Abstract
Objective
There is emerging evidence from animal studies suggesting a key role for methylation in the pathogenesis of essential hypertension. However, to date, very few studies have investigated the role of methylation in the development of human hypertension, and none has taken a genome-wide approach. Based on the recent studies that highlight the involvement of inflammation in the development of hypertension, we hypothesize that changes in DNA methylation of leukocytes are involved in the pathogenesis of hypertension.
Method & Results
We conducted a genome-wide methylation analysis on 8 hypertensive cases and 8 normotensive age-matched controls aged 14–23 years and performed validation of the most significant CpG sites in 2 genes in an independent sample of 36 hypertensive cases and 60 normotensive controls aged 14–30 years. Validation of the CpG sites in the SULF1 gene was further conducted in a second replication sample of 36 hypertensive cases and 34 controls aged 15.8–40 years. A CpG site in the SULF1 gene showed higher methylation levels in cases than in healthy controls in the genome-wide step (p = 6.2×10−5), which was confirmed in the validation step (p = 0.011) for subjects ≤30 years old but was not significant for subjects of all ages combined (p = 0.095).
Conclusion
The identification of a difference in a blood leukocyte DNA methylation site between hypertensive cases and normotensive controls suggests that changes in DNA methylation may play an important role in the pathogenesis of hypertension. The age dependency of the effect further suggests complexity of epigenetic regulation in this age-related disease.
Introduction
Essential hypertension (EH) is a major health problem worldwide with approximately one in three adults suffering from the disease. Although twin and family studies highlight a clear inherited component to EH [1], the current purely sequence-based approach only accounts for a fraction of the genetic risk of the disease as evidenced by the recent genome-wide association studies in which the identified genetic variants explain less than 1% of the blood pressure (BP) variation in the population [2]. Several epidemiological and clinical peculiarities of EH such as the incomplete concordance between monozygotic (MZ) twins (ranges from 38% to 52%) [3], [4] and its late onset and progressive nature, are difficult to explain with traditional DNA sequence-based approaches. These observations may indicate the involvement of epigenetic factors in EH development. Epigenetics refers to all meiotically and mitotically heritable changes in gene expression that are not coded in the DNA sequence itself. DNA methylation is an important epigenetic modification and can play a significant regulatory role in both normal and pathological cellular processes. Emerging evidence from animal studies [5], [6], [7], [8] suggests a key role for methylation in the pathogenesis of EH. However, to date, very few studies [9], [10] have investigated the role of methylation in the development of human EH, and none has taken a genome-wide approach. Based on recent studies [11], [12] that highlight the involvement of inflammation in the development of EH, we hypothesized that changes in the DNA methylation of leukocytes are involved in the pathogenesis of EH. The goal of this study was to characterize the DNA methylation profile in peripheral blood leukocytes in EH cases versus normotensive controls using a 3-stage genome-wide approach.
Methods
Subjects
A total of 44 EH cases and 68 age (±2 years) matched controls were selected from 4 existing cohorts in Georgia Prevention Institute, Georgia Health Sciences University using the following inclusion criteria: (1) African American (AA) ancestry; (2) male; (3) having leukocyte DNA available; (4) EH cases have age, sex, and height adjusted systolic BP (SBP) ≥95th percentile (if the age of the subject is less than 20 years), or SBP≥140 mmHg, while controls have age, sex and height adjusted SBP≤20th percentile, or have SBP levels<120 mmHg. These 4 cohorts include the BP stress study (n = 603) [13], the Georgia Cardiovascular twin study (n = 1183) [14], the Lifestyle, Adiposity, and Cardiovascular Health in Youth (LACHY) study (n = 740) [15], and the Prevention of Hypertension in African American Teens (PHAT) study (n = 262) [16]. Both the BP Stress study and the twin study are on-going longitudinal studies which have followed the subjects for more than 10 years. Both studies included roughly equal numbers of AAs and European Americans (EA) or males and females. The BP stress study was established in 1989 with subjects aged 7–16 years at baseline [13] and the Georgia Cardiovascular Twin study was established in 1996 with subjects aged 7–25 years at baseline [14]. LACHY and PHAT are cross-sectional studies. The LACHY study [15]consisted of roughly equal numbers of AA and EA adolescents aged 14–18 years of both sexes and the PHAT study [16] consisted of AA males and females aged 14–20 years. For the subjects from the BP Stress and the Georgia Cardiovascular Twin study, if multiple visits (with multiple leukocyte DNA) were available for a subject, the leukocyte DNA collected at the visit when the subject had the highest (for cases) or lowest (for controls) SBP was used. For the subjects from the twin study, only one twin from a pair was selected if both twin and co-twin met the criteria.
Subjects in all the 4 studies were recruited from Augusta, GA area. For all four cohorts self identification by self-reports of each subject or by a parent if the subject was under 18 years of age was used to classify ethnicity according to previously described criteria [17]. Subjects in all the 4 studies were overtly healthy, free of any acute or chronic illness on the basis of parental reports and were not on anti-hypertensive, lipid lowering, anti-diabetic and anti-inflammatory medications [13], [14], [15], [16]. The Institutional Review Board at the Medical College of Georgia approved the studies. Written informed consent was obtained from all subjects and by parents if subjects were younger than 18 years of age.
The discovery panel included 7 EH cases and 7 age matched controls selected from the 44 EH cases and 68 controls from the Georgia cohorts. To increase the power, we also included 1 AA male MZ pair discordant for EH selected from the Georgia CV twin study in which the EH case had age, sex, and height adjusted SBP≥95th percentile while the control had SBP levels 20 mmHg less than the case.
We included the remaining 37 cases and 61 controls not used in the discovery panel into the first replication panel. However, one EH case and one control failed the pyrosequencing assay. In total, the first replication stage included 36 cases and 60 controls.
The subjects in the second replication panel were selected from two existing cohorts at Thomas Jefferson University, Philadelphia, Pennsylvania. The first cohort included 412 young AA adults aged 18–49 years [18] and the second cohort included 300 AA youth aged 12–19 years [19]. A total of 38 AA male cases (SBP≥140 mmHg or DBP≥90 mmHg with or without medication) and 38 age (±2 years) and BMI (normal/overweight/obese) matched male controls (SBP<120 mmHg and DBP<80 mmHg) were selected from the young adult cohort and 6 AA male EH cases (SBP≥140 mmHg) and 6 age (±2 years) and BMI (normal/overweight/obese) matched controls (SBP<110 mmHg and DBP<70 mmHg) were selected from the youth cohort. Four more controls from the young adult cohort were further included to increase the overall number to 92, which is the number of samples that can be measured by pyrosequencing in one plate. Unfortunately, 22 subjects turned out to have insufficient DNA available to conduct the pyrosequencing. That is, in this 2nd replication cohort data were only available in 70 subjects (63 from the young adult cohort and 7 from the youth cohort).
The participants in both the young adult cohort and the youth cohort were living in urban Philadelphia. All protocols were approved by the Institutional Review Board of Thomas Jefferson University and written informed consent was obtained from each participant at enrollment or from parents if subjects were less than 18 years of age. The participants in the youth cohort were overtly healthy, free of any acute or chronic illness on the basis of parental reports and were not on anti-hypertensive, lipid lowering, anti-diabetic and anti-inflammatory medications. For the young adult cohort, the exclusion criteria included secondary HBP, history of diabetes, renal disease, heart failure, autoimmune disease, sickle cell anemia, or endocrine disorders.
Measurements
For all the four Georgia cohorts, height and weight were measured by standard methods using a wall-mounted stadiometer and a scale, respectively. Body mass index (BMI) was calculated as weight/height2. SBP and diastolic BP (DBP) were measured with Dinamap monitors, using an appropriately sized BP cuff placed on the subject’s right arm. BP measurements were taken at 11, 13, and 15 minutes, during a 15-minute supine relaxation period. The average of the last 2 readings was used to represent SBP and DBP values [13], [14], [15], [16].
Fasting peripheral blood samples in the LACHY cohort and non-fasting peripheral blood samples in the other three cohorts were collected. The buffy coat and plasma samples were separated and stored at −80°C. DNA was extracted from the buffy coat.
For the two cohorts in Pennsylvania, BP was measured, in the seated position, by auscultation. The average of eight separate BP measurements obtained at two separate visits (four measurements at each visit) was used to represent BP values. Fasting peripheral blood samples were collected and DNA was extracted from the buffy coat [18], [19].
Genome-wide Methylation Chip
The HumanMethylation27 BeadChip from Illumina (Illumina, San Diego, CA, USA) was used. This chip can quantitatively measure 27, 000 CpG sites, covering more than 14,000 well-annotated genes at single-CpG resolution. Each chip can accommodate 12 samples. After bisulfite treatment, 200 ng of the converted DNA was whole genome amplified (WGA) and enzymatically fragmented. The bisulfite-converted WGA-DNA samples were purified and applied to the BeadChips. Image processing and intensity data extraction were performed according to Illumina’s instructions (www.illumina.com/products/infinium_humanmethylation27_beadchip_kits.ilmn). Each methylation data point is represented by fluorescent signals from the methylated and unmethylated alleles. DNA methylation beta values are continuous variables between 0 (completely unmethylated) and 1 (completely methylated), representing the ratio of the intensity of the methylated bead type to the combined locus intensity. Initial array processing and quality control were performed with BeadStudio software. The microarray data discussed in this paper have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE42774.
Pyrosequencing
The methylation levels of the top CpG sites from the 2 genes selected for replication were determined by pyrosequencing technology, a rapid and robust method for quantitative methylation analysis. After bisulfite treatment, 10 ng of the converted DNA was used in a PCR reaction to amplify the target region. One of the PCR primers was biotin labeled. Single-stranded biotinylated PCR products were prepared for sequencing by use of the Pyrosequencing Vacuum Prep Tool according to the manufacturer’s instructions. The PCR products (each 10 µl) were sequenced by Pyrosequencing PSQ96 HS System (Pyrosequencing-Qiagen) following the manufacturer’s instructions. The methylation status of each locus was analyzed individually as a T/C SNP using QCpG software (Biotage, Kungsgatan, Sweden). PCR primers and sequencing primers for the 2 genes selected for replication are available upon request.
Statistical Analysis
To identify genome wide methylation differences between EH cases and controls, the LIMMA (Linear Models for Microarray Analysis) package from the Bioconductor project [20] was used. LIMMA uses an empirical Bayes approach that uses the variability in all genes for testing for significant differences, which results in more stable inferences for a relatively small number of arrays. We used a design matrix of a paired test to analyze each CpG site for differential methylation. Each CpG site was assigned a raw p-value based on a moderated t statistic. To correct for multiple testing, the set of raw p-values were converted to false discovery rates (FDR) according to Benjamini and Hochberg [21]. For the replication cohort, the methylation levels of the CpG sites were square root-transformed to obtain a better approximation of the normal distribution prior to analysis. A Student’s t-test was used to investigate whether their methylation levels differed between cases and controls. Linear regression was further used to adjust for the potential effect of age and BMI. We combined the replication steps as well as the genome wide step on the CpG sites carried to the validation stages with meta-analysis using the weighted z score–based approach implemented in the package METAL [22]. Preliminary analyses, t-tests and regression analyses were done using STATA 8 (StataCorp, College Station, Texas).
Gene ontology analysis was conducted with the FatiGO tool [23]. FatiGO takes two lists of genes and converts them into two lists of GO terms. Then a Fisher's exact test for 2×2 contingency tables is used to check for significant over-representation of GO terms in one of the sets with respect to the other one. Multiple testing correction (indexed by adjusted p values) to account for the multiple hypotheses tested (one for each GO term) was applied to reduce the likelihood of false positives. Since at least two CpG sites were included for the majority of genes in the genome-wide chip, for each gene we only used the CpG site with the lowest p value.
Results
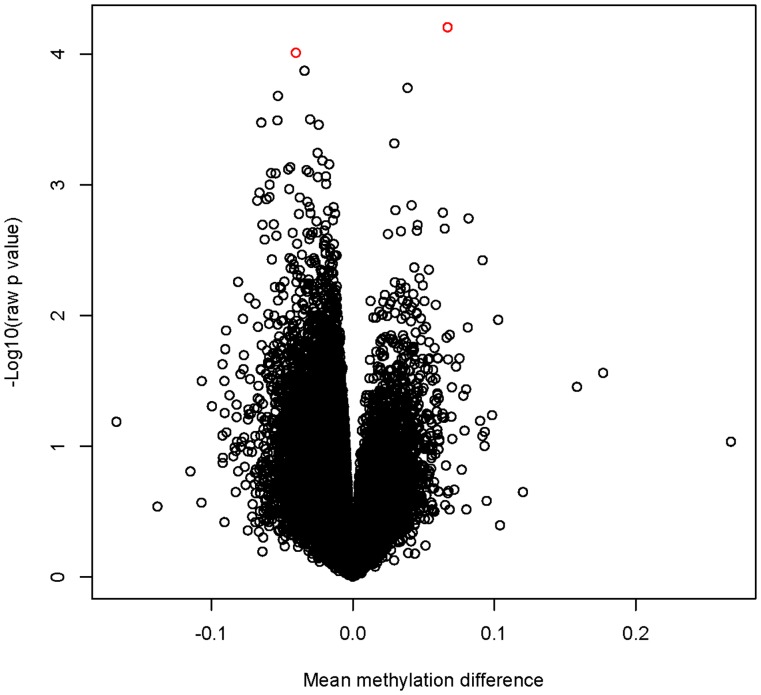
Table 1 displays the general characteristics of the 7 EH cases and 7 controls in the discovery panel and Table S1 displays the general characteristics of the 1 MZ pair discordant for EH. Figure 1 is a volcano plot showing the raw p-values for all CpG sites versus mean methylation difference between the case and the control group. We did not observe any CpG sites survive multiple testing corrections with the most significant CpG site showing a FDR of 0.75 and a raw p value of 6.2×10−5. Table 2 lists the top 10 most significant CpG sites. Out of the 10 CpG sites, we selected the top 2 CpG sites (one CpG site in the sulfatase 1 gene [SULF1] and one CpG site in the prolylcarboxypeptidase gene [PRCP]) for validation in the replication cohort. The general characteristics of the first replication cohort are displayed in Table 3. Although the pyrosequencing assays were designed to target one specific CpG site for each gene (Illumina ID cg04845579 for SULF1 and cg09772827 for PRCP), both assays covered several surrounding CpG sites. For the SULF1 gene, methylation levels on 4 CpG sites were obtained with CpG2 as the target CpG site. All these 4 CpG sites locate in the promoter region with a distance of 214, 186, 119, and 99 base pairs upstream to the transcription starting site. The differences in methylation status of these 4 CpG sites between cases and controls are shown in Table 4. The methylation levels of CpG1 and CpG2 (the target CpG site) were significantly higher in cases than in controls (p = 0.040 and 0.046, respectively). The results remained significant after adjustment for age (p = 0.041 and 0.038, respectively) but became non-significant after further adjustment for BMI (p = 0.074 and 0.081, respectively). For the PRCP gene, methylation levels on 8 CpG sites were obtained with CpG6 as the target CpG site. All these 8 CpG sites locate in the first intron with a distance of 293, 302, 305, 320, 323, 346, 353 and 365 base pairs downstream of the transcription starting site. The differences in methylation status of these 8 CpG sites between cases and controls are shown in Table S2. None of these CpG sites showed a significant difference in their methylation levels between cases and controls (p ranged from 0.22–0.87). The correlations within samples among the multiple CpG sites measured within each of these two genes are listed in Tables S3 and S4, respectively.
Table 1. General characteristics of cases and controls for genome-wide methylation analysis.
| Cases | Controls | |
| N | 7 | 7 |
| Age, years | 18.0±3.1 [14.8–23.3] | 18.3±3.2 [14.8–23.0] |
| BMI, kg/m2 | 27.3±3.5 [21.2–31.5] | 23.8±5.5 [17.8–33.2] |
| SBP, mmHg | 145.8±4.7 [137.3–149.5] | 107.6±3.3 [103.3–114] |
| SBP percentile | 0.99±0.01 [0.96–1.0] | 0.16±0.04 [0.10–0.19] |
| DBP, mmHg | 69.1±6.8 [62.0–78.3] | 55.4±5.4 [49.5–63.3] |
| DBP percentile | 0.45±0.22 [0.25–0.89] | 0.14±0.10 [0.01–0.26] |
Means±SD [Range].
Figure 1. Volcano plot showing raw p values versus mean methylation difference between cases and controls.
The two most significant CpG sites are highlighted in red.
Table 2. Top 10 differentially methylated CpG sites.
| Gene | Illumina ID | Distance to TSS | CpG island | Methylation, % | P | FDR | ||
| Case | Control | Difference | ||||||
| SULF1 | cg04845579 | 186 | NO | 29.49 | 22.79 | 6.70 | 0.000062 | 0.75 |
| PRCP | cg09772827 | 346 | YES | 13.85 | 17.87 | −4.03 | 0.000098 | 0.75 |
| NEUROG1 | cg14958635 | – | YES | 7.82 | 11.24 | −3.41 | 0.000134 | 0.75 |
| PITPNA | cg11719157 | 630 | YES | 61.38 | 57.51 | 3.87 | 0.000182 | 0.75 |
| SLC26A10 | cg14371590 | 222 | NO | 22.04 | 27.33 | −5.30 | 0.000209 | 0.75 |
| CDC34 | cg27431859 | 691 | YES | 10.43 | 13.45 | −3.02 | 0.000315 | 0.75 |
| C9orf95 | cg07962315 | 1375 | NO | 37.50 | 42.83 | −5.33 | 0.000321 | 0.75 |
| YWHAQ | cg06701500 | 565 | YES | 13.79 | 20.26 | −6.47 | 0.000334 | 0.75 |
| SIRT7 | cg15118204 | 191 | YES | 19.48 | 21.89 | −2.41 | 0.000348 | 0.75 |
| CLDN5 | cg04463638 | 148 | NO | 75.20 | 72.27 | 2.93 | 0.000483 | 0.75 |
TSS, transcription starting site.
Table 3. General characteristics of the subjects in GA cohort (1st replication panel).
| Case | Control | |
| N | 36 | 60 |
| Age, years | 20.6±5.3 [14.3–30.7] | 19.6±4.5 [14.1–30.9] |
| BMI, kg/m2 | 29.9±8.7 [21.0–52.4] | 24.7±8.0 [16.5–59.9] |
| SBP, mmHg | 143.6±7.5 [133.3–175] | 106.9±5.2 [93.7–117] |
| SBP percentile | 0.98±0.01 [0.96–1.00] | 0.12±0.06 [0.02–0.20] |
| DBP, mmHg | 71.9±10.4 [56.5–96.5] | 58.6±5.6 [46–74.5] |
| DBP percentile | 0.50±0.23 [0.18–0.98] | 0.20±0.13 [0.03–0.67] |
Means±SD [Range].
Table 4. Pyrosequencing results of SULF1 gene in GA cohort.
| Case | Control | P | P, adjusteda | P, adjustedb | |
| CpG1 | 16.8±9.0 | 13.4±7.5 | 0.040 | 0.041 | 0.074 |
| CpG2 | 22.7±9.4 | 19.0±8.6 | 0.046 | 0.038 | 0.081 |
| CpG3 | 5.94±3.04 | 4.8±3.22 | 0.105 | 0.098 | 0.204 |
| CpG4 | 20.6±9.1 | 17.8±7.9 | 0.106 | 0.101 | 0.238 |
Adjusted for age.
Adjusted for age and BMI.
The CpG sites in the SULF1 gene were further validated in the second replication panel. The general characteristics of this cohort are displayed in Table 5. To keep the second comparable to the first replication which only comprised subjects ≤30 years old, we further split the sample by age (≤30 years or >30 years). The general characteristics of the split samples were also listed in Table 5. The differences in methylation status of these 4 CpG sites in the SULF1 gene between cases and controls in the second replication panel are shown in Table 6. None of these 4 CpG sites showed a significant difference in their methylation levels between cases and controls either in the overall sample or in the samples split by age. Meta-analysis on the CpG1 and CpG2 with the two replication panels was conducted and the results are shown in Table 7. Significant higher methylation levels of CpG1 & CpG2 were observed in cases (p = 0.014 and p = 0.011, respectively) in the meta-analysis on the first replication cohort and the young age group of the second replication cohort. The significant result remained after adjustment of age (p = 0.017 and p = 0.015, respectively) or age and BMI (p = 0.030 and p = 0.037, respectively). Further meta-analysis with the discovery panel on CpG2 showed a p value of 0.0051 (with p = 0.0027 after adjustment of age and p = 0.0171 after adjustment of age and BMI) in all the subjects and a p value of 0.0004 (with p = 0.0004 after adjustment of age and p = 0.0054 after adjustment of age and BMI) in subjects younger or equal to 30 years old.
Table 5. General characteristics of the subjects in PA cohort (2nd replication panel).
| All subjects | Subjects older than 30 | Subjects younger or equal to 30 | ||||
| Case | Control | Case | Control | Case | Control | |
| N | 36 | 34 | 31 | 25 | 5 | 9 |
| Age, years | 39.3±7.6 | 35.6±9.7 | 41.8±3.8 | 40.8±3.7 | 23.6±6.2 | 21.1±5.1 |
| Age range, years | 16.8–49 | 15.8–47 | 33–49 | 34–47 | 16.8–30 | 15.8–28 |
| BMI, kg/m2 | 29.5±6.2 | 28.5±5.7 | 29.4±6.2 | 28.4±5.6 | 30.4±7.0 | 28.9±2.1 |
| SBP, mmHg | 149.5±16.3 | 108.3±6.4 | 149.8±17.4 | 108.1±6.7 | 147.6±5.6 | 109.0±2.0 |
| DBP, mmHg | 94.6±12.3 | 68.1±6.1 | 96.8±10.3 | 69±6.1 | 80.9±16.2 | 65.7±6.0 |
| Antihypertensive Medication | 25% | 0% | 29% | 0% | 0% | 0% |
Means±SD.
Table 6. Pyrosequencing results of SULF1 gene in the PA cohort.
| Case | Control | P | P, adjusteda | P, adjustedb | ||
| Overall | ||||||
| CpG1 | 12.5±6.2 | 12.1±5.6 | 0.77 | 0.61 | 0.65 | |
| CpG2 | 19.7±7.0 | 18.9±6.7 | 0.64 | 0.51 | 0.53 | |
| CpG3 | 5.3±2.1 | 5.6±1.9 | 0.59 | 0.77 | 0.69 | |
| CpG4 | 20.9±7.3 | 21.0±7.0 | 0.97 | 0.94 | 0.96 | |
| Older than30 | ||||||
| CpG1 | 11.9±6.4 | 12.0±5.9 | 0.81 | 0.88 | 0.80 | |
| CpG2 | 19.1±7.3 | 18.6±7.3 | 0.85 | 0.81 | 0.86 | |
| CpG3 | 5.1±2.1 | 5.6±2.2 | 0.39 | 0.41 | 0.35 | |
| CpG4 | 20.3±7.3 | 21.1±7.9 | 0.70 | 0.70 | 0.66 | |
| Younger or equal to 30 | ||||||
| CpG1 | 16.3±2.7 | 12.5±4.9 | 0.13 | 0.18 | 0.16 | |
| CpG2 | 23.6±3.3 | 19.7±4.6 | 0.12 | 0.20 | 0.25 | |
| CpG3 | 6.9±1.9 | 5.7±1.0 | 0.13 | 0.17 | 0.22 | |
| CpG4 | 24.9±6.4 | 20.8±4.2 | 0.17 | 0.23 | 0.21 | |
Adjusted for age.
Adjusted for age and BMI.
Table 7. Meta -analysis on two CpG sites in the SULF1 gene.
| P | P, adjusteda | P, adjustedb | ||
| CpG1 | GA cohort+PA cohort | 0.080 | 0.059 | 0.098 |
| GA cohort+PA cohort (age≤30) | 0.014 | 0.017 | 0.030 | |
| CpG2 | GA cohort+PA cohort | 0.095 | 0.058 | 0.105 |
| GA cohort+PA cohort (age≤30) | 0.011 | 0.015 | 0.037 | |
| Discovery+Replication | 0.0051 | 0.0027 | 0.0171 | |
| Discovery+Replication (age≤30) | 0.0004 | 0.0004 | 0.0054 |
Adjusted for age.
Adjusted for age and BMI.
Gene Ontology analysis was performed to test whether some common functional trends in molecular functions and biological processes were associated with the genes exhibiting differences between cases and controls for the genome-wide methylation analyses. We included those genes with a raw p≤0.01 in the first list (n = 226) and included all the other genes in the second list. As expected from a pilot study in 8 cases and 8 controls, we did not observe any GO categories survive multiple testing. Table S5 list the GO categories with raw P value less than 0.05. Interestingly, we observed enriched functional processes that are potentially relevant for inflammation with response to biotic stimulus (GO:0009607), response to other organism (GO:0051707), interleukin-1 production (GO:0032612), and interleukin-13 production (GO:0032616) among the top GO categories. The results are consistent with the involvement of inflammation and oxidative stress in the development of EH.
Discussion
In this study we aimed to identify methylation differences in peripheral blood leukocytes between EH cases and controls using a genome-wide approach in male AA youth and young adults. We observed increased methylation levels at two CpG sites in the SULF1 gene in EH cases in comparison with normotensive controls in subjects equal or younger than 30 years.
Our study is the first genome wide methylation study on EH. In fact, there are very few human studies that explored the role of epigenetics on the risk of EH. In one study, Friso et al [9] measured promoter methylation of the HSD11B2 gene in peripheral blood mononuclear cells from patients with EH and 32 subjects on prednisone therapy. Elevated HSD11B2 promoter methylation was associated with decreased HSD11B2 activity and EH development in glucocorticoid-treated patients. In a recent study by Smolarek et al [10] global DNA methylation level indexed by the genome level of 5-methylcytosine was significantly lower in patients with EH in comparison with controls.
The protein encoded by the SULF1 gene is Sulfatase 1 (Sulf1). It is a cell surface polypeptide that can rapidly modify the sulfation status of heparin sulfate proteoglycans (HSPGs), resulting in changes in HSPG-related signal transduction pathways [24]. Sulf1 has been reported to be down-regulated in several human cancers [25], [26]. Absence of Sulf1 in cancer cell lines is associated with increased cell growth, proliferation and reduced cell apoptosis [25]. In addition to cancer, Sulf1 was also studied with respect to normal development including neural, muscular, vascular and skeletal development. However, there is no direct study on Sulf1 and EH. SULF1 and SULF2 (another protein in this family) double knockout mice show impairment in skeletal development [27], but whether they display high blood pressure has never been explored.
In this study, we observed that the methylation levels of 2 CpG sites in the promoter region of the SULF1 gene were higher in EH cases than in controls. This is in line with the previous studies [28], [29] in cancers in which epigenetic silencing is involved in the down-regulation of Sulf1. The SULF1 gene spans a ∼211 kb genomic fragment on chromosome 8q13.3 with 23 exons [25]. Staub et al. [29] observed that methylation of 12 CpG sites within SULF1 exon 1A was associated with ovarian cancer cells and primary ovarian cancer tissues lacking Sulf1 expression. This region is about 10 kb downstream of the promoter region we targeted. The relationship between the methylation level of this region and the promoter region is unknown. No CpG island exists in this gene and the two CpG sites showing significant association with EH in our study locate at a distance of 214bp and 186 bp upstream from the transcription start site. There is a possibility that methylation of these two CpG sites or other CpG sites with methylation levels correlated with them inhibits the interactions between DNA sequence and nuclear proteins, resulting in changes in gene expression. In-silico analysis of the region of these two CpG site using TFSEARCH software [30] did not find they located at any known transcription factor binding sites. However, methylation of these two CpG sites may suppress gene transcription by recruiting methylcytosine-binding proteins that in turn associate with large protein complexes containing corepressors and histone deacetylases. The binding of these complexes to DNA may lead to a change in the chromatin structure from an active to an inactive form [31]. This speculation needs to be confirmed.
The age dependency of the effect of SULF1 gene methylation on EH may be related to the different pathogenesis of EH in youth in comparison with middle aged or older people. For examples, sympathetic nervous system hyperactivity is most apparent in youth with EH while abnormal development of aortic elasticity or reduced development of the microvascular network is more related to high BP later in life [32]. Furthermore, there is evidence showing that BP regulation may be controlled by different set of genes at different ages [1]. Two longitudinal twin studies [33], [34] have observed age-specific genetic variation for blood pressure, this is, there are new genes being switched on or off at different time points. On the other hand, it is also possible that the current study did not have enough power to find the effect of methylation on EH in middle aged or older people in consideration of the fact that disease-specific epigenetic alterations may be masked by the background of age-related and medication-arising epigenetic “drift” [35], [36].
The observed DNA methylation differences between EH cases and controls were relatively small. They were 6.7% in the genome-wide step and 4.1% in the replication step for the SULF1 gene CpG2. This modest level of differences raises an important question: what is the biological significance of changes in methylation to this degree? Although transcriptional profiling studies will be very valuable in understanding this question, cellular RNA is not available for the samples used in the current study, which were selected from several existing cohorts. We searched the GEO database and identified a dataset [37] (GSE3846) which included genome wide gene expression data in peripheral blood samples in six healthy volunteers tested by Affymetrix microarrays. In all the 6 samples, the expression of SULF1 was detectable. The average expression value of SULF1 is 4.38 at baseline. The average rank order of expression measurement in each subject is 69%, which means that about 31% genes have lower gene expression levels compared with SULF1 in peripheral blood samples of healthy individuals. This confirms that the SULF1 gene is expressed in peripheral blood cells. However, we failed to find any GEO dataset which included both methylation and gene expression data from peripheral blood samples. Therefore, we cannot test whether DNA methylation in SULF1 affects its expression in peripheral blood cells.
In this study, we used the DNA from leukocytes, which represent different cell populations with distinct epigenetic profiles [38]. A recent study by Kelsey’s group [39] indicated that shifts in leukocyte subpopulations may account for a considerable proportion of variability in peripheral blood DNA methylation of diseases such as head and neck squamous cell carcinoma and ovarian cancer. This study also provided a list of the top 50 differentially methylated CpG sites among 6 leukocyte subtypes including CD4+ T cells, CD8+ T cells, CD56+ NK cells, B cells, monocytes and granulocytes. The SULF1 gene CpG site is not within the list. Another study [40] from the same group developed an algorithm which predicts the distributions of the 6 leukocyte subtypes using illumina 27 k methylation data from peripheral blood DNA. We applied this algorithm to our data and did not observe difference in the distributions of these 6 cell types between EH cases and controls (Table S6). Therefore, it is highly unlikely that our significant finding on the SULF1 gene is caused by shifts in these 6 leukocyte subpopulations. On the other hand, it is plausible that only the change in the epigenetic profile of one specific cell type is related to EH. In this case, the actual epigenetic differences might be more substantial than reported here but only present in this specific blood leukocyte cell type. Future studies on epigenetic profiling of various types of cell populations of leukocytes are warranted to gain a greater understanding of the epigenetic dysregulation in EH.
Two strengths of our study deserve mentioning. First, we selected controls with low blood pressure, which maximizes the power to make discoveries. Second, a hypothesis free genome-wide approach was used. This approach supersedes the limitations imposed by candidate gene methylation studies and allows searching the whole genome in an unbiased manner.
Interpretation of these data is also limited by several additional constraints. First, in this study we aimed to identify EH related methylation changes. However, our study design cannot determine whether the identified methylation changes are the cause or the consequence of EH. Future studies on subjects with baseline DNA and follow-up for de novo development of EH will be needed to resolve causality [41]. Second, because obesity is an important risk factor for EH, patients often have higher BMI levels than normotensive controls. Obesity might be a confounder explaining the relation between methylation levels of the SULF1 gene and EH. In the first replication cohort, the association of the SULF1 gene CpG1 and CpG2 with EH attenuated and became non-significant after adjustment of BMI. Therefore, in the second replication cohort, controls were selected to match with cases on obesity status (normal weight/overweight/obese). We could not replicate the findings from the first 2 stages in the overall analysis. In addition to BMI, this discrepancy might also be due to the age difference between the second replication cohort and the cohorts used in the first 2 stages. Moreover, it is also possible that SULF1 gene methylation is a mediator of obesity related EH. In this case, including BMI as a covariate in the analysis or matching cases and controls on obesity status will lead to over-adjustment. Future studies on hypertensive subjects with normal weight are needed to clarify whether SULF1 gene methylation independently affects EH. Third, in the current study, the Infinium HumanMethylation27 Beadchip was selected because of its quantitative measure at each CpG site. However, the limited coverage of this genome-wide chip will restrict the findings to certain CpG sites within certain genes. Future studies should use chips with more complete coverage of the genome such as the recently released 450 K Infinium Methylation BeadChip from Illumina. Fourth, the current study is a pilot study with the genome-wide step conducted only in 7 EH cases and 7 normotensive controls and one MZ pair discordant for EH. Future studies with much larger sample size are warranted to discover a more complete profile of EH related methylation changes.
In conclusion, we identified a reproducible change in DNA methylation of peripheral blood leukocytes between EH cases and controls in subjects ≤30 years. It provides preliminary evidence that DNA methylation may play an important role in the pathogenesis of EH. Further studies are warranted to determine the causal direction of this relationship.
Supporting Information
General characteristics of the MZ pair discordant for EH.
(DOCX)
Replication results for PRCP gene.
(DOCX)
Correlation among the CpG sites in the SULF1 gene.
(DOCX)
Correlation among the CpG sites in the PRCP gene.
(DOCX)
Gene-Ontology analysis.
(DOCX)
Cell population estimates in cases vs. controls.
(DOCX)
Funding Statement
The participants in this study were recruited by several National Institutes of Health funded projects including HL69999, HL56622, HL077230, HL64157, DK046107 & HL092030. The current study is funded by the NLHBI HL105689. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang X, Snieder H (2010) Familiar aggregation of blood pressure. In: Flynn JT, Ingelfinger JR, Portman RJ, editors. Clinical Hypertension and Vascular Diseases: Pediatric Hypertension. 2nd ed: Totowa Humana Press Inc. 241–258.
- 2. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carmelli D, Cardon LR, Fabsitz R (1994) Clustering of hypertension, diabetes, and obesity in adult male twins: same genes or same environments? Am J Hum Genet 55: 566–573. [PMC free article] [PubMed] [Google Scholar]
- 4. Carmelli D, Robinette D, Fabsitz R (1994) Concordance, discordance and prevalence of hypertension in World War II male veteran twins. J Hypertens 12: 323–328. [PubMed] [Google Scholar]
- 5. Alikhani-Koopaei R, Fouladkou F, Frey FJ, Frey BM (2004) Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J Clin Invest 114: 1146–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogdarina I, Welham S, King PJ, Burns SP, Clark AJ (2007) Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res 100: 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frey FJ (2005) Methylation of CpG islands: potential relevance for hypertension and kidney diseases. Nephrol Dial Transplant 20: 868–869. [DOI] [PubMed] [Google Scholar]
- 8. Zhang D, Yu ZY, Cruz P, Kong Q, Li S, et al. (2009) Epigenetics and the control of epithelial sodium channel expression in collecting duct. Kidney Int 75: 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friso S, Pizzolo F, Choi SW, Guarini P, Castagna A, et al. (2008) Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis 199: 323–327. [DOI] [PubMed] [Google Scholar]
- 10. Smolarek I, Wyszko E, Barciszewska AM, Nowak S, Gawronska I, et al. (2010) Global DNA methylation changes in blood of patients with essential hypertension. Med Sci Monit 16: CR149–155. [PubMed] [Google Scholar]
- 11. Pauletto P, Rattazzi M (2006) Inflammation and hypertension: the search for a link. Nephrol Dial Transplant 21: 850–853. [DOI] [PubMed] [Google Scholar]
- 12. Savoia C, Schiffrin EL (2006) Inflammation in hypertension. Curr Opin Nephrol Hypertens 15: 152–158. [DOI] [PubMed] [Google Scholar]
- 13. Wang X, Poole JC, Treiber FA, Harshfield GA, Hanevold CD, et al. (2006) Ethnic and gender differences in ambulatory blood pressure trajectories: results from a 15-year longitudinal study in youth and young adults. Circulation 114: 2780–2787. [DOI] [PubMed] [Google Scholar]
- 14. Ge D, Dong Y, Wang X, Treiber FA, Snieder H (2006) The Georgia Cardiovascular Twin Study: influence of genetic predisposition and chronic stress on risk for cardiovascular disease and type 2 diabetes. Twin Res Hum Genet 9: 965–970. [DOI] [PubMed] [Google Scholar]
- 15. Gutin B, Johnson MH, Humphries MC, Hatfield-Laube JL, Kapuku GK, et al. (2007) Relationship of visceral adiposity to cardiovascular disease risk factors in black and white teens. Obesity (Silver Spring) 15: 1029–1035. [DOI] [PubMed] [Google Scholar]
- 16. Zhu H, Chao J, Guo D, Li K, Huang Y, et al. (2009) Urinary prostasin: a possible biomarker for renal pressure natriuresis in black adolescents. Pediatr Res 65: 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snieder H, Dong Y, Barbeau P, Harshfield GA, Dalageogou C, et al. (2002) Beta2-adrenergic receptor gene and resting hemodynamics in European and African American youth. Am J Hypertens 15: 973–979. [DOI] [PubMed] [Google Scholar]
- 18. Martinez Cantarin MP, Ertel A, Deloach S, Fortina P, Scott K, et al. (2010) Variants in genes involved in functional pathways associated with hypertension in African Americans. Clin Transl Sci 3: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falkner B, Deloach S, Keith SW, Gidding SS (2012) High blood pressure and obesity increase the risk for left ventricular hypertrophy in African American Adolescents. J Peadiatr in press. [DOI] [PMC free article] [PubMed]
- 20. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful appraoch to multiple testing. J Roy Statist Soc Ser B (Methodological) 57: 289–300. [Google Scholar]
- 22. Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, et al. (2008) Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet 40: 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Shahrour F, Diaz-Uriarte R, Dopazo J (2004) FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20: 578–580. [DOI] [PubMed] [Google Scholar]
- 24. Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, et al. (2005) HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem 280: 40066–40073. [DOI] [PubMed] [Google Scholar]
- 25. Lai J, Chien J, Staub J, Avula R, Greene EL, et al. (2003) Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem 278: 23107–23117. [DOI] [PubMed] [Google Scholar]
- 26. Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, et al. (2005) Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, et al. (2008) Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev Dyn 237: 339–353. [DOI] [PubMed] [Google Scholar]
- 28. Chen Z, Fan JQ, Li J, Li QS, Yan Z, et al. (2009) Promoter hypermethylation correlates with the Hsulf-1 silencing in human breast and gastric cancer. Int J Cancer 124: 739–744. [DOI] [PubMed] [Google Scholar]
- 29. Staub J, Chien J, Pan Y, Qian X, Narita K, et al. (2007) Epigenetic silencing of HSulf-1 in ovarian cancer:implications in chemoresistance. Oncogene 26: 4969–4978. [DOI] [PubMed] [Google Scholar]
- 30. Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, et al. (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Attwood JT, Yung RL, Richardson BC (2002) DNA methylation and the regulation of gene transcription. Cell Mol Life Sci 59: 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapur G, Mattoo TK (2010) Primary Hypertension. In: Flynn JT, Ingelfinger JR, Portman RJ, editors. Clinical Hypertension and Vascular Diseases: Pediatric Hypertension. 2nd ed: Totowa Humana Press Inc. 241–258.
- 33. Colletto GM, Cardon LR, Fulker DW (1993) A genetic and environmental time series analysis of blood pressure in male twins. Genet Epidemiol 10: 533–538. [DOI] [PubMed] [Google Scholar]
- 34. Kupper N, Ge D, Treiber FA, Snieder H (2006) Emergence of novel genetic effects on blood pressure and hemodynamics in adolescence: the Georgia Cardiovascular Twin Study. Hypertension 47: 948–954. [DOI] [PubMed] [Google Scholar]
- 35. Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS (2009) Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 5: 401–408. [DOI] [PubMed] [Google Scholar]
- 36. Groom A, Elliott HR, Embleton ND, Relton CL (2011) Epigenetics and child health: basic principles. Arch Dis Child 96: 863–869. [DOI] [PubMed] [Google Scholar]
- 37. Baty F, Facompre M, Wiegand J, Schwager J, Brutsche MH (2006) Analysis with respect to instrumental variables for the exploration of microarray data structures. BMC Bioinformatics 7: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, et al. (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7: e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, et al. (2012) Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol Biomarkers Prev 21: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, et al. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rakyan VK, Down TA, Balding DJ, Beck S (2011) Epigenome-wide association studies for common human diseases. Nat Rev Genet 12: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
General characteristics of the MZ pair discordant for EH.
(DOCX)
Replication results for PRCP gene.
(DOCX)
Correlation among the CpG sites in the SULF1 gene.
(DOCX)
Correlation among the CpG sites in the PRCP gene.
(DOCX)
Gene-Ontology analysis.
(DOCX)
Cell population estimates in cases vs. controls.
(DOCX)