where 46 disrupted transposases were also discovered (Figure 3, see Additional file 1: Table S2). IS transposition disrupted not only host-specific ORFs but also the ORFs of other IS element transposases (Figure 3).
Table 2.
Categorized IS subfamilies in the Anabaena sp. 90 genome
| Name | Family and Subgroup | Size (bp) | Number |
|---|---|---|---|
| ISAsp2 |
ISAs1 |
1627 |
6 |
| ISAsp3 |
IS4 ssgr ISPepr1 |
1462 |
5 |
| ISAsp4 |
IS1380 |
1005 |
5 |
| ISAsp5 |
IS1634 |
2038 |
6 |
| ISAsp6 |
IS607 |
1353 |
3 |
| ISAsp7 |
IS607 |
862 |
1 |
| ISAsp8 |
IS607 |
1242 |
2 |
| ISAsp9 |
IS607 |
1575 |
7 |
| ISAsp10 |
IS4 ssgr IS10 |
1270 |
2 |
| ISAsp11 |
IS200/IS605 ssgr IS1341 |
1790 |
4 |
| ISAsp12 |
IS200/IS605 ssgr IS1341 |
1685 |
3 |
| ISAsp13 |
IS200/IS605 |
1607 |
2 |
| ISAsp14 |
IS200/IS605 |
1798 |
3 |
| ISAsp15 |
IS607 |
1660 |
2 |
| ISAsp16 |
IS630 |
1228 |
3* |
| ISAsp17 |
IS200/IS605 ssgr IS1341 |
1060 |
1 |
| ISAsp18 |
IS200/IS605 ssgr IS1341 |
1223 |
1 |
| ISAsp19 |
IS200/IS605 ssgr IS608 |
1652 |
2 |
| ISAsp20 |
IS200/IS605 ssgr IS200 |
682 |
1 |
| ISAsp21 |
IS5 ssgr IS5 |
1275 |
2 |
| ISAsp22 |
IS607 |
1866 |
2 |
| ISAsp23 | IS5 ssgr IS1031 | 413 | 2* |
* all members of the subfamily are disrupted.
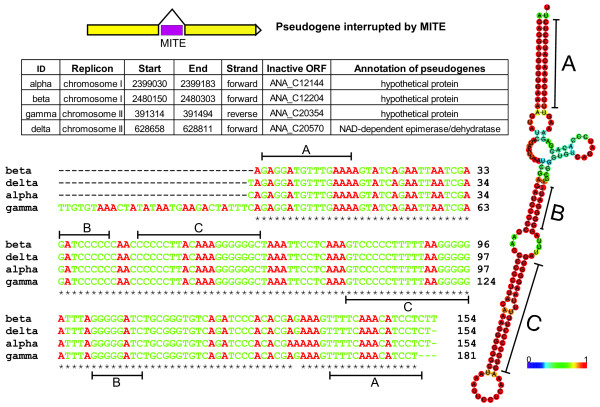
In addition, a total of 147 complete or partial miniature inverted-repeat transposable elements (MITEs) were located in the genome. Their lengths ranged from 77 to 541 bp (see Additional file 1: Table S4). These small genetic elements are usually present in multiple copies and are characterized by terminal inverted repeat regions [25]. The MITEs identified in this study could be grouped into type II MITEs. A total of 132 MITEs were distributed in intergenic regions, while 15 were found within disrupted chromosomal ORFs and led to the prediction of pseudogenes (Figure 4).
Figure 4.
Sample group of MITEs causing pseudogenes in the Anabaena sp. 90 genome. Four MITEs in group 1 that interruptedly caused pseudogenes were demonstrated. The secondary structure was predicted with the sequence data using RNAfold [84], while similarity of the paired bases was indicated by the colour scheme shown. Three sets of inverted repeat regions (A, B and C) were located from the secondary structure of the sequences, and labelled in the multisequence alignment
Bioactive peptide synthesis
Anabaena sp. 90 produces many bioactive peptides by nonribosomal or ribosomal pathways. In addition to the previously identified nonribosomal biosynthetic gene clusters for anabaenopeptilides [26], anabaenopeptins [27] and microcystins [28], a large gene cluster responsible for production of glycolipopeptides (hassallidins) was found (Figure 5). In addition to an anacyclamide-encoding cyanobactin gene cluster [29], seven putative bacteriocin gene clusters were also discovered (Figure 5). All these biosynthetic gene clusters were located in chromosome I. Together they amount to a total of ~250 kb, thus at least 5% of the genome is dedicated to the production of bioactive peptides.
Figure 5.
Biosynthetic gene clusters of nonribosomal and ribosomal peptides in the Anabaena sp. 90 genome. The genetic organization of the biosynthetic gene clusters for producing nonribosomal (a) and ribosomal (b) peptides. The putative bacteriocin gene clusters were classified according to methods described previously [15]
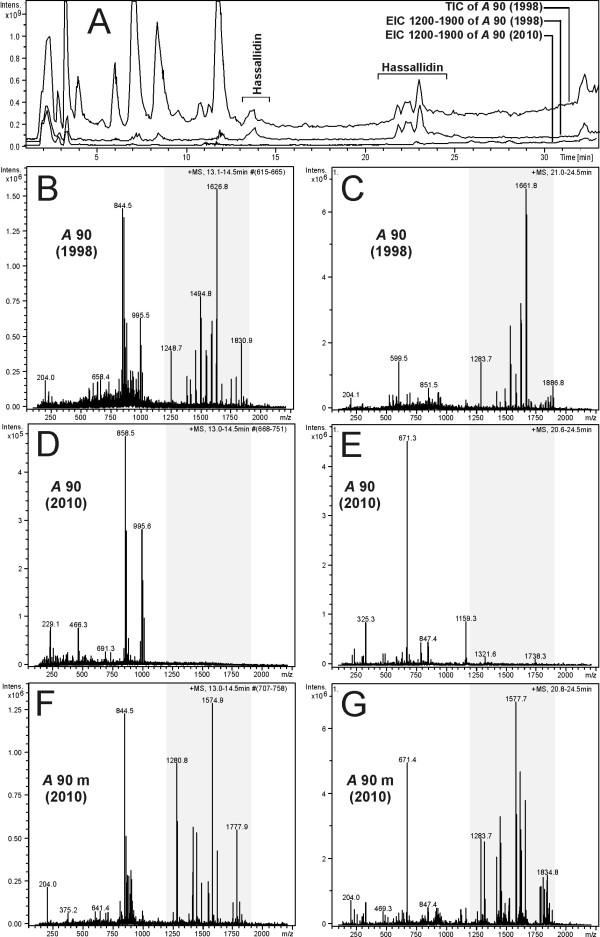
The hassallidin biosynthetic gene cluster is the largest gene cluster in Anabaena sp. 90. It encodes four nonribosomal peptide synthetase enzymes that catalyse the incorporation of amino acid residues into cyclic hassallidin peptides (Vestola et al., unpublished data). Surprisingly, the genome assembly revealed two different contigs within the same sequence, which led us to the discovery of a 526-bp deletion in the peptide synthetase gene hasV (ANA_C13069). The deletion introduced a frameshift and rendered the gene cluster nonfunctional. In PCRs performed with Anabaena sp. 90 DNA originating from the years 1998–2009, the deletion first appeared around 2003–2006, but had not fully segregated by 2009 (Figure 6). Due to the deletion, hassallidins can no longer be detected in current cultures of Anabaena sp. 90. However, hassallidins could still be identified from cells extracted prior to the deletion in 1998 and from an anabaenopeptilide synthetase gene mutant (apdA-) strain of Anabaena sp. 90 constructed in 1999 [26] (Figure 7).
Figure 6.
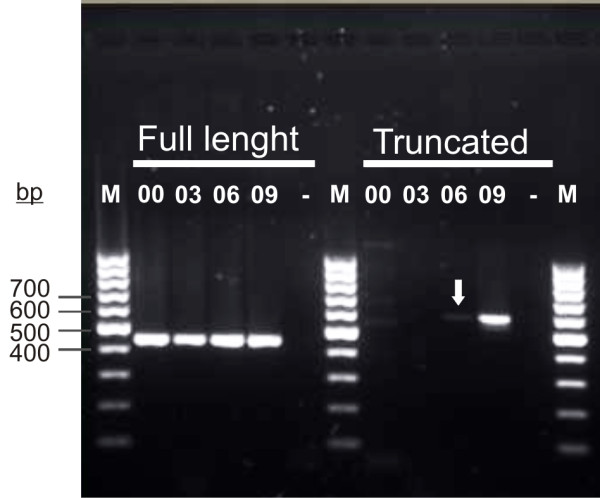
PCR screening of the deletion in the hasV gene with Anabaena sp. 90 cells originating from different years. PCR analysis of genomic DNA demonstrates occurrences of the deletion in hasV in 2000, 2003, 2006 and 2009. PCRs with primers across the site of deletion show that the deletion appeared in 2006 (arrow). Recent cultures of Anabaena sp. 90 carry a deletion in hasV and this strain no longer produces hassallidin
Figure 7.
Chromatograms and mass spectra show the presence and absence of hassallidin variants in Anabaena sp. 90. A: Total ion chromatogram (TIC) of Anabaena sp. 90, originating in 1998, and extracted ion chromatograms (EICs) of Anabaena sp. 90, originating from 1998 and 2010, show the elution of ions with m/z 1200 – 1900, representing the mass range of hassallidin ions. Two eluting time ranges of the hassallidin variants are marked in the chromatograms. B-G: Average mass spectra from the first (B, D, F) and the second (C, E, G) time range of the chromatograms in which the hassallidin variants were eluting. The grey areas show the hassallidin ions in the spectra. B, C: Spectra from Anabaena sp. 90 originating from 1998. D, E: Spectra from Anabaena sp. 90 originating from 2010. F, G: Spectra from Anabaena sp. 90 anabaenopeptilide minus mutant originating from 2010
Restriction-modification systems
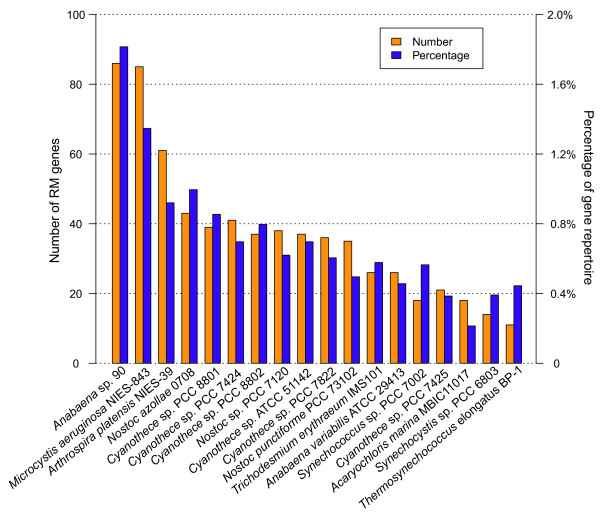
The Anabaena sp. 90 genome contains a very high number (88) of restriction-modification (RM) system-related ORFs, amounting to 1.8% of the gene repertoire. Another bloom-forming cyanobacterium, Microcystis aeruginosa NIES-843, had nearly the same number of RM system-related ORFs, but other cyanobacteria had lower numbers of RM genes (Figure 8). There are 31 Type I, II, III and IV RM systems, constituting 56 chromosomal ORFs in Anabaena sp. 90 (see Additional file 1: Table S5). The majority (22 out of 31) were categorized as type II systems. In addition, there are 14 separate restriction enzymes and 18 unaccompanied DNA methyltransferases. However, 13 of them are pseudogenes with disrupted ORFs, including five enzymes in type I, II and III RM systems, three restriction enzymes and five DNA methylases.
Figure 8.
Number and percentage of RM genes found in the Anabaena sp. 90 and other cyanobacterial genomes. The RM genes in other cyanobacterial genomes were collected from the REBASE Genomes [85]
Photosynthesis
The photosynthetic gene clusters in the genome retain the conserved organizational pattern found in other cyanobacteria [30]. A quarter of the photosynthetic genes are distributed in chromosome II, such as psbA, psbB, and operons of atpIHGFDAC encoding ATP synthase, petBD cytochrome b6/f complex and apcABC light antenna. The rest of the photosynthetic genes were found in chromosome I.
Nitrogen fixation
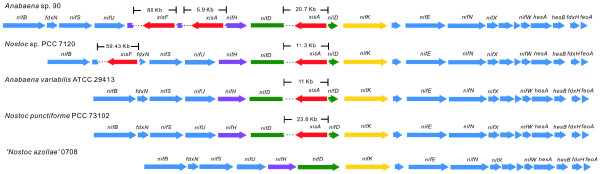
Anabaena sp. 90 was grown in the laboratory in nitrogen-free medium and is capable of active nitrogen fixation. The nif operon, encoding for the dinitrogenase and dinitrogenase reductase enzyme complexes, was located in chromosome I with conserved gene organization (Figure 9). Three excision elements of 20.7, 5.9 and 80 kb were found within the nif operon in Anabaena sp. 90. Each element is adjacent to a single site-specific recombinase in the opposite strand that removes the elements during heterocyst development [31]. The 80- and 5.9-kb excision elements that split the nifH gene into three parts with sizes of 153, 273 and 444 bp have not been described previously in cyanobacteria. The third element, which commonly occurs in heterocyst-forming cyanobacteria (Figure 9), splits the nifD gene into two parts with 1,356 and 147 bp. The nif operon spans 122 kb including the three excision elements. The 80-kb element contains one of the prophage remnants. In addition, the fourth 11-kb excision element was detected within the nitrogen-fixation associated hupL gene. Surprisingly, the counterparts of patS and hetN, both involved in pattern formation by preventing neighbouring cells from undergoing heterocyst differentiation [32], could not be detected in this genome.
Figure 9.
Comparative illustration of genetic organization of nif operon in heterocyst-forming cyanobacteria. The gene organizations of nif operons in heterocyst-forming cyanobacteria are demonstrated in colour as nifH in purple, nifD in green, nifK in yellow and others in blue. Each excision element was indicated by a site-specific recombinase labelled in red and three dots that represent the remaining part of the element. The lengths of the excision elements are shown above
Transporter proteins
The genome shows a remarkable density of genes encoding transporter proteins. There are four porin genes that encode channels for passive nutrient diffusion. A variety of ATP transport systems, often in operons, are present in both chromosomes (see Additional file 1: Table S6) for active uptake of various substrates, such as cations and anions, nucleosides, amino acids, sugars, glycolipids and polyamines. Six copies (three complete, two partial and one disrupted) of the ABC transporter operon devBCA, which encode essential exporters for heterocyst envelope formation [33], are present. Two of these are encoded in chromosome II.
Signal transduction and gene regulation
In all, 153 ORFs, including those carrying insertions, were annotated for signal transduction and regulation. A total of 69 ORFs (see Additional file 1: Table S7), scattered over both chromosomes, were predicted to be involved in two-component signal transduction systems. They include 19 histidine kinases (of which three were pseudogenes), 31 response regulators and 19 hybrid kinases according to their domain composition. These also include five pseudogenes. There are 32 Ser/Thr type protein kinases, six protein phosphatases and other regulation or sensor domain-containing proteins that form the one-component systems that coordinate with the two-component systems. Moreover, all group 1 (sigA) and group 2 (sigB, sigC, sigD, sigE) sigma factors are present in the Anabaena sp. 90 genome. The common group 3 (sigF, sigG) and one extracytoplasmic function sigma factors were found as well. Four proteins with anti-sigma-factor antagonist domain were also identified. They work together with the sigma factors in regulating various cell processes at the transcriptional level.
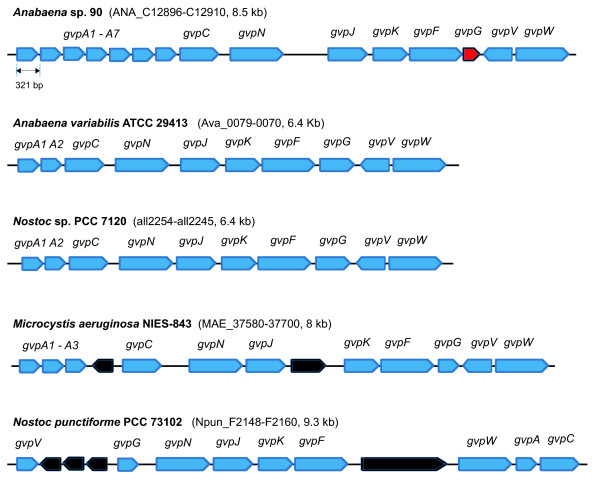
Gas vesicle gene cluster
An 8.5-kb gvp gene operon encoding the building blocks of gas vesicles was located in chromosome I. The operon organization (gvpA7CNJKFGVW) is similar to that in other sequenced cyanobacterial strains [34], but with seven tandem gvpA genes (Figure 10). A truncated gvpG gene was found. This coincided with loss of buoyancy from cells in the present culture, while the original culture showed the buoyant phenotype.
Figure 10.
Genetic organization of cyanobacterial gas vesicle (gvp) operons. The gas vesicle formation genes were indicated in blue. The truncated gvpG gene in Anabaena sp. 90 was indicated in red. The other genes in the gvp operons were indicated in black
Metabolic pathway analysis
We annotated 206 putative metabolic pathways in the genome of Anabaena sp. 90 (see Additional file 1: Table S8), in addition to those for bioactive peptide biosynthesis. These pathways are composed of 777 enzymes that catalyse 1,211 enzymatic reactions. However, nearly half of these pathways are incomplete, because 227 (29%) enzyme-encoding genes are missing or were not found. Many of the incomplete pathways are responsible for catabolic processes, such as nutrient degradation, utilization and assimilation, whereas the essential pathways are complete, e.g. amino acid metabolism, photosynthesis and glycolysis. The energy-related pathways have fewer missing enzymes than others. Nearly 40% of the genes in this genome are hypothetical or with unknown functions, due most likely to their low homology to counterparts in model organisms. This may leave some of the enzymes in annotated pathways unrecognized. Moreover, our analysis revealed that some enzyme-encoding genes are pseudogenes with disrupted ORF. For instance, one (ANA_C20606) of the two deoxycytidine triphosphate deaminases, which catalyse the conversion of dCTP to dUTP in the pyrimidine deoxyribonucleotide de novo biosynthesis pathway, was interrupted by an inserted DNA recombinase. In addition, an alcohol dehydrogenase-encoding gene (ANA_C20081) was found with a deletion. This enzyme catalyses the reduction of acetaldehyde to ethanol in fermentation pathways. Three copies of alcohol dehydrogenases were found in this genome in an intact form.
Discussion
Here, we report the complete genome of Anabaena sp. 90, an ecologically important hepatotoxic bloom-forming cyanobacterium. The genome has a multichromosome composition with essential metabolic core genes encoded in the two circular chromosomes. This study was the first to report such a multichromosome composition in the order of Nostocales. Previously, the strain of Cyanothece sp. ATCC 51142 was known with two chromosomes, one circular and one linear [35]. We succeeded in completing the genome with high sequence quality, even though it contains a plethora of repetitive mobile genetic elements with diverse sizes (prophage remnants: ~11 kb, ISs: 1–3 kb, MITEs: 100–300 bp) and five nearly identical rRNA operons. A large number of repeats makes genome assembly difficult and cannot be resolved simply by increasing the sequencing depth e.g. using the next-generation low-cost sequencing platforms, due in large part to their short read lengths [36]. As a result, the majority of genomes remain in the ‘draft’ state, including a number of bloom-forming cyanobacteria [17-19]. In this study, we tackled this problem by including data from mate-pair libraries with large inserts [36]. This strategy could be used in sequencing bloom-forming cyanobacterial genomes that are rich in mobile genetic element-derived repeats [16,17,19]. Complete and high quality genomes are crucial to comparative genomics and genome evolution studies [37], pathway reconstruction for metabolic engineering [38] and postgenomic analysis [39-41].
The Anabaena sp. 90 genome contains various types of mobile genetic elements, including plasmids, prophage remnants, ISs and MITEs. They are collectively termed as mobile genetic elements, since they are capable of moving within genomes (duplication) and between prokaryotic organisms (horizontal gene transfer) [42]. These elements may have contributed to genome plasticity, genomic rearrangements and most likely the multichromosome composition. ISs are mobile genetic elements transferred within and between species through a cut-and-paste mechanism, which is driven by the internally encoded transposases [43,44]. The percentage of IS elements in Anabaena sp. 90 (~2%) was comparable to that in other cyanobacteria, but less than in the bloom-forming hepatotoxin-producing Microcystis aeruginosa strains, in which IS elements comprised about 10% of the genomes [45]. Recent genome studies showed that the planktonic cyanobacteria Cylindrospermopsis raciborskii CS505 and Crocosphaera watsonii WH8501 also contain high numbers of repetitive elements, partly contributed by ISs [19]. MITEs are small mobile sequences that have only terminal inverted repeats [25]. The MITEs found in this study were classified as type II, because there is no evident similarity found with inverted repeats of the ISs in this genome [45]. The mobilization of type I MITEs was hypothesized to be mediated by the transposase of IS that holds the same terminal inverted repeats [46]. This may imply an autonomous mechanism for the movement and duplication of type II MITEs.
Surprisingly, we found that nearly 5% of the gene repertoires are pseudogenes that include not only disrupted transposases but also disrupted ORFs from many other different functional categories (Figure 3). However, the pseudogenes found in the Anabaena sp. 90 genome were most likely derived from the transposition activities of ISs and MITEs. Mutations and genome rearrangements induced by transposable elements have also been described in Microcystis aeruginosa strains [16,17]. However, an abundance of pseudogenes has rarely been reported in genomes of cyanobacteria, except the endosymbiotic strain ‘Nostoc azollae’ 0708, which has an extremely high number (31.2%) of pseudogenes that were attributed to the genome erosion process [47]. Here, we accurately labelled a number of disrupted ORFs, many of which could be associated with the transpositions of mobile genetic elements, through detailed manual annotation (Figures 1 and 4). Recent metagenomic data analysis also demonstrated the incidence of genes inactivated by IS transposition, including some with essential functions, within the population of Synechococcus strains in hot spring mats [48]. High frequencies of mobile genetic element-derived pseudogenes are likely to be common among transposable element-rich cyanobacteria, especially in strains of bloom-forming genera, but as yet remain undocumented.
Anabaena sp. 90 dedicates 5% of its genome to biosynthesis of small peptides, such as hepatotoxic microcystins and the protease inhibitors anabaenopeptins and anabaenopeptilides. Between 3% and 4% of the genomes is typically devoted to the biosynthesis of secondary metabolites among sequenced cyanobacterial genomes [17,18,49]. However, previous works only counted nonribosomal gene clusters and missed the ribosomal biosynthesis pathways. Here, we took into account both ribosomal and nonribosomal gene clusters. Considering the widespread occurrences of ribosomal pathways in cyanobacteria [14,15] and that nonribosomal gene clusters also amount to 3.5% of Anabaena sp. 90 genome, it appears that about 5% of the genome is commonly assigned to natural product biosynthesis in cyanobacteria. An additional nonribosomal gene cluster responsible for production of the antifungal compounds hassallidins was discovered during genome assembly, mainly because the gene cluster was inactivated by a deletion event. Moreover, the genome sequence led us to discover the ribosomal production pathway for cyanobactins and their common occurrence in Anabaena[29] as well as in other planktonic cyanobacteria [50]. We also identified seven putative bacteriocin gene clusters in the genome. Their end products are presently unknown, but the high number of these types of gene clusters in cyanobacterial genomes [15] suggests that yet new compound families from cyanobacteria await identification and structural determination. To date, bacteriocins have only been identified in Prochlorococcus marinus MIT9313 [51].
The toxin microcystin synthetase (mcy) gene cluster was shown to be of ancient origin [52]. Previous environmental studies showed the stable presence of mutant cyanobacterial mcy gene clusters, which were inactivated both in freshwater samples by ISs [53] and in brackish water by MITEs [54]. Note that the intact hassallidin gene cluster is still retained in parallel cultures of mutant Anabaena sp. 90, which has an inactivated anabaenopeptilide synthetase gene cluster [26]. Our results showed that those mutants with an inactive hassallidin biosynthetic pathway prevailed over cells with functional genes in the culture. This may indicate a growth advantage for cells with mutated bioactive compound synthetase gene clusters under laboratory conditions, perhaps due to a lower metabolic burden.
Similarly, we detected a loss of gas vesicles, the subcellular structures responsible for buoyancy, which are essential for bloom-forming planktonic cyanobacteria [3]. This phenotype is derived from the truncated gvpG gene in the gvp operon (Figure 10). The truncation most probably was selected when the strain was purified (axenic) on solid media [55]. Loss of buoyancy due to rearranged gas vesicle gene clusters by IS transpositions was also previously described in Microcystis strains kept in laboratory culture [56].
Anabaena sp. 90 has been under continuous culture since 1986 and was introduced into pure culture in 1992 [55]. The mobile genetic elements were probably acquired by the strain prior to this time. Some metabolic properties of Anabaena sp. 90 were altered by the activities of these mobile genetic elements. We found mobile element-derived pseudogenes among the genes encoding enzymes for the metabolic pathways of the strain. This may indicate that Anabaena sp. 90 has lost genes for metabolic pathways with nonessential functions under laboratory conditions, where optimal, nutrient-rich and competitor-free environments are provided. Our analysis revealed that the laterally acquired mobile genetic elements may have played a role in the process. Perhaps growing cyanobacteria under conditions that reflect those found in nature would reduce genetic changes.
The Anabaena sp. 90 genome contains 31 RM systems, which function as microbial defence systems against foreign DNAs [57]. Type I, II, III and IV RM systems were annotated from the Anabaena sp. 90 genome, type II restriction enzymes being the most numerous RMs (22). Previously, restriction enzyme activities, such as AflIII, in Anabaena sp. 90 were experimentally confirmed [58]. An RM system usually contains a restriction endonuclease that recognizes a specific sequence for cleavage and a DNA methyltransferase that modifies the same sequence and protects it from cleavage [59]. Bacteria that possess multiple RM systems are thought to be virtually impregnable. This seems to hold true for Anabaena sp. 90 since it is resistant to genetic manipulation. Over the years we have succeeded in producing only one mutant of this strain [26]. This is a familiar feature with many filamentous cyanobacteria [60], which were proposed more intensively protected by RM systems than unicellular strains [61]. However, this has been contradicted by the frequent occurrence of RM systems in genomes of unicellular toxic bloom-forming Microcystis aeruginosa strains [16,17]. To date the two highest numbers of restriction enzymes are found in two planktonic microcystin-producing cyanobacteria, Anabaena sp. 90 (this study) and Microcystis aeruginosa NIES-843 (Figure 8). This perhaps is a reflection of the ecological or evolutionary pressures exerted on planktonic cyanobacteria. The Anabaena sp. 90 genome also contains abundant mobile genetic elements. The question that naturally arises is how these mobile elements have invaded the planktonic cyanobacterial genomes with the presence of pronounced RM systems. The mobile genetic elements are, however, relatively short sequences (see Additional file 1: Tables S3 and S4) and may lack many restriction cleavage sites of restriction endonucleases. For Anabaena sp. 90, this may be partly explained by the disrupted genes encoding enzymes in RM systems (see Additional file 1: Table S5). In addition, it is known that RM systems may be inefficient in blocking single-stranded or modified foreign DNA and work only temporarily [62]. However, it was suggested that RM systems may also be mobile genetic elements that cause genome rearrangements [63].
In filamentous cyanobacteria, the cellular processes of photosynthesis and nitrogen fixation are spatially separated and compartmentalized into vegetative cells and heterocysts, respectively [10]. Anabaena strains have long been used as model organisms for studying heterocyst development and nitrogen fixation in cyanobacteria [1,64]. The differentiation processes from vegetative cells to heterocysts involve programmed DNA rearrangements at multiple sites in Anabaena genomes [65-67]. To date, three known elements, ranging from 11 to 55 kb, have been reported in nifD, fdxN and hupL genes in heterocystous cyanobacteria [67]. Here, for the first time we report the simultaneous presence of four excision elements in the genome of Anabaena sp. 90. Moreover, two novel elements for the nifH gene were found and one of these is the largest known in size (80 kb) (Figure 9). The nifH gene is commonly used to detect nitrogen-fixing organisms in environmental samples. The presence of excision elements is likely to cause problems with detection of nifH genes if they occur commonly among heterocystous cyanobacteria. Our results showed that the heterocyst differentiation process in cyanobacteria involves precise genomic splicing over distances as long as 122 kb. The excision elements interrupting nif genes in heterocystous cyanobacteria appear to play a role in protecting nitrogenase and hydrogenase from the effects of oxygen during heterocyst development [68]. A compact and conserved nif operon was recently discovered without any excision elements in a symbiotic strain ‘Nostoc azollae’ 0708 (Figure 9) [47]. This evidence supports the proposed loss of photosynthetic activity in cells of ‘Nostoc azollae’ 0708, which may have differentiated only to perform nitrogen fixation for the hosts [47]. A similar nif operon without excision elements was presented in the heterocyst-forming cyanobacterium Cylindrospermopsis raciborskii CS-505, but lost in its closely related strain Raphidiopsis brookii D9 [19]. This may suggest that the growth of these strains is less dependent on the nitrogen fixation process. The presence of multiple copies of devBCA operons in Anabaena sp. 90 may increase the efficiency of nitrogen fixation, whereas lack of the patS and hetN genes in Anabaena sp. 90 (this study) and Cylindrospermopsis raciborskii CS-505 [19] may suggest a new pattern of heterocyst spatial development. Simultaneous inactivation of patS and hetN genes in Nostoc sp. PCC 7120 is known to be lethal, due to overproduction of HetR and heterocysts [69]. Genome mining of Cylindrospermopsis raciborskii CS-505 suggested that a protein with the C-terminal pentapeptide RGSGR may take over the function of PatS [19]. Similarly, we also identified an ORF (ANA_C10293) with the C-terminal pentapeptide in Anabaena sp. 90, which may play a role as a HetR suppressor.
The 5.3-Mb Anabaena sp. 90 genome contains a lower number of genes for signal transduction than the 7.2 Mb-genome of Nostoc PCC 7120 [64] and nearly 10-Mb genome of Nostoc punctiforme[70]. However, the number of signal transduction pathways is positively correlated with genome size. It is also well known that soil microbes such as Nostoc punctiforme invest more heavily in sensing changes in environmental conditions than organisms living in more stable aquatic environments. Usually, a one-to-one relationship exists between the cyanobacterial sensors (histidine kinases) and response regulators [71], but in Anabaena sp. 90 the ratio is lower, indicating either integration of multiple signalling pathways or perhaps loss of sensoring systems when grown in protected laboratory environments.
Conclusions
This study gives a snapshot of the Anabaena sp. 90 genome. It shows a high potential of genetic variation by virtue of the occupation of a wide range of mobile genetic elements. Our results indicated that mobile genetic element-imposed selective pressure led to genome adaption to the strain by trimming nonessential genes and pathways during cultivation in the laboratory. In addition, due to the array of biosynthesis gene clusters for multiple peptides in Anabaena sp. 90, the complete sequence provides a valuable research subject in studying the regulation of natural product biosynthesis, which may have potential pharmaceutical and biotechnology applications.
Methods
Strain isolation and culture
Anabaena sp. 90 was isolated as a microcystin producer in 1986 from Lake Vesijärvi, Finland [72]. The axenic culture was originally purified from a single filament that was placed over a solid medium and then has been continuously maintained at the University of Helsinki cyanobacterial culture collection in Z8 nitrogen-free medium at room temperature (20–23°C) with continuous illumination of 10–20 μmol m-2 s-1[8]. The phylogeny of this strain was previously published [73].
DNA extraction and genomic library construction
The DNA extraction was described earlier [26]. Three sizes of genomic libraries were utilized for end sequencing. The large insert library was a cosmid library with an insert size of approximately 40 kb [26]. Two shotgun libraries with 2-kb and 6-kb inserts ligated into the pUC18 plasmid vector were constructed using standard protocols.
Genome sequencing, assembly and finishing
All reads were generated from clone ends sequencing by the Sanger sequencing platforms Megabase 1000 (GE Healthcare, Buckinghamshire, UK) and ABI 3730 (Applied Biosystems, CA, USA). An initial 9.9X sequencing data was sequenced from a combination of 2%, 8% and 90% of the reads from the 40-kb, 6-kb and 2-kb libraries, respectively. The Phred/Phrap/Consed software package [74] was used for genome assembly and gap closure according to the paired ends from the large-insert (6 and 40 kb) libraries. The remaining physical gaps that were derived from the unclonable regions were linked through combinatorial multiplex PCR screening of primers designed from the contig ends. Autofinish [75] was used for guiding, either by clone-end resequencing or primer walking over the clones or PCR products to attain the standard that each base was covered by at least two independent high-quality reads and with a Phred [76,77] quality value ≥ Q40. Large repetitive regions (i.e. RNA operons, prophage remnants) were resolved by primer-walking over long PCR products amplified from the corresponding regions. In all, 119316 reads were produced, which amounted to a final sequencing depth of 12.5X.
Genome annotation and analysis
Gene finding and function assignment
ORFs were initially predicted by Glimmer 3.02 [78] with a threshold of 100 bp. The intergenic regions were subjected to blastx searching [79] against the nonredundant database for unrecognized ORFs. All predicted genes were translated into amino acid sequences for homologue searches with the InterPro [80], Cluster of Orthologous Groups [81] and nonredundant databases. Functional assignments and start sites for each ORF were determined manually by combining the search results from these sources. Transfer RNA genes were predicted with tRNAscan-SE [82] and rRNA genes were located through homologue searches. The annotated proteins were further assigned to functional groups according to the Comprehensive Microbial Resource role category [20]. The putative bacteriocin gene clusters were classified, according to methods described previously [15]. Hypothetical proteins were defined as conserved if they had at least 30 homologues with full-length matching in other genomes, while unique hypothetical proteins had no full-length matching in other genomes.
Pseudogenes
The pseudogenes (see Additional file 1: Table S2) were examined manually using Artemis [83] for frameshift and premature stop codons, as well as the boundaries of the truncation, deletion and insertion. The boundaries of truncated pseudogenes were determined through iterative BLAST searches for the surrounding regions. The pseudogenes were assigned a function according to the hits of the homologue search with significant similarity.
IS and MITE
The ISs were identified and classified using the ISfinder database [24]. Fifteen MITEs were originally discovered as insertions from interrupted pseudogenes. Additional MITEs were found by blastn searching the 15 insertions against the complete genome sequence (E < 1e-5). Fragmented MITEs with over 50% coverage were counted as partial. The secondary structures of the MITEs were predicted using the RNAfold webserver [84].
RM systems
A protein database of RM system genes was locally prepared by collecting data from the REBASE [85]. All annotated proteins initially searched against this database, and protein hits of blastP (E < 1e-4) were chosen as candidates for manual checking. RM systems were confirmed with the associated presence of restriction enzymes, DNA methylase and other RM genes or domains. Separate RM genes were kept while the E value was 1e-10 or less. The disrupted RM genes were reconfirmed by manually inspecting the alignment of the remaining parts.
Metabolic pathway analysis
A pathway/genome database of Anabaena sp. 90 was constructed using Pathway Tools [86] from the annotation. Pathway Hole Filler [87] was further used for retrieving the missing enzymes.
Hassallidin
DNA extraction and PCR
Both Anabaena sp. 90 and a knockout mutant constructed in 1999 [26] were grown as previously described. Genomic DNA from the two Anabaena strains was extracted using either the DNeasy® Plant Mini Kit (Qiagen Gmbh, Hilden, Germany) or the E.Z.N.A™ SP Plant DNA Mini Kit (Omega Bio-Tek Inc., Norcross, GA, USA). PCR amplifications of DNA from Anabaena were performed in iCycler (Bio-Rad, Hercules, CA, USA) using the primers hasV-fw (5-TCTAGATGGTTGGAGTGTGGC-3) and hasV-rev (5-AGGATGCGGTAGCTTTGAGGAGGCG-3), which were designed to amplify across the site of deletion in the hasV gene. The primer pairs a1 (5-TGGTAACAGGTAACGTAATTAAAAC-3) and hasV-rev, and y1 (5-ATCAGTAGTTTCGGGTCTGG-3) and hasV-fw were designed to allow specific detection of deletion and full-length versions of hasV, respectively. Each PCR was carried out in 1 × DyNAzymeTM buffer (Tris–HCl, MgCl2, KCl, Triton® X-100) (Finnzymes, Espoo, Finland) with 100 μM of each dNTP (dNTP mix, 10 mM, Finnzymes), 0.25 μM of each primer (Sigma Genosys, Sigma-Aldrich, St. Louis, MO, USA), 0.4 U DyNAzyme II DNA polymerase (F-501S, 2 U/μl, Finnzymes) and 10–170 ng of template DNA in a final volume of 20 μl. The following PCR thermocycle was used to amplify the hasV gene: initial denaturation at 94°C for 3 min, 32 cycles of 94°C for 30 s, 57°C for 30 s and 72°C for 1.5 min, and a final extension at 72°C for 10 min. For detection of the deletion, a similar PCR programme was used, with the exception of annealing at 64°C for 30 s and elongation at 72°C for 1 min.
Chemical analysis
Cells of the cyanobacterial strains were collected from the 20–40-ml cultures by centrifugation at 7000 × g for 7 min and freeze-dried (Maxi dry plus, Heto-Holten A/S, Allerød, Denmark). The dried cells were extracted with 1 ml of methanol in 2-ml plastic tubes containing glass beads (Cell Disruption Media, 0.5-mm glass beads, Scientific Industries Inc., Bohemia, NY, USA) using a FastPrep cell disrupter (FP120, Bio101, Thermo Electron Corporation, Qbiogene, Inc., CA, USA) for 10 s at a speed of 6.5 m s-1. The extracts were centrifuged at 10 000 × g 5 min prior to LC/MS analysis. The LC/MS analyses of extracts were carried out in an Agilent 1100 Series LC/MSD Trap XCT Plus System (Agilent Technologies, CA, USA) using a Phenomenex Luna C18(2) LC column (100 mm × 4.6 mm, particle size 5 μm, 100 Å; Phenomenex, CA, USA). The column was protected with a C18 precolumn (4 mm × 2 mm; Phenomenex). The LC/MS parameters were optimized with hassallidin ion m/z 1862 with negative mode of polarity. The mobile phase consisted of 0.1% aqueous formic acid (50% solution in water; Fluka, Sigma Aldrich, Steinheim, Germany) (solvent A) and acetonitrile (Chromasolv® for LC/MS, Fluka, Sigma-Aldrich, Steinheim) (solvent B). A gradient (solvent B) from 10% to 100% was run over 30 min at a flow rate of 0.15 ml min-1 with 5-μl injection at 40°C. Hassallidins were detected with MS using electrospray ionization set in the positive ion mode. The nebulizer gas (N2) pressure was 35.0 pounds per square inch (psi), the drying gas flow rate was 8.0 l min-1 and the temperature was 350°C. The capillary voltage was set to 3650 V, and the capillary offset value was 250 V. A skimmer potential of 58.0 V and a trap drive value of 112.0 was used. The spectra were recorded at a scanning range of 100–2200 m/z on average of three spectra using an ion charge control (ICC). Identification of hassallidins was based on the similarities between the spectra of the extracts analysed and known hassallidins A and B (Alexis Biochemicals, Ezno Life Science Inc., Lausen, Switzerland). The structures of the hassallidins were identified by fragmentation analysis MSn (n = 1–3) using the SmartFrag function.
Accession number
The complete annotated genome sequence was submitted to GenBank under accession number [GenBank: CP003284 (ChromosomeI), CP003285 (ChromosomeII), CP003286 (PlasmidA), CP003287 (PlasmidB), CP003288 (PlasmidC)].
Abbreviations
bp: Base pair; kb: Kilobase pairs; ORF: Open reading frame; IS: Insertion sequence; MITE: Miniature inverted-repeat transposable element; RM: Restriction-modification; CRISPR: Clustered regularly interspaced short palindromic repeat.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HW carried out the genome assembly, gap closure and detailed genome analysis. ZL and BL were responsible for sequencing of the genome. HW, LR, CL, ARY and KR manually annotated the genome. JV, LR, DPF analysed the hassallidin gene cluster and JJ conducted the chemical analysis. HW, KS, DPF, ARY, LR, CL, JV and JJ drafted the manuscript. KS conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Hypothetical proteins in the Anabaena sp. 90 genome. Hypothetical proteins with disrupted ORF are labelled in yellow. Table S2. Pseudogenes annotated from the Anabaena sp. 90 genome. Table S3. Summary of ISs identified in the Anabaena sp. 90 genome. Table S4. List of MITEs discovered in the Anabaena sp. 90 genome. The MITEs that disrupted chromosome ORFs by insertion are labelled in green. Table S5. RM system genes in the Anabaena sp. 90 genome. Table S6. Summary of transporter proteins in the Anabaena sp. 90 genome. Table S7. Two-component system genes annotated from the Anabaena sp. 90 genome. The two-component genes were classified as HK, RR and HR, representing histidine kinase, response regulator, and hybrid kinase, respectively. Pseudogenes with disrupted ORF are labelled in yellow. Table S8. Predicted metabolic pathways of Anabaena sp. 90.
Contributor Information
Hao Wang, Email: wang.hao@helsinki.fi.
Kaarina Sivonen, Email: kaarina.sivonen@helsinki.fi.
Leo Rouhiainen, Email: leo.rouhiainen@helsinki.fi.
David P Fewer, Email: david.fewer@helsinki.fi.
Christina Lyra, Email: christina.lyra@helsinki.fi.
Anne Rantala-Ylinen, Email: anne.ylinen@helsinki.fi.
Johanna Vestola, Email: johanna.vestola@alumni.helsinki.fi.
Jouni Jokela, Email: jouni.jokela@helsinki.fi.
Kaisa Rantasärkkä, Email: kaisa.rantasarkka@helsinki.fi.
Zhijie Li, Email: lizhj@bgilt.com.
Bin Liu, Email: binliu_gh@yahoo.com.
Acknowledgments
We thank Dr. Elina Roine for her helpful advice concerning prophage elements and Lyudmila Saari for maintaining and growing the organism. This work was financially supported by the Academy of Finland and the University of Helsinki to KS (Research Centre of Excellence grants 53305 and 118637), to ARY (128480), and by the Viikki Doctoral Programme in Molecular Biosciences to HW.
References
- Bryant DA. The molecular biology of cyanobacteria. Dordrecht: Kluwer Academic Publishers; 1994. [Google Scholar]
- Hudnell HK, editor. Cyanobacterial harmful algal blooms: State of the science and research needs. 2008. pp. 1–912. (Adv Exp Med Biol). [PubMed] [Google Scholar]
- Walsby AE. Gas vesicles. Microbiol Rev. 1994;58(1):94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivonen K. In: Encyclopedia of Microbiology. 3. Schaechter M, editor. Oxford: Academic; 2009. Cyanobacterial toxins; pp. 290–307. [Google Scholar]
- Ressom R, Soong FS, Fitzgerald J, Turczynowicz L, El Saadi O, Roder D, Maynard T, Falconer I. Health effects of toxic cyanobacteria (blue-green algae) Canberra, Australia: National Health and Medical Research Council & Australian Government Publishing Service; 1994. [Google Scholar]
- Kuiper-Goodman T, Falconer I, Fitzgerald J. In: Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. Chorus I, Bartram J, editor. London: Taylor & Francis; 1999. Human health aspects; pp. 113–153. [Google Scholar]
- Rippka R, Castenholz RW, Iteman I, Herdman M. In: Bergey's manual of systematic bacteriology. Volume 1. 2. Boone DR, Castenholz RW, Garrity GM, editor. New York: Springer; 2001. Form-genus I. Anabaena; pp. 566–568. [Google Scholar]
- Sivonen K, Niemelä SI, Niemi RM, Lepistö L, Luoma TH, Räsänen LA. Toxic cyanobacteria (blue-green algae) in Finnish fresh and coastal waters. Hydrobiologia. 1990;190(3):267–275. doi: 10.1007/BF00008195. [DOI] [Google Scholar]
- Halinen K, Fewer DP, Sihvonen LM, Lyra C, Eronen E, Sivonen K. Genetic diversity in strains of the genus Anabaena isolated from planktonic and benthic habitats of the Gulf of Finland (Baltic Sea) FEMS Microbiol Ecol. 2008;64(2):199–208. doi: 10.1111/j.1574-6941.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8(1):39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- Sivonen K, Börner T. In: The cyanobacteria: molecular biology, genomics, and evolution. 1. Herrero A, Flores E, editor. Norfolk, UK: Caister Academic Press; 2008. Bioactive compounds produced by cyanobacteria; pp. 159–197. [Google Scholar]
- Velasquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol. 2011;15(1):11–21. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker M, von Döhren H. Cyanobacterial peptides - nature's own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol. 2010;86(5):1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fewer DP, Sivonen K. Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria. PLoS One. 2011;6(7):e22384. doi: 10.1371/journal.pone.0022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Nakajima N, Okamoto S, Suzuki I, Tanabe Y, Tamaoki M, Nakamura Y, Kasai F, Watanabe A, Kawashima K, Kishida Y, Ono A, Shimizu Y, Takahashi C, Minami C, Fujishiro T, Kohara M, Katoh M, Nakazaki N, Nakayama S, Yamada M, Tabata S, Watanabe MM. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 2007;14(6):247–256. doi: 10.1093/dnares/dsm026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangeul L, Quillardet P, Castets AM, Humbert JF, Matthijs HC, Cortez D, Tolonen A, Zhang CC, Gribaldo S, Kehr JC, Zilliges Y, Ziemert N, Becker S, Talla E, Latifi A, Billault A, Lepelletier A, Dittmann E, Bouchier C, de Marsac NT. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics. 2008;9:274. doi: 10.1186/1471-2164-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounge TB, Rohrlack T, Nederbragt AJ, Kristensen T, Jakobsen KS. A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain. BMC Genomics. 2009;10:396. doi: 10.1186/1471-2164-10-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucken K, John U, Cembella A, Murillo AA, Soto-Liebe K, Fuentes-Valdes JJ, Friedel M, Plominsky AM, Vasquez M, Glockner G. The smallest known genomes of multicellular and toxic cyanobacteria: comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS One. 2010;5(2):e9235. doi: 10.1371/journal.pone.0009235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53(1):9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29(1):123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Database issue):D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N. Small mobile sequences in bacteria display diverse structure/function motifs. Mol Microbiol. 2008;67(3):475–481. doi: 10.1111/j.1365-2958.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhiainen L, Paulin L, Suomalainen S, Hyytiäinen H, Buikema W, Haselkorn R, Sivonen K. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol Microbiol. 2000;37(1):156–167. doi: 10.1046/j.1365-2958.2000.01982.x. [DOI] [PubMed] [Google Scholar]
- Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria) Chem Biol. 2010;17(3):265–273. doi: 10.1016/j.chembiol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Rouhiainen L, Vakkilainen T, Siemer BL, Buikema W, Haselkorn R, Sivonen K. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl Environ Microbiol. 2004;70(2):686–692. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikoski N, Fewer DP, Jokela J, Wahlsten M, Rouhiainen L, Sivonen K. Highly diverse cyanobactins in strains of the genus Anabaena. Appl Environ Microbiol. 2010;76(3):701–709. doi: 10.1128/AEM.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Bibby TS, Jiang L, Irwin AJ, Falkowski PG. Protein interactions limit the rate of evolution of photosynthetic genes in cyanobacteria. Mol Biol Evol. 2005;22(11):2179–2189. doi: 10.1093/molbev/msi216. [DOI] [PubMed] [Google Scholar]
- Golden JW, Robinson SJ, Haselkorn R. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature. 1985;314(6010):419–423. doi: 10.1038/314419a0. [DOI] [PubMed] [Google Scholar]
- Golden JW, Yoon HS. Heterocyst development in Anabaena. Curr Opin Microbiol. 2003;6(6):557–563. doi: 10.1016/j.mib.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Fiedler G, Arnold M, Hannus S, Maldener I. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol. 1998;27(6):1193–1202. doi: 10.1046/j.1365-2958.1998.00762.x. [DOI] [PubMed] [Google Scholar]
- van Keulen G, Hopwood DA, Dijkhuizen L, Sawers RG. Gas vesicles in actinomycetes: old buoys in novel habitats? Trends Microbiol. 2005;13(8):350–354. doi: 10.1016/j.tim.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Welsh EA, Liberton M, Stockel J, Loh T, Elvitigala T, Wang C, Wollam A, Fulton RS, Clifton SW, Jacobs JM, Aurora R, Ghosh BK, Sherman LA, Smith RD, Wilson RK, Pakrasi HB. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc Natl Acad Sci USA. 2008;105(39):15094–15099. doi: 10.1073/pnas.0805418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel J, Kingsford C, Pop M. Assessing the benefits of using mate-pairs to resolve repeats in de novo short-read prokaryotic assemblies. BMC Bioinformatics. 2011;12:95. doi: 10.1186/1471-2105-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8(1):61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H, Zilliges Y, Lockau W, Steuer R. The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiol. 2010;154(1):410–422. doi: 10.1104/pp.110.157198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol. 2007;189(14):5247–5256. doi: 10.1128/JB.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battchikova N, Vainonen JP, Vorontsova N, Keränen M, Carmel D, Aro EM. Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J Proteome Res. 2010;9(11):5896–5912. doi: 10.1021/pr100651w. [DOI] [PubMed] [Google Scholar]
- Wegener KM, Singh AK, Jacobs JM, Elvitigala T, Welsh EA, Keren N, Gritsenko MA, Ghosh BK, Camp DG 2nd, Smith RD, Pakrasi HB. Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol Cell Proteomics. 2010;9(12):2678–2689. doi: 10.1074/mcp.M110.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Siguier P, Filee J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 2006;9(5):526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62(3):725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Haas S, Zemojtel T, Xiao P, Vingron M, Li R. Genome-wide comparison of cyanobacterial transposable elements, potential genetic diversity indicators. Gene. 2011;473(2):139–149. doi: 10.1016/j.gene.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Oggioni MR, Claverys JP. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology. 1999;145(Pt 10):2647–2653. doi: 10.1099/00221287-145-10-2647. [DOI] [PubMed] [Google Scholar]
- Ran L, Larsson J, Vigil-Stenman T, Nylander JA, Ininbergs K, Zheng WW, Lapidus A, Lowry S, Haselkorn R, Bergman B. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One. 2010;5(7):e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WC, Wollerman L, Bhaya D, Heidelberg JF. Analysis of insertion sequences in thermophilic cyanobacteria: exploring the mechanisms of establishing, maintaining, and withstanding high insertion sequence abundance. Appl Environ Microbiol. 2011;77(15):5458–5466. doi: 10.1128/AEM.05090-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AC, Monroe EA, Podell S, Hess WR, Klages S, Esquenazi E, Niessen S, Hoover H, Rothmann M, Lasken RS, Yates JR 3rd, Reinhardt R, Kube M, Burkart MD, Allen EE, Dorrestein PC, Gerwick WH, Gerwick L. Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proc Natl Acad Sci USA. 2011;108(21):8815–8820. doi: 10.1073/pnas.1101137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikoski N, Fewer DP, Sivonen K. Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria. Appl Environ Microbiol. 2009;75(3):853–857. doi: 10.1128/AEM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci USA. 2010;107(23):10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Börner T, Sivonen K. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc Natl Acad Sci USA. 2004;101(2):568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Kurmayer R, Liu Q, Börner T. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl Environ Microbiol. 2006;72(1):117–123. doi: 10.1128/AEM.72.1.117-123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewer DP, Halinen K, Sipari H, Bernardová K, Mänttäri M, Eronen E, Sivonen K. Non-autonomous transposable elements associated with inactivation of microcystin gene clusters in strains of the genus Anabaena isolated from the Baltic Sea. Env Microbiol Rep. 2011;3(2):189–194. doi: 10.1111/j.1758-2229.2010.00207.x. [DOI] [PubMed] [Google Scholar]
- Rouhiainen L, Sivonen K, Buikema WJ, Haselkorn R. Characterization of toxin-producing cyanobacteria by using an oligonucleotide probe containing a tandemly repeated heptamer. J Bacteriol. 1995;177(20):6021–6026. doi: 10.1128/jb.177.20.6021-6026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlouka A, Comte K, Castets AM, Bouchier C, Tandeau de Marsac N. The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to loss of cell buoyancy. J Bacteriol. 2004;186(8):2355–2365. doi: 10.1128/JB.186.8.2355-2365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ershova AS, Karyagina AS, Vasiliev MO, Lyashchuk AM, Lunin VG, Spirin SA, Alexeevski AV. Solitary restriction endonucleases in prokaryotic genomes. Nucleic Acids Res. 2012;40(20):10107–10115. doi: 10.1093/nar/gks853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyra C, Halme T, Torsti AM, Tenkanen T, Sivonen K. Site-specific restriction endonucleases in cyanobacteria. J Appl Microbiol. 2000;89(6):979–991. doi: 10.1046/j.1365-2672.2000.01206.x. [DOI] [PubMed] [Google Scholar]
- Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Vioque A. Transformation of cyanobacteria. Adv Exp Med Biol. 2007;616:12–22. doi: 10.1007/978-0-387-75532-8_2. [DOI] [PubMed] [Google Scholar]
- Zhao F, Zhang X, Liang C, Wu J, Bao Q, Qin S. Genome-wide analysis of restriction-modification system in unicellular and filamentous cyanobacteria. Physiol Genomics. 2006;24(3):181–190. doi: 10.1152/physiolgenomics.00255.2005. [DOI] [PubMed] [Google Scholar]
- Matic I, Taddei F, Radman M. Genetic barriers among bacteria. Trends Microbiol. 1996;4(2):69–72. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome – restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9(6):649–656. doi: 10.1016/S0959-437X(99)00026-X. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8(5):205–213. doi: 10.1093/dnares/8.5.205. 227–53. [DOI] [PubMed] [Google Scholar]
- Golden JW, Whorff LL, Wiest DR. Independent regulation ofnifHDKoperon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacteriumAnabaena sp. strain PCC 7120. J Bacteriol. 1991;173(22):7098–7105. doi: 10.1128/jb.173.22.7098-7105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco CD, Buettner JA, Golden JW. Programmed DNA rearrangement of a cyanobacterial hupL gene in heterocysts. Proc Natl Acad Sci USA. 1995;92(3):791–795. doi: 10.1073/pnas.92.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson BJ, Hartman L, Watson LE, Barnum SR. Evolution and variation of the nifD and hupL elements in the heterocystous cyanobacteria. Int J Syst Evol Microbiol. 2011;61(Pt 12):2938–2949. doi: 10.1099/ijs.0.028340-0. [DOI] [PubMed] [Google Scholar]
- Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev. 1992;56(2):340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthakur PB, Orozco CC, Young-Robbins SS, Haselkorn R, Callahan SM. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 2005;57(1):111–123. doi: 10.1111/j.1365-2958.2005.04678.x. [DOI] [PubMed] [Google Scholar]
- Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki P, Atlas R. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth Res. 2001;70(1):85–106. doi: 10.1023/A:1013840025518. [DOI] [PubMed] [Google Scholar]
- Ashby MK, Houmard J. Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol Mol Biol Rev. 2006;70(2):472–509. doi: 10.1128/MMBR.00046-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivonen K, Namikoshi M, Evans WR, Carmichael WW, Sun F, Rouhiainen L, Luukkainen R, Rinehart KL. Isolation and characterization of a variety of microcystins from seven strains of the cyanobacterial genus Anabaena. Appl Environ Microbiol. 1992;58(8):2495–2500. doi: 10.1128/aem.58.8.2495-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaniemi P, Hrouzek P, Kastovska K, Willame R, Rantala A, Hoffmann L, Komarek J, Sivonen K. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales, Cyanobacteria) Int J Syst Evol Microbiol. 2005;55(Pt 1):11–26. doi: 10.1099/ijs.0.63276-0. [DOI] [PubMed] [Google Scholar]
- Phred, Phrap, and Consed. http://www.phrap.org/phredphrapconsed.html.
- Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11(4):614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE−a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38(Database issue):D234–236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, Altman T, Paulsen I, Keseler IM, Caspi R. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 2010;11(1):40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ML, Karp PD. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Hypothetical proteins in the Anabaena sp. 90 genome. Hypothetical proteins with disrupted ORF are labelled in yellow. Table S2. Pseudogenes annotated from the Anabaena sp. 90 genome. Table S3. Summary of ISs identified in the Anabaena sp. 90 genome. Table S4. List of MITEs discovered in the Anabaena sp. 90 genome. The MITEs that disrupted chromosome ORFs by insertion are labelled in green. Table S5. RM system genes in the Anabaena sp. 90 genome. Table S6. Summary of transporter proteins in the Anabaena sp. 90 genome. Table S7. Two-component system genes annotated from the Anabaena sp. 90 genome. The two-component genes were classified as HK, RR and HR, representing histidine kinase, response regulator, and hybrid kinase, respectively. Pseudogenes with disrupted ORF are labelled in yellow. Table S8. Predicted metabolic pathways of Anabaena sp. 90.