Abstract
The Id family of helix–loop–helix (HLH) transcriptional regulatory proteins does not possess a basic DNA-binding domain and functions as a negative regulator of basic HLH transcription factors. Id proteins coordinate cell growth and differentiation pathways within mammalian cells and have been shown to regulate G1-S cell-cycle transitions. Although much recent data has implicated Id1 in playing a critical role in modulating cellular senescence, no direct genetic evidence has been reported to substantiate such work. Here we show that Id1-null primary mouse embryo fibroblasts undergo premature senescence despite normal growth profiles at early passage. These cells possess increased expression of the tumor-suppressor protein p16/Ink4a but not p19/ARF, and have decreased cyclin-dependent kinase (cdk) 2 and cdk4 kinase activity. We also show that Id1 is able to directly inhibit p16/Ink4a but not p19/ARF promoter activity via its HLH domain, and that Id1inhibits transcriptional activation at E-boxes within the p16/Ink4a promoter. Our data provide, to our knowledge, the first genetic evidence for a role for Id1 as an inhibitor of cellular senescence and suggest that Id1 functions to delay cellular senescence through repression of p16/Ink4a. Because epigenetic and genetic abrogation of p16/Ink4a function has been implicated in the evolution of several human malignancies, we propose that transcriptional regulation of p16/Ink4a may also provide a mechanism for the dysregulation of normal cellular growth controls during the evolution of human malignancies.
Basic helix–loop–helix (HLH) DNA-binding proteins regulate tissue-specific transcription within multiple cell lineages (1). The Id family of HLH proteins does not possess a basic DNA-binding domain and inhibits lineage commitment within multiple cell types through sequestration of basic HLH transcription factors (reviewed in ref. 2). The Id family member Id1 has been implicated in regulating cellular lifespan, and we and others have reported delayed senescence of primary human keratinocytes that constitutively express Id1 (3, 4). Because recent studies have illustrated the importance of the p16ink4a/retinoblastoma (pRb) tumor-suppressor pathway and telomerase activity in regulating primary mammalian cell growth and senescence (5–7), it has been postulated that Id1 regulation of cellular growth and senescence may function through direct regulation of these pathways. Previous studies have demonstrated Id1 reactivation of DNA synthesis in senescent human fibroblasts in cooperation with a mutant SV40 T antigen that is unable to bind pRb, suggesting that Id1 can antagonize the functions of pRb or other members of this tumor-suppressor pathway, including p16/Ink4a (8). More recently, Id1 has been demonstrated to oppose transcription factor (Ets)-mediated activation of p16/Ink4a via Ras-Raf-MEK signaling (9).
The p16/pRb pathway has been shown to be deregulated in a large majority of human tumors either through loss of p16 or pRb function or deregulated expression of cyclin D or cdk4 (reviewed in ref. 10). Several mechanisms of inactivation of the p16/retinoblastoma pathway have been noted in various tumors and transformed cell lines. Among the most common ways in which these gene products are dysregulated in tumors is through gene deletion, inactivating mutations, epigenetic changes such as promoter methylation, protein sequestration, and inactivation (e.g., viral oncoproteins), and posttranslational modification (e.g., inactivating phosphorylation events; ref. 11). To date, little is known about the direct transcriptional regulation of genes within the p16/pRb pathway. In the present study, we investigate the role of the HLH protein Id1 in transcriptional regulation of cellular growth and senescence by using embryo fibroblasts derived from Id1-null mice. We demonstrate that loss of Id1 promotes cellular senescence and that this is associated with increased expression of the cell-cycle inhibitor p16/Ink4a. We also provide evidence for DNA-binding motif (E-box)-mediated repression of the p16/Ink4a promoter by Id1, which may be important for Id1 regulation of cellular senescence.
Materials and Methods
Cell Culture.
Id1 −/− mice were generated as described (12) and are of identical genetic background to Id1 +/+ mice. Mouse embryo fibroblasts were prepared in the following manner. Heads and organs were dissected away from day 13.5 embryos and used for DNA genotype analysis. Embryos were rinsed twice in PBS, then minced in 500 μl of 0.25% trypsin/2 mM EDTA, and sheared in a 22-gauge syringe 3 times. Embryos then were incubated at 37°C for 15–20 min to disperse cells that were resuspended in DMEM supplemented with 10% FBS, 2 mM glutamine, and 100 units/ml penicillin and streptomycin, and plated in 60-mm tissue-culture dishes overnight. The next day, dead cells and debris were removed and cells were refed with the above media. Cells obtained at this stage were considered to be of the passage zero (P-0) generation. When these cells reached 80% confluency (2–4 days), they were trypsinized and plated into 100-mm dishes (P-1). Once these cells reached 80% confluency, they were counted and 1 × 106 cells were plated into a 75-cm2 flask (P-2). Cells at 80% confluency from the 75-cm2 flask were considered to be at P-3 and were passaged subsequently on a 3T9 protocol. In brief, 9 × 105 cells were plated in a 60-mm dish and counted every 3 days with subsequent replating of 9 × 105 cells at each passage until senescence was reached. Growth curves were initiated at P-3 with replicate cultures of 1 × 105 cells per 35-mm diameter dish. Cells were counted each day for 8 consecutive days. Growth curves and the 3T9 protocol were established from data generated by using mouse embryo fibroblasts (MEFs) from six different embryos for each genotype. These experiments were repeated 3 times (total of 18 embryos per genotype) from 3 different litters. Senescence-associated β-galactosidase (β-gal) staining was performed as previously described. Cells were stained at pH 4.0 for lysosomal β-gal (positive control), at pH 6.0 for senescence-associated β-gal, and at pH 7.5 for bacterial β-gal (negative control). Human foreskin keratinocytes were isolated, transfected, and cultured as described (3).
Immunohistochemistry.
Sections were blocked with 10% (vol/vol) normal horse serum (Vector Laboratories) in PBS-thromboxane (TX) and incubated with monoclonal anti-p16 (Santa Cruz Biotechnology) antibody diluted 1:250 or polyclonal anti-p19 (AEC40, gift of N. Sharpless and R. DePinho, Dana–Farber Cancer Institute, Boston) antibody diluted 1:200 in PBS-TX with 1% normal horse serum. Sections then were incubated with the biotinylated horse anti-mouse secondary antibody (Vector) diluted 1:200 in PBS with 1.5% normal horse serum. The Vectastain Elite ABC kit (Vector) was used.
Promoter Analysis.
Constructs possessing 1.2 kb of the human p16 promoter or the minimal murine p19/ARF promoter driving the luciferase-expression plasmid pGL2-Basic (Promega) were provided kindly (G. Peters, Imperial Cancer Research Fund, London, and C. Sherr, St. Jude Children's Research Hospital, Memphis, TN). NIH 3T3 cells at 60% confluence were transfected by using standard calcium-phosphate procedures with 1 μg of reporter and 0.3, 1.5, or 3 μg of a plasmid containing unique EcoRI cloning site and Maloney Sarcoma Virus long terminal repeat (EMSV)-Id1, EMSV-Id1ΔHLH, EMSV-Id1V91P, or EMSV-Id1ΔC-term (13). EMSV scribe was used as vector control and carrier DNA in order for total transfected DNA to equal 4 μg per 35-mm dish. Cells were harvested 48 h after transfection and luciferase activity was measured according to suppliers recommendations (Promega). All transfections were performed in triplicate and repeated at least 3 times. Transfections were normalized for efficiency by using either 0.01 μg of pRL vector (renilla luciferase assay) per transfection and measuring activity as recommended by the supplier (Promega) or by using 1 μg of pGreen Lantern plasmid/transfection and counting the percent of fluorescently labeled cells. The p16 promoter E-box mutants were prepared by using site-directed mutagenesis of the canonical E-box motif (CANNTG) to a nonfunctional motif (CANNTT; ref. 14). Mutants were verified by sequence analysis. Mutant constructs were transfected into NIH 3T3 cells as described above by using 1 μg of reporter plasmid and 1 μg of EMSV-Id1 or EMSV plasmid and FuGENE 6 reagent (Roche) according to the manufacturer's protocols. Transfections were normalized for efficiency by using 0.01 μg of pRL and assayed as described previously.
Western Blotting and Kinase Assays.
MEFs were lysed in 0.5% Nonidet P-40 lysis buffer (50 mM Tris, pH 7.5/250 mM NaCl/5 mM EDTA/1 mM DTT/0.5 mM PMSF/1 μg/ml aprotinin/leupeptin/2 mM NaF/0.5 mM sodium-vanadate) for 30 min at 0°C. Western blot analysis was according to standard methods, using enhanced chemiluminescence (Amersham Pharmacia). A list of antisera is available on request. Cdk2 kinase assays were performed on cell lysates as described above. After immunoprecipitation, samples were washed 3 times in lysis buffer and 2 times in kinase buffer (50 mM Hepes, pH 7.4/5 mM MnCl2/10 mM MgCl2/1 mM DTT). Samples then were resuspended in 10 μl of kinase buffer containing 10 μCi (1 Ci = 37 GBq) of [32P]ATP and 1 μM ATP (Amersham Pharmacia). Samples were incubated for 30 min at 37°C in the presence of 1 μg of histone H1. Kinase assays specific for cdk4 were performed as follows. Cells were lysed in DIP buffer [50 mM Hepes/150 mM NaCl/1 mM EDTA/2.5 mM EGTA/10% (vol/vol) glycerol/0.1% Tween-20/1 mM DTT with added phosphatase and protease inhibitors] and cleared by microcentrifugation for 5 min at 8,000 × g. After immunoprecipitation, samples were washed 3 times in lysis buffer and twice in kinase buffer. A C-terminal fragment of the retinoblastoma protein (pRb) fused to glutathione S-transferase (gift from E. Harlow, Massachusetts General Hospital, Charlestown, MA) was used as a substrate. Kinase assays were performed as described above.
Expression Analysis.
Total RNA was extracted by using guanidinium isothiocyanate (separated on 1.2% agarose) transferred to nitrocellulose and hybridized according to standard procedures with 32P-labeled probes specific for exon 1a of mouse p16, for exon 1β of mouse p19/ARF, for mouse Id1, or for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Semiquantitative reverse transcription–PCR was performed as described (15).
Results
Id1-Null MEFs Senesce Prematurely.
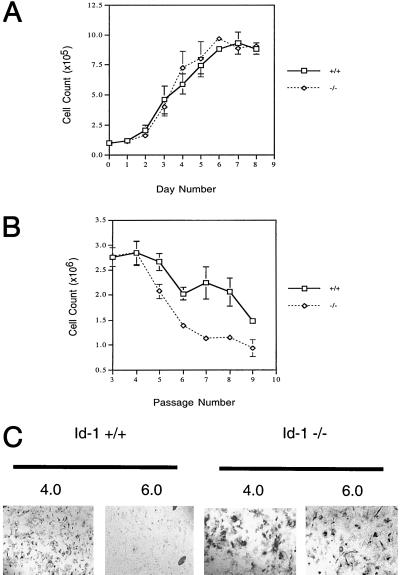
To investigate the role of Id1 in cellular proliferation, MEFs were prepared from Id1 +/+ and Id1 −/− embryos. Early passage (P-3) +/+ and −/− MEFs were evaluated under growth conditions with 10% (vol/vol) FCS and found to have identical growth profiles (Fig. 1A); however, evaluation of these cells on a 3T9 protocol revealed premature growth inhibition of Id1 −/− MEFs beginning at P-5 (Fig. 1B). Morphologic evaluation of these cells demonstrated cytoplasmic enlargement and flattening of Id1 −/− MEFs and 80–90% positive staining of these cells with senescence-associated β-gal (16) at P-8, indicating premature cellular senescence in contrast to normal (+/+) MEFs that were 10–20% positive (Fig. 1C).
Figure 1.
Id1 −/− MEFs exhibit premature cellular senescence. (A) Growth curves for Id-1 +/+ and −/− MEFs at P-3. Replicate cultures of 1 × 105 cells per 35-mm diameter dish were plated on day zero and cells were counted each day for 8 consecutive days (diao). (B) Senescence profile of Id-1 +/+ and −/− MEFs. P-3 MEFs were passaged on a 3T9 protocol in which 9 × 105 cells were plated in a 60-mm dish and counted every 3 days with subsequent replating of 9 × 105 cells at each passage until senescence was reached. (C) β-gal activity of P-8 MEFs. Cells were stained at pH 4.0 for lysosomal β-gal activity (positive control) and pH 6.0 for senescence-associated β-gal activity. Positive control is included to detail cellular morphology of Id1 +/+ vs. −/− cells at P-8 (×100).
Loss of Id1 Results in Inhibition of cdk2 and cdk4 Kinase Activity.
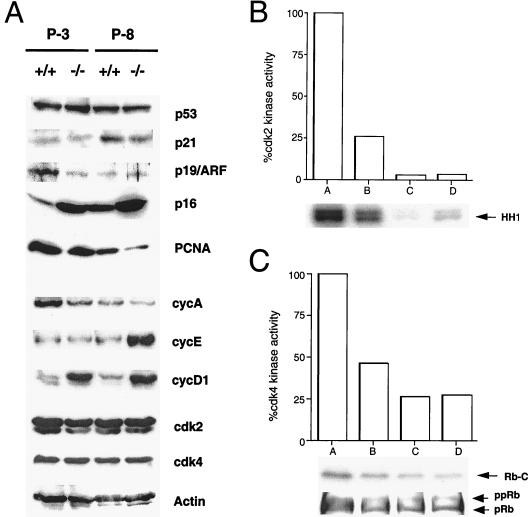
To determine the nature of premature senescence in Id1 −/− MEFs we evaluated the expression of various cell-cycle regulatory proteins previously implicated in regulating the senescence phenotype (reviewed in ref. 17). Consistent with a senescence phenotype, we observed up-regulation of p16 and cyclin D1 expression as well as down-regulation of cdk2, cyclin A, and PCNA expression in early passage (P-3) Id1 −/− MEFs (Fig. 2A). We also noted late (P-8) up-regulation of cyclin E in Id1 −/− MEFs. Although Id1 has been implicated in the regulation of cell-cycle control through inhibition of p21 expression by E2A (18), no significant differences were observed in p53 or p21 expression between normal and Id1−/− MEFs. Evaluation of cdk2 and cdk4 functions revealed 75% and 50% inhibition of kinase activity, respectively, in early passage MEFs lacking Id1 (Fig. 2 B and C) with hypophosphorylation of pRb in P-3 Id1 −/− MEFs (Fig. 2C) consistent with a premature senescence phenotype. By P-8, however, no significant differences in kinase activity or pRb phosphorylation state between Id1 +/+ and −/− MEFs were observed.
Figure 2.
Loss of Id1 affects the expression of cell-cycle regulatory proteins and cdk activity in early passage MEFs. (A) Western blot analysis of cell lysates from Id1 +/+ and −/− MEFs at early (P-3) and late (P-8) passages. Actin lane serves as loading control. (B and C) cdk2 and cdk4 kinase activity of Id1 +/+ and Id1 −/− MEFs. A, P-3 +/+ MEFs; B, P-3 −/− MEFs; C, P-8 +/+ MEFs; D, P-8 −/− MEFs. HH1 represents histone H1 phosphorylation and Rb-C represents phosphorylation of a C-terminal fragment of the retinoblastoma protein. Western blot for hyperphosphorylated (ppRb) or hypophosphorylated (pRb) is depicted below corresponding cdk4 kinase activity (29).
Id1 Represses p16/Ink4a Promoter Activity.
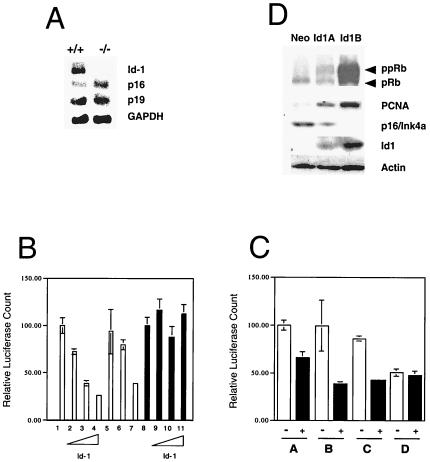
We next sought to determine whether loss of Id1 could transcriptionally regulate the expression of senescence-associated genes. Northern blotting and semiquantitative reverse transcription–PCR were performed (Fig. 3A and data not shown) on transcripts derived from P-3 Id1 +/+ and −/− MEFs. Because the p16 and p19/ARF transcripts are alternative splice variants derived from the Ink4a locus, and because both are capable of inducing cellular growth arrest (reviewed in refs. 19 and 20), we evaluated the expression of both genes in Id1-deficient cells. We found that +/+ MEFs expressed high levels of Id1 at P-3 and that p16 expression was low in these cells but significantly up-regulated in the −/− MEFs. Expression of p19 was not altered in Id1 +/+ vs. −/− MEFs.
Figure 3.
Id1 is a repressor of p16/Ink4a but not p19/ARF promoter activity. (A) Northern analysis of p16, p19, and Id1 expression at early (P-3) passage in Id1 wild-type (+/+) and Id1-null (−/−) MEFs. (B) Id1 repression of the p16/Ink4a promoter but not the p19/ARF promoter. 1, p16 promoter activity; 2–4, p16 promoter activity with 0.3, 1.5, or 3.0 μg of Id1; 5, p16 promoter + 3.0 μg of HLH dimerization domain of Id1 mutant of Id1; 6, p16 promoter + 3.0 μg of V91P mutant of Id1; 7, p16 promoter + 3.0 μg of ΔC-terminal mutant of Id1; 8, p19 promoter activity; 9–11, p19 promoter activity with 0.3, 1.5, or 3.0 μg of Id1. (C) Both the E-box at -349 and -615 bp of the p16/Ink4a promoter are critical for repression by Id1. White box, promoter alone; black box, promoter plus 1 μg of Id1; A, full-length p16 promoter (1.2 kb); B, p16 promoter with mutated E-box at -349 bp; C, p16 promoter with mutated E-box at -615 bp; D, p16 promoter with mutated E-boxes at both -349 and -615 bp. (D) Expression of p16 is repressed in Id1-transfected primary human keratinocytes. Western blot for expression of retinoblastoma protein, proliferating cell nuclear antigen, p16/Ink4a, and Id1 in transfected human foreskin keratinocytes expressing control plasmid (Neo) or Id1 for 1 month (Id1A) or 1 year (Id1B).
Class A basic HLH proteins have been shown to activate E-boxes within the p16/Ink4a promoter (21), suggesting that Id1 might function in directly repressing the p16/Ink4a promoter in vivo. More recently, Id1 has been demonstrated to oppose ras-mediated activation of the p16/Ink4a promoter by means of interactions with Ets-2 (9). Because the p16/Ink4a and p19/ARF transcripts are alternative splice variants derived from the Ink4a locus, and because both are capable of inducing cellular senescence, we sought to determine whether Id1 regulation of cellular senescence could be mediated by direct transcriptional effects on the expression of either of these genes. Of the p16 promoter, which possesses two E-box motifs (-349 and -615 bp), 1.2 kb was transfected into NIH 3T3 cells for these studies, whereas the minimal p19/ARF promoter, which does not possess any E-boxes (22), was used to evaluate Id1 effects on p19 promoter activity. We observed potent (75%) repression of the p16 promoter by full-length Id1 or a C-terminal Id1 deletion mutant (with an intact HLH domain), but no repression of the p19/ARF promoter in transient transfection assays in NIH 3T3 cells (Fig. 3A). Deletion of the HLH dimerization domain of Id1 abrogated the repressive effects on the p16 promoter as did a point mutation (V91P) within the HLH domain of Id1 that abolishes HLH dimerization (23). These data suggest that Id1 repression of p16 expression in this system functions by means of its ability to heterodimerize with other HLH proteins via its HLH domain. By using site-directed mutagenesis of the p16 promoter, we determined that mutation of either the first or second E-box within the p16 promoter had a minimal effect on the repression by Id1, but that mutation of both E-boxes simultaneously could completely abolish Id1 repression (Fig. 3B). We therefore concluded that a redundant role for E-boxes 1 and 2 in activation of the p16 promoter by E-proteins must exist, as has been described (21).
Because we had previously noted inactivation of the retinoblastoma protein in Id1-transfected primary human keratinocytes (3), we sought to determine whether this might be caused by the repression of the p16/Ink4a protein in Id1-expressing keratinocytes and subsequent inactivation of pRb through cdk4 kinase activity. We analyzed human foreskin keratinocytes that constitutively expressed Id1 for p16 expression levels and found that p16 expression inversely correlated with Id1 expression in these cells. Furthermore, we detected no p16 expression by Western analysis in Id1-immortalized human foreskin keratinocytes (Fig. 3D.).
p16/Ink4a Expression Is Elevated Within the Ventral Telencephalon of Id1-Null Embryos.
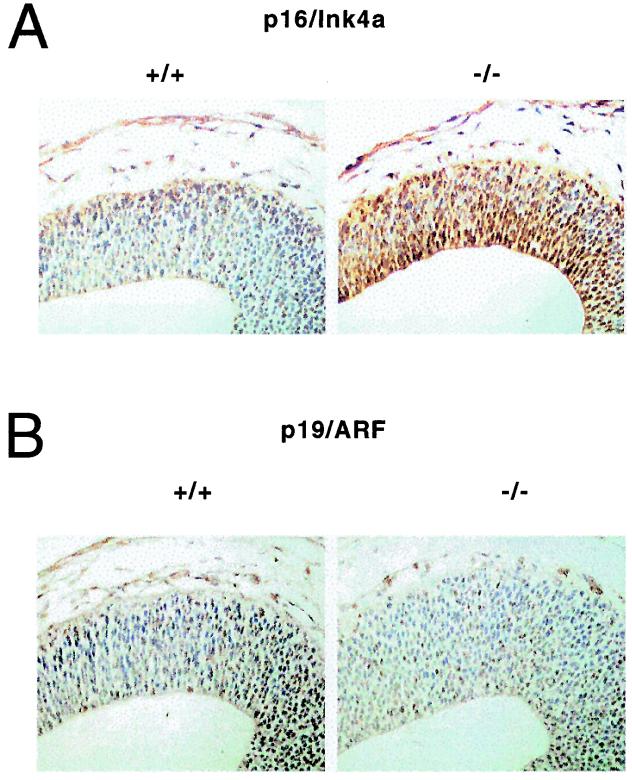
To determine the in vivo significance of Id1 repression of p16/Ink4a, we examined Id1 wild-type and Id1-null embryos for altered expression of p16/Ink4a and p19/ARF. Immunohistochemical analysis of p16/Ink4a and p19/ARF in embryonic day (E)11.5 embryonic tissue revealed increased p16/Ink4a expression in the subventricular zone of the ventral telencephalon of E11.5 −/− mouse embryos vs. wild-type animals, whereas p19/ARF expression was virtually undetectable in either Id1 wild-type or Id1-null embryos in this region (Fig. 4). Further examination of all other organs revealed low to undetectable expression of p16/Ink4a and p19, which did not differ significantly between wild-type and Id1-null embryos. Similar up-regulation of p16/Ink4a expression has been identified in the ventral telencephalon of Id1/Id3-double null E11.5 mouse embryos that possess altered neural differentiation and are lethal at E13.5 (24).
Figure 4.
Expression p16 is up-regulated in the ventral telencephalon of Id1 −/− mouse embryos. Immunohistochemical analysis of E11.5 mouse embryo forebrains stained for p16/Ink4a (A) or p19/ARF (B) (brown). Blue represents counterstain.
Discussion
Our data demonstrate a role for Id1 in regulating cellular growth and senescence that may be mediated by repressive effects on p16/Ink4a expression. Previous studies have demonstrated premature senescence in MEFs lacking the polycomb transcriptional repressor bmi-1 because of derepression at the ink4a locus and dysregulated expression of both p16Ink4a and p19/ARF (15). Because the Ink4a locus encodes two independent tumor-suppressor proteins with independent transcriptional regulatory regions (19), it is not surprising that Id1, a sequence-specific transcriptional repressor of E-box binding proteins (25), is only a functional inhibitor of a single Ink4a transcript, p16. Bmi-1, however, as a more globally functional transcriptional repressor, can function over long stretches of chromatin and influence the expression of both Ink4a-derived transcripts, which may account for the more severe phenotype in bmi-1 −/− mice and the more profound effect of bmi-1 loss on MEF lifespan, since these cells senesce at P-3. Although transiently induced p16 expression has been demonstrated to promote cellular senescence in human tumor cell lines (26)—the specific role of p16/Ink4a in regulating Id1-null premature senescence remains to be determined. Future studies in Id1/Ink4a-Exon 1a (p16/Ink4a-specific) double knockout MEFs will allow us to determine whether the premature senescence in Id1-null MEFs is mediated specifically by p16/Ink4a.
Our data also demonstrate Id1 repression of the p16/Ink4a promoter by means of E-box-mediated effects, whereas recent work has demonstrated Id1-mediated repression of the p16/Ink4a promoter through inactivation of Ets-2 functions (9). Although these data propose alternate mechanisms of p16 repression by Id1, the cell systems and conditions under which these studies were undertaken allow for both mechanisms to exist. Because Ohtani et al. (9) demonstrate significant activation of p16/Ink4a promoter activity by Ets proteins or Ras/Raf/MEK only in the setting of SV40-immortalized human fibroblasts or cotransfection of MEK with Ets proteins in primary human cells, it is unclear what the precise mechanism of p16 activation is by these individual factors in vivo. Additionally, Ets and Id proteins were never demonstrated to physically interact in vivo and therefore the inactivation of Ets functions by Id1 on the p16 promoter may be through indirect mechanisms. Therefore, we suggest that Ras-Raf-MEK kinase signals promote Ets-2-mediated activation of p16 and oncogene-induced senescence (27), which can be opposed by Id1. Additionally, endogenous p16 promoter activity induced through cumulative cell divisions can be inactivated directly by Id1 through E-box-mediated functions as demonstrated here. Although previous studies have shown that only Id2 expression and not Id1 or Id3 expression is able to antagonize the cell-cycle arrest induced by the inhibitors p16/Ink4a and p21 in human osteosarcoma cell lines (28), neither p16 nor p21 were being expressed by their endogenous promoters in those studies and therefore Id1 repression by means of the p16 promoter would not have been anticipated.
The p16Ink4a gene has been demonstrated to be inactivated in familial and sporadic melanomas by a variety of mechanisms (reviewed in ref. 29), and it has been demonstrated that p16 expression decreases as a function of increasing malignant potential in melanocytic lesions (30). Because the frequencies of loss of heterozygosity at the p16/Ink4a locus, p16/Ink4a intragenic mutations, and promoter methylation are extremely low in thin sporadic melanomas (<10% in lesions <4 mm), although p16 expression in these lesions is lower than that of premalignant melanocytic lesions (31), it is likely that other mechanisms allow for decreased p16 expression in early melanomas. Here we demonstrate that Id1 is able to repress p16 promoter activity, and we propose that Id1 transcriptional repression of p16/Ink4a may represent a mechanism for early dysregulation of p16 expression in human tumors. Interestingly, Id1 expression has been noted to be up-regulated in pancreatic tumors that, like melanomas, frequently demonstrate inactivation of the tumor suppressor p16/Ink4a (32). Further studies will need to be undertaken to determine the precise role of Id1 in the regulation of p16/Ink4a expression and its functional significance in regulating cell growth, senescence, and tumorigenesis.
Acknowledgments
We thank R. Benezra, C. Sherr, G. Peters, R. DePinho, and N. Sharpless for generously providing reagents. We are also grateful to R. Benezra, S. Baylin, B. Vogelstein, and K. Kinzler for their critical review of this manuscript and helpful discussions. This work was supported by the National Institute for Arthritis, Musculoskeletal, and Skin Diseases Grant AR01975 (to R.M.A.).
Abbreviations
- HLH
helix–loop–helix
- MEF
mouse embryo fibroblast
- pRb
p16ink4a/retinoblastoma
- P-n
passage n
- cdk
cyclin-dependent kinase
- EMSV
a plasmid containing unique EcoRI cloning site and Maloney Sarcoma Virus long terminal repeat
- β-gal
β-galactosidase
References
- 1.Massari M E, Murre C. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton J D. J Cell Sci. 2000;113:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 3.Alani R M, Hasskarl J, Grace M, Hernandez M-C, Israel M A, Munger K. Proc Natl Acad Sci USA. 1999;96:9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nickoloff B J, Chaturvedi V, Bacon P, Qin J Z, Denning M F, Diaz M O. J Biol Chem. 2000;275:27501–27504. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- 5.Lundberg A S, Hahn W C, Gupta P, Weinberg R A. Curr Opin Cell Biol. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- 6.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 7.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 8.Hara E, Uzman J A, Dimri G P, Nehlin J O, Testori A, Campisi J. Dev Genet. 1996;18:161–172. doi: 10.1002/(SICI)1520-6408(1996)18:2<161::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Ohtani N, Zebedee Z, Huot T, Stinson J, Sugimoto M, Ohashi Y, Sharrocks A, Peters G, Hara E. Nature (London) 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 10.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 11.Dynlacht B D. Nature (London) 1997;389:149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- 12.Yan W, Young A Z, Soares V C, Kelley R, Benezra R, Zhuang Y. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pesce S, Benezra R. Mol Cell Biol. 1993;13:7874–7880. doi: 10.1128/mcb.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malecki M T, Jhala U S, Antonellis A, Fields L, Doria A, Orban T, Saad M, Warram J H, Montminy M, Krolewski A S. Nat Genet. 1999;23:323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs J J, Kieboom K, Marino S, DePinho R A, van Lohuizen M. Nature (London) 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 16.Dimri G P, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubelj I, Pereira-Smith O, et al. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campisi J. Eur J Cancer. 1997;33:703–709. doi: 10.1016/S0959-8049(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 18.Prabhu S, Ignatova A, Park S T, Sun X H. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber D A. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 21.Pagliuca A, Gallo P, De Luca P, Lania L. Cancer Res. 2000;60:1376–1382. [PubMed] [Google Scholar]
- 20.Sherr C J, DePinho R A. Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 22.Inoue K, Roussel M F, Sherr C J. Proc Natl Acad Sci USA. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tournay O, Benezra R. Mol Cell Biol. 1996;16:2418–3240. doi: 10.1128/mcb.16.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyden D, Young A Z, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader B L, Hynes R O, Zhuang Y, Manova K, Benezra R. Nature (London) 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 25.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 26.Dai C Y, Enders G H. Oncogene. 2000;19:1613–1622. doi: 10.1038/sj.onc.1203438. [DOI] [PubMed] [Google Scholar]
- 27.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 28.Lasorella A, Iavarone A, Israel M A. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liggett W H J, Sidransky D. J Clin Oncol. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 30.Reed J A, Loganzo F J, Shea C R, Walker G J, Flores J F, Glendening J M, Bogdany J K, Shiel M J, Haluska F G, Fountain J W, et al. Cancer Res. 1995;55:2713–2718. [PubMed] [Google Scholar]
- 31.Fujimoto A, Morita R, Hatta N, Takehara K, Takata M. Oncogene. 1999;18:2527–2532. doi: 10.1038/sj.onc.1202803. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama H, Kleeff J, Wildi S, Friess H, Buchler M W, Israel M A, Korc M. Am J Pathol. 1999;155:815–822. doi: 10.1016/S0002-9440(10)65180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]