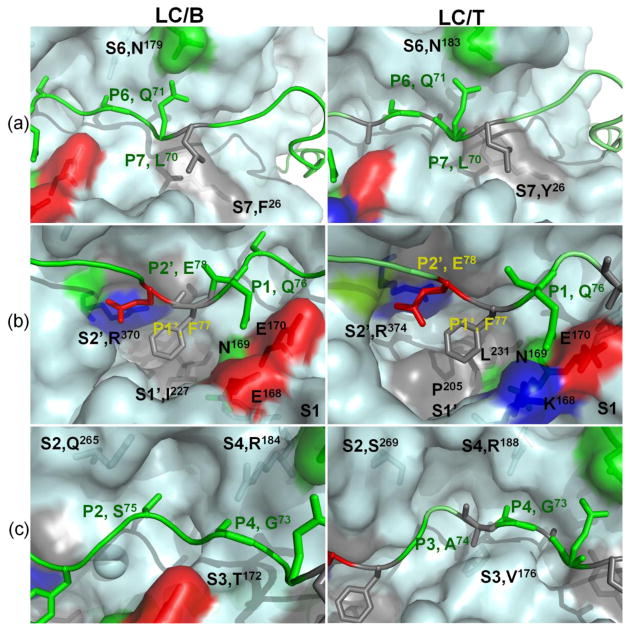
Fig 1. Recognition of VAMP2 P sites by LC/B and LC/T substrate recognition pockets.
Computational and experimental data from the current study provides a model for the interaction of LC/B (left panel) and LC/T (right panels) with VAMP2. (a) Recognition of P7 and P6 sites by S7 and S6 pockets of LC/B and LC/T. The S7 pockets of LC/B was formed by F26, which recognized P7, L70 and the S6 pocket of LC/B was formed by N179, which recognized P6, Q71. The S7 of LC/T was formed by Y26, which recognized P7, L70 and the S6 pocket of LC/B was formed by N183, which specifically recognize P6, Q71. (b) Recognition of P1, P1′ and P2′ sites of VAMP2 by S1, S1′ and S2′ pockets of LC/B and LC/T. The S1 pocket of LC/B and LC/T was formed by residues E168N169E 170 and K168N169E 170 respectively, recognized P1, Q76 of VAMP2. The S1′ pocket of LC/B and LC/T was formed by I227 and L231, P205, respectively, which recognized F77 of VAMP2. The S2′ of LC/B and LC/T was formed by R370 and R374, respectively, which recognized P2′, E78 of VAMP2. (c) Fine alignment of P4, P3 and P2 sites of VAMP2 into S4, S3, and S2 pockets of LC/B and LC/T. The large side chain of S4 pocket residue, R, allows the alignment of smaller side chain residue, G. The different shapes of S2 and S3 pockets of LC/B and LC/T, while not the residue composition of the pockets enable S2 and S3 to tolerate different P2 and P3 site residues.