Abstract
Biomarkers are increasingly important in the clinical management of complex diseases, yet our ability to discover new biomarkers remains limited by our dependence on endogenous molecules. Here we describe the development of exogenously administered `synthetic biomarkers' composed of mass-encoded peptides conjugated to nanoparticles that leverage intrinsic features of human disease and physiology for noninvasive urinary monitoring. These protease-sensitive agents perform three functions in vivo: target sites of disease, sample dysregulated protease activities and emit mass-encoded reporters into host urine for multiplexed detection by mass spectrometry. Using mouse models of liver fibrosis and cancer, we show that they can noninvasively monitor liver fibrosis and resolution without the need for invasive core biopsies and can substantially improve early detection of cancer compared with clinically used blood biomarkers. This approach of engineering synthetic biomarkers for multiplexed urinary monitoring should be broadly amenable to additional pathophysiological processes and to point-of-care diagnostics.
Biomarker discovery is motivated by the desire to identify reliable indicators of disease for risk assessment, early detection, predicting patient responses to therapies and surveillance of recurrent disease1,2. To date, a broad range of distinct biological species such as metabolites3, peptides4, proteins2,5, cell-free nucleic acids6, exosomes7 and circulating tumor cells8 have been developed into biomarkers of varying performance. However, the use of naturally occurring biomarkers to indicate disease is limited by fundamental technical and biological challenges because biomarkers are frequently found in low levels in circulation8,9, are difficult to resolve in complex biological fluids5 and can be rapidly degraded both in vivo and ex vivo10.
An alternative to endogenous biomarkers is the systemic administration of exogenous agents to interrogate biological states. These approaches offer the potential to tailor agents to exploit host physiology or interface with disease-specific molecular processes as alternative indicators of disease. Examples include the polysaccharide inulin to assess glomerular filtration rates, FDG-PET to unveil regions of increased glucose metabolism and a suite of molecular and activity-based probes for imaging biological activities in vivo11–13. Because these agents can be designed and tested in vitro and in preclinical models, they can be iteratively optimized and can be administered at concentrations significantly above biological background. The limitations of these approaches include the inability to monitor large family of probes simultaneously due to limited multiplexing capabilities and substantial infrastructure for in vivo analysis, which require patients to be on-site (e.g. PET, MRI) and preclude remote data or sample collection.
Here we describe a class of engineered nanoscale agents that passively accumulate in diseased tissues from host circulation via organ- or disease-specific vascular fenestrations (e.g. liver sinusoid endothelium or angiogenic tumor vessels respectively)14,15. Upon arrival in the diseased microenvironment, they are cleaved by aberrantly active proteases, releasing surface-conjugated, mass-encoded peptide substrates into the host urine for detection by mass spectrometry (MS) as synthetic biomarkers of disease (Fig. 1). Because dysregulated protease activities are implicated in a wide range of human diseases, including cancer, fibrosis, atherosclerosis, inflammation, Alzheimer's and many others16, highly multiplexed monitoring of aberrant protease activities has the potential to distinguish diverse disease states. Here we describe the application of this technology to noninvasively monitor liver fibrosis17 and to detect early-stage cancers18.
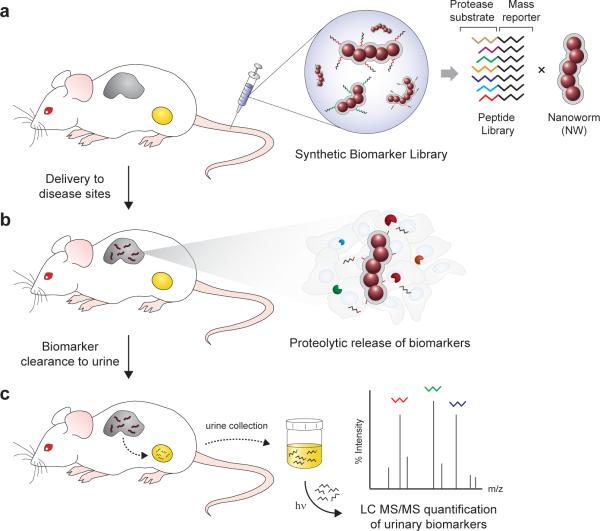
Figure 1. Schematic of approach.
(a) Synthetic biomarker library comprised of mass-encoded tandem peptides conjugated onto nanoworm nanoparticles. (b) NW accumulation in disease tissues following intravenous administration in living animals. Dysregulated proteases cleave peptides from NWs, allowing fragments to filter into the urine. (c) Recovery of photo-caged mass reporters from urinary cleavage fragments by UV-light irradiation and their quantification by liquid chromatography tandem mass spectrometry.
RESULTS
Protease-sensitive nanoparticles for urinary monitoring
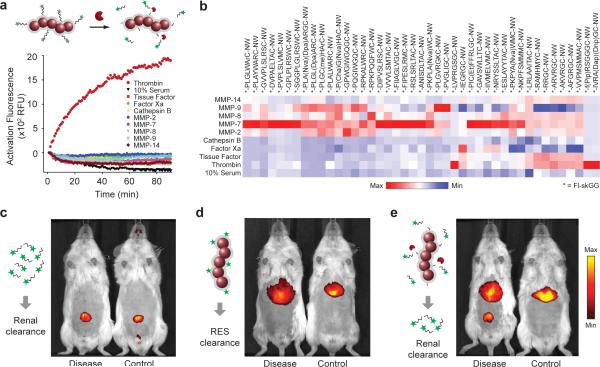
To develop a protease-sensing platform, we first set out to identify peptide substrates of proteases implicated in liver fibrosis and cancer. Fluorescein-labeled derivatives of ~50 candidate peptide substrates19–23 were conjugated to PEG-coated, long-circulating iron oxide nanoworm (NW) nanoparticles24 (Supplementary Fig. 1a,b,c) and incubated with recombinant proteases commonly overexpressed in disease (e.g. Matrixmetalloproteases (MMPs), cathepsins) as well as blood-borne proteases to assess cross-reactivity (FXa, Tissue factor (TF), thrombin). Relative substrate activities for each protease-substrate combination were determined by monitoring increases in sample fluorescence resulting from peptidolysis that allowed previously homoquenched fluorophores to freely emit in solution (Fig. 2a). Initial reaction velocities were compiled for comparative analysis in a heat map (Fig. 2b) from which 10 peptide substrates (S1–S10, Table) with broad protease susceptibility were selected as our peptide-NW library.
Figure 2. Urinary detection of in vivo protease activity with peptide-NWs.
(a) Shown are representative activation profiles of peptide-NWs following treatment with recombinant proteases. Specific protease-substrate combinations led to rapid increases in sample fluorescence. (b) Heat map comparison of cleavage velocities for different substrate-protease combinations grouped according to activity and specificity. (c) Fluorescence in vivo images of DDC-treated and control animals following intravenous injection of VivoTag-680 labeled Glu-fib peptides, (d) peptide-free NWs or (e) peptide-conjugated NWs.
Table. 10-plex synthetic biomarker library.
List of synthetic biomarkers (G1–G10), protease substrates (S1–S10) and isobaric mass reporters (R1–R10) used in study.
| Synthetic Biomarker Library[a],[b] (G1–G10) | Substrate (S1–S10) | Isobaric mass code[c],[d] (R1–R10) | y6 reporter | [y6+H+] |
|---|---|---|---|---|
| e+3G+6VndneeGFfsAr-X-K(FAM)GGPQGIWGQC-NW | PQGIWGQ | e+3G+6VndneeGFfsAr | GFfsAr | 683.4 |
| e+2G+6Vndnee+1GFfsAr-X-K(FAM)GGLVPRGSGC-NW | LVPRGSG | e+2G+6Vndnee+1GFfsAr | +1GFfsAr | 684.4 |
| e+1G+6Vndnee+2GFfsAr-X-K(FAM)GGPVGLIGC-NW | PVGLIG | e+1G+6Vndnee+2GFfsAr | +2GFfsAr | 685.4 |
| eG+6Vndnee+2GFfs+1Ar-X-K(FAM)GGPWGIWGQGC-NW | PWGIWGQG | eG+6Vndnee+2GFfs+1Ar | +2GFfs+1Ar | 686.4 |
| eG+5VndneeGFfs+4Ar-X-K(FAM)GGPVPLSLVMC-NW | PVPLSLVM | eG+5VndneeGFfs+4Ar | GFfs+4Ar | 687.4 |
| e+3G+1Vndnee+1GFfs+4Ar-X-K(FAM)GGPLGLRSWC-NW | PLGLRSW | e+3G+1Vndnee+1GFfs+4Ar | +1GFfs+4Ar | 688.4 |
| e+3GVndneeG+6FfsAr-X-K(FAM)GGPLGVRGKC-NW | PLGVRGK | e+3GVndneeG+6FfsAr | G+6FfsAr | 689.4 |
| e+2GVndneeG+6Ffs+1Ar-X-K(FAM)GGf(Pip)RSGGGC-NW | f(Pip)RSGGG | e+2GVndneeG+6Ffs+1Ar | G+6Ffs+1Ar | 690.4 |
| e+1GVndnee+2G+6FfsAr-X-K(FAM)GGfPRSGGGC-NW | fPRSGGG | e+1GVndnee+2G+6FfsAr | +2G+6FfsAr | 691.4 |
| eGVndnee+3G+6FfsAr-X-K(FAM)GGf(Pip)KSGGGC-NW | f(Pip)KSGGG | eGVndnee+3G+6FfsAr | +3G+6FfsAr | 692.4 |
X (3-amino-3-(2-nitrophenyl)propionic acid), FAM (carboxyfluorescein), Pip (pipecolic acid), NW (nanoworm)
lower case = d-amino acid
photocleaved C-terminus = CONH2
mass= 1589.8 Da
To establish the potential to probe disease microenvironments remotely from urine, we next investigated the in vivo behavior of each system component (i.e. peptide and NW). Here, we selected a xenobiotic mouse model of liver fibrosis in which FVB/NJ mice fed with 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) develop progressive liver disease as a result of chronic bile duct injury25, leading to liver fibrosis and upregulation of local MMPs (Fig. 4c–e, Supplementary Fig. 8b). First, to determine the efficiency of peptide clearance into urine, we selected the peptide glutamate-fibrinopeptide B (Glu-fib, EGVNDNEEGFFSAR) as a prototypic urinary marker because its endogenous derivative (fibrinopeptide B) is biologically inert and filters freely into urine when released during coagulation26. As expected, fluorophore-labeled Glu-fib administered intravenously (i.v.) cleared rapidly into urine in both fibrosis and healthy animals with no evidence of hepatic uptake at the site of disease (Fig. 2c). By contrast, peptide-free NWs predominantly localized to the liver (Fig. 2d) but were unable to filter into urine, consistent with the renal clearance threshold of ~ 5 nm for inorganic nanoparticles27 (NWs ~ 40 nm, Supplementary Fig. 1a) as well as our previous pharmacokinetic studies with NWs28. Infusion of peptide-NWs conjugated with fluorescent derivatives of substrates S1–S10 (Table) resulted in strong liver uptake and elicited a marked urinary response in fibrotic animals from renal filtration of cleaved peptide fragments (Fig. 2e). Collectively, these studies demonstrated the ability of NWs to redirect peptides to sites of disease to facilitate production of urinary biomarkers.
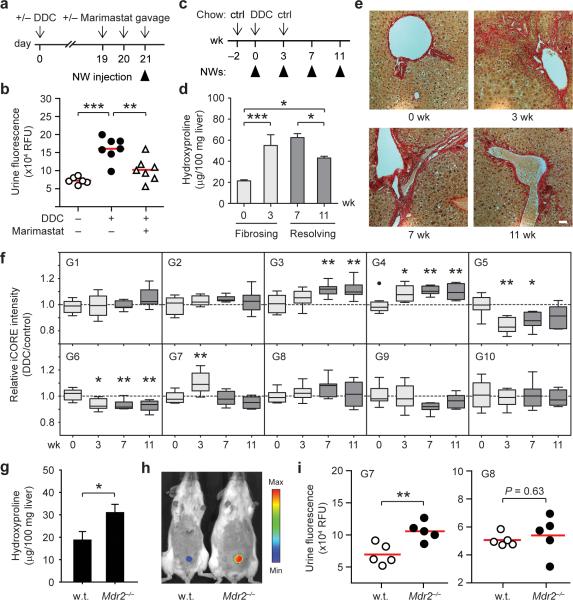
Figure 4. Urinary biomarkers of hepatic fibrosis and resolution in DDC-treated mice.
(a) Timeline for DDC and Marimastat treatment. (b) Urinary biomarker levels by α-FITC immunoprecipitation following G1–G10 administration (ANOVA, *** P < 0.001, ** P < 0.01). (c) Induction of fibrogenesis and resolution timeline. (d) Quantification of total liver collagen by hydroxyproline analysis. DDC treatment led to ~3 fold increase in liver collagen by week 3 (*** P < 0.001) and a ~30% reduction between week 7 and 11 (* P < 0.05) that remained above pretreatment values (* P < 0.05) (One-way ANOVA, Tukey post test, n = 3, s.e.m.). (e) Shown are Sirius Red staining of representative liver sections indicating the presence of fibrotic extensions emanating from portal triads at week 3, persisting to week 7 and reversing by 11 weeks. (scale bar = 50 μm) (f) Box and whisker plots of individual iCORE peak intensities plotted as DDC over control at 0, 3, 7 and 11 weeks (* P < 0.05, ** P < 0.01; repeated measures ANOVA, Tukey post test; n = 8–10). (g) Cross-validating G7 as a urinary biomarker of fibrosis in Mdr2−/− mice. Quantification of liver collagen by hydroxyproline analysis (* P < 0.05, n = 3). (h) IVIS fluorescence in vivo imaging of urinary responses in Mdr2−/− mice following NW infusion. (i) Administration of G7 led to elevations in urinary fluorescence while G8 did not track with fibrosis (** P < 0.01).
Profiling protease activities by mass spectrometry
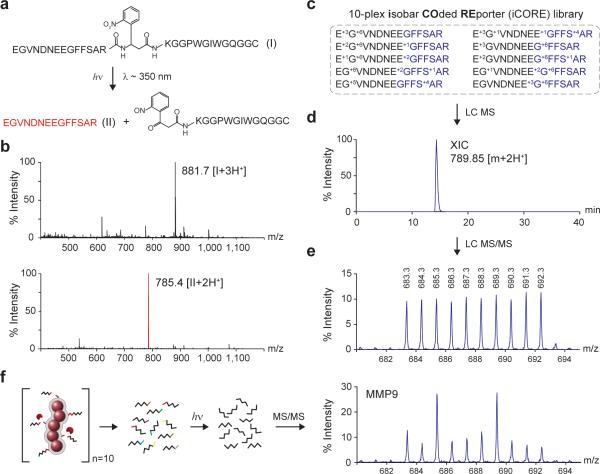
Despite the multiplexing advantages of mass-encoding, one challenge of detecting protease activity by MS is that peptide substrates in complex proteolytic environments can be cleaved at multiple sites by promiscuous proteases and truncated by exoproteases29,30 to produce diverse pools of poorly defined fragments that confound mass analysis. Here, we set out to create well-defined mass reporters to encode our substrate library. In light of the favorable renal clearance properties of Glu-fib, we chose to append d-isomer rich derivatives of Glu-fib to the N-termini of each protease substrate to serve as protease resistant mass reporters and to promote renal filtration upon substrate cleavage and release from NWs. These tandem peptides were further modified with internal photo-labile residues31 to enable the recovery of Glu-fib peptides by photolysis from complex urinary cleavage fragments following in vivo proteolysis. To test this construct, we synthesized a model photo-caged tandem peptide (compound I, Fig. 3a). Consistent with previously published reports on nitrophenyl groups, exposure of compound I (triply charged, 881.7 m/z; Fig. 3b, top panel) to UV light triggered peptide cleavage, resulting in the appearance of doubly charged, acetamide-terminated Glu-fib (785.4 m/z; Fig. 3b, bottom panel).
Figure 3. Photo-caged iCORE libraries for multiplexed profiling of protease activities by LC MS/MS.
(a) Structure of tandem peptide (compound I) containing an internal UV-sensitive linker. Structure of free Glu-fib (compound II) generated after photolysis (~350 nm). (b) LC MS spectra of compound I before (top, triply-charged m/z: 881.7) and after (bottom, doubly-charged m/z: 785.4) exposure to UV light. (c) 10-plex isobaric peptide library derived from Glu-fib (R1–R10). (d) Extracted ion chromatogram of an equimolar 10-plex iCORE mixture (789.80–789.90 m/z). The entire multiplexed set was chromatographically indistinguishable. (e) iCORE MS/MS spectrum following collision induced disassociation. Individual reporters were identified via unique y6 reporter ions (683.3–692.3 m/z) each differentiable by a single mass unit. (f) iCORE MS/MS spectrum following incubation of a 10-plex, iCORE-encoded peptide-NW cocktail with recombinant MMP9.
To design an extensible encoding strategy for our library of protease substrates, we adapted principles of isobaric mass encoding32,33 to produce a family of mass reporters from Glu-fib. The distinguishing feature of an isobaric encoding strategy is that individual members within a family of reporters share a parent mass to facilitate efficient peptide collection by MS, but can be subsequently identified via unique MS/MS ions upon fragmentation. Observing that Glu-fib fragments into C-terminal y-type ions (Supplementary Fig. 2a), we constructed 10 mass codes centered on the y6 ion (GFFSAR) by enriching the hexamer with heavy amino acids to produce variants differentiated by 1 Da each (Supplementary Fig. 2b). This introduced mass shift was then balanced by isotope enrichment within the remaining residues (EGVNDNEE) to produce peptides with identical parent masses but distinct y6 fragment ions. We call this encoding method “isobar COded REporters” (iCORE). To validate this approach, we analyzed an equimolar 10-plex iCORE library (R1–R10, Table) by LC MS/MS and found the entire peptide library to initially appear as a single, unresolved peak (extracted ion chromatogram, 789.95 ± 0.5 m/z, Fig. 3c,d) but following fragmentation, resolve as predicted into a 10 peak spectrum with no fragmentation bias (683.4–692.4 m/z, Supplementary Fig. 3, Fig. 3e). Confounding peak overlap from naturally occurring isotopes (e.g. 13C) were removed by collecting iCORE peptides with a 1 m/z window centered on the precursor ion (Supplementary Fig. 4a), minimizing signal from naturally occurring isotopes to ~5% of the parent peak (Supplementary Fig. 4b). Consequently, in samples spiked with reporters at defined ratios (1:2:3:5:10:10:5:3:2:1), we observed a linear correlation between peak intensity and stoichiometry in both unmodified and peak-subtracted analysis (n = 3, R2 = 0.99 and R2 = 0.99 respectively, Supplementary Fig. 5a,b,c). All subsequent samples were peak-adjusted to reflect contributions from naturally occurring isotopes.
To test the ability of iCORE reporters for monitoring peptide cleavage, protease substrates S1–S10 were extended with iCORE mass tags R1–R10 via photo–sensitive amino acids and coupled to NWs to produce synthetic biomarkers G1–G10 (Table). Following treatment of an equimolar cocktail of G1–G10 with recombinant MMP9, cleavage products were isolated by size filtration and exposed to UV-light to release reporters R1–R10 for MS/MS quantification. Collective substrate activities appeared as distinct iCORE landscapes with individual y6 peak intensities corresponding to substrate preference for MMP9 (Fig. 3F). We applied this library to several additional proteases (Supplementary Fig. 6a) and found that individual iCORE protease profiles were unique as determined from Pearson's correlation analysis (i.e. MMP2, MMP9, MMP12 and thrombin; Supplementary Fig. 6b), illustrating the ability of iCORE-encoded NWs to monitor many protease-substrate combinations simultaneously.
Monitoring hepatic fibrogenesis and resolution
Liver fibrosis is a wound healing response to chronic liver injury and results in the deposition of scar tissue that can lead to cirrhosis, liver failure and cancer17. The dynamics of extracellular matrix (e.g. collagen) accumulation is largely driven by activated hepatic stellate cells and matrix remodeling proteases such as MMPs and their inhibitors. The current gold standard for monitoring this process is a needle biopsy followed by histological analysis; however, this technique is invasive, confounded by high sampling heterogeneity, carries a finite risk of complications and cannot be performed as frequently as needed (e.g. for assessing antifibrotic therapies)34. Noninvasive assays including ultrasound imaging, elastography and serum biomarkers are limited by their low accuracies and limited prognostic utility35. Thus, there remains an urgent need for noninvasive biomarkers to replace biopsy-based monitoring, to facilitate the identification and validation of new antifibrotic agents and to support clinical decision making36. Here, we sought to identify synthetic biomarkers with the capacity to monitor liver fibrosis and resolution and extended our DDC model to include both aspects of the disease (Fig. 4a,b,c).
We first evaluated the potential toxicity of NWs to determine whether serial monitoring could be performed safely. NWs are composed of iron oxide cores that are FDA-approved for use in humans (Feridex ®); however, we sought to further examine whether fibrotic livers could be sensitized to nanomaterial toxicity. To investigate nanomaterial safety, we administered peptide-NWs (1 mg/kg) or PBS weekly (day 0, 7 and 14) to animals fed DDC or control chow for 3 consecutive weeks (Supplementary Fig. 7a) and found that NWs did not exacerbate fibrosis, decrease body weights nor induce hepatotoxicity over PBS-injected animals (Supplementary Fig. 7b–e). Serial NW infusions could also introduce experimental artifacts if residual urinary reporters from prior administrations are insufficiently cleared. Analysis of urine samples following the last NW injection (day 14) revealed that both residual fluorescent and mass reporters were cleared within 5 days (day 19) (Supplementary Fig. 7f,g). Collectively, these experiments showed that NWs are well-tolerated at the dosage selected and require 5 days for full clearance.
We next investigated whether urinary responses are specifically produced by fibrosis-associated proteases such as MMPs by testing urinary sensitivity to pharmacological inhibition of MMPs (Fig. 4a). Whereas infusion of our 10-plex iCORE-encoded NW cocktail (G1–G10) in animals given DDC chow for 3 weeks resulted in a strong increase in ensemble urinary fluorescence over controls (ANOVA, *** P < 0.001, Fig. 4b), urinary responses were significantly attenuated in mice additionally treated with the broad-spectrum MMP inhibitor Marimastat via oral gavage for two days prior to G1–G10 administration, resulting in over 70% inhibition of reporter levels (** P < 0.01, Fig. 4b). To determine the accessibility of NWs to sites of fibrosis, immunofluorescence analysis of liver sections revealed that most NWs infiltrated freely into the parenchyma and further penetrated periportal zones of active fibrosis, escaping sequestration by resident macrophages (Supplementary Fig. 8a). Compared to control sections, these regions showed significant upregulation of MMP9, a representative fibrosis-associated MMP (Supplementary Fig. 8b,c). Similar punctate patterns appeared in fibrotic sections treated with DQ-gelatin substrates (Supplementary Fig. 8d), confirming the enzymatic activity of collagen-degrading proteases (e.g. MMP2/9). These results showed that MMPs upregulated during fibrosis are proteolytically active and are largely responsible for urinary responses.
We next monitored the processes of fibrosis and resolution by iCORE mass analysis in order to determine the response of individual biomarkers apart from their collective fluorescence in urine. Mice treated transiently with DDC for 3 weeks followed by restoration of DDC-free chow develop distinct active fibrosis and resolution windows (0–3 and 7–11 weeks respectively, Fig. 4c) as verified macroscopically by Sirius red collagen staining of liver sections and hydroxyproline quantification (Fig. 4d,e). With this treatment regime, liver collagen increased ~ 3-fold compared to pretreatment levels after 3 weeks on DDC, persisted from week 3–7 after initial removal of DDC and significantly decreased from week 7–11 after sustained DDC withdrawal (ANOVA, * P < 0.05, *** P < 0.005, n = 3). Thus, to monitor the transitions between fibrosing and resolving disease, we administered NWs at 0, 3, 7 and 11 weeks into DDC-treated and age-matched control animals followed by iCORE MS/MS analysis.
The resulting activities of the ten synthetic biomarkers displayed markedly divergent kinetics (Fig. 4f). Biomarkers G3 and G4 both strongly increased relative to pretreatment baselines, reaching a plateau by week 11 despite staggered onset at week 7 and 3 respectively. G5 and G6 showed opposing kinetics, significantly decreasing at week 3 before either gradually returning to pretreatment intensities (G5) or persisting to week 11 (G6). G7 tracked with the kinetics of DDC treatment, elevating sharply at week 3 followed by a rapid reversal at week 7. All remaining biomarkers (G1, G2, G8, G9 and G10) did not deviate from initial pretreatment activities (G1–G10; * P < 0.05, ** P < 0.01, repeated measures ANOVA, Tukey post test, n = 10) as well as all biomarkers in control animals (Supplementary Fig. 9).
Having identified a set of putative biomarkers for liver fibrosis in the context of DDC, we next sought to cross-validate promising biomarkers in Mdr2−/− mice, a mechanistically-distinct model of liver fibrosis. Cross-validating biomarker responses in an independent cohort not involved in hypothesis generation is crucial for eliminating potential model-specific artifacts from the use of inbred mice as well as data overfitting37. Mdr2−/− mice lack a critical phospholipid transporter required for bile stabilization and develop chronic liver injury from birth as a result of bile leakage to the portal tract38.
At 8 weeks of age, Mdr2−/− animals showed evidence of periportal fibrosis as well as significant upregulation of MMP9 compared to age-matched wild type animals (Fig. 4g, Supplementary Fig. 10a,b). From our library of 10 probes, we selected G7 for cross-validation because it was highly specific for liver fibrosis in our DDC study, tracking with fibrogenesis and declining upon fibrotic resolution (Fig. 4f). Similar to our initial observations, we detected significant elevations in urinary fluorescence in 8-week Mdr2−/− rodents over controls (Fig. 4h,i). By contrast, G8, a biomarker revealed to be unresponsive in our DDC studies, did not show potential for monitoring fibrosis in Mdr2−/− animals (Fig. 4i). Collectively, these results further corroborated the ability of G7 to track liver fibrosis and underscored the potential for monitoring fibrogenesis with distinct molecular etiologies.
To explore potential improvements in disease classification that could be gained by employing more than one biomarker, we further analyzed our biomarker responses in our DDC study using receiver operating characteristic (ROC) curves. ROC curves characterize the predictive power of a biomarker by returning the area under the curve (AUC) as a metric with a baseline AUC of 0.5 representing a random biomarker classifier (dashed line, Supplementary Fig. 11–13). Within the 0–3 week fibrogenesis window, biomarkers G4, G5, G6 and G7 each discriminated disease with high sensitivity and specificity with associated AUCs ranging from 0.83–0.96 (n = 10, Supplementary Fig. 11) and combinatorial panels such as the best dual (G5+G7) and triple biomarker (G5+G6+G7) combinations led to improvements in predictivity (0.98 and 1.0 respectively, Supplementary Fig. 13). Conversely during fibrotic resolution, the ability of candidate biomarkers to track disease such as G1 (AUC = 0.73) was improved in dual (G1+G9, AUC = 0.9) or triple biomarker combinations (G1+G7+G9, AUC = 0.91) (Supplementary Fig. 13).
Collectively, these experiments demonstrated that liver fibrosis and resolution are revealed by distinct collections of synthetic biomarkers and that multiplexed combinations allowed the highest diagnostic performance—illustrating the ability of this platform to noninvasively illuminate otherwise inaccessible aspects of liver disease evolution.
Early detection of colorectal cancer
When diagnosed prior to systemic dissemination, many primary tumors can be effectively treated with conventional clinical interventions39. However, the rates at which most biomarkers are shed from tumors are prohibitively low and cannot be readily augmented40, effectively precluding early detection9,18. Thus, there remains a stark mismatch between the smallest tumors detectable by blood biomarkers (>2–5 cm) and the size of tumors that would best respond to treatment (<1–5 mm), resulting in delayed detection, low drug response rates and reduced overall patient survival. Here, we hypothesized that because nanoparticles can passively target tumors to sample proteases through fenestrated angiogenic tumor vessels15, cancer-specific proteases could be co-opted to amplify tumor detection via sustained enzymatic release of synthetic urinary biomarkers.
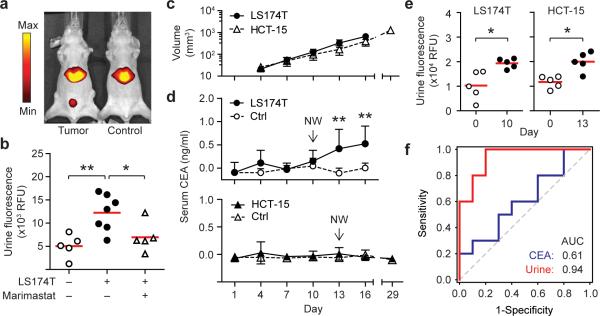
To explore this hypothesis, we chose to compare carcinoembryonic antigen (CEA), a clinically-utilized blood biomarker for colorectal cancer (CRC), with our NWs. Because plasma CEA levels in CRC patients are highly variable, we first compiled ATCC-documented CEA production rates from 24 established human CRC lines (14 additional lines were uncharacterized) and found rates to range well over 4 log units with a median value of 1.65 ng/106 cells/10 days (Supplementary Fig. 14). We selected the cell line LS174T to represent colorectal tumors capable of producing CEA near the maximum observed rates (~100× above the median) and validated its ability to secrete CEA in vitro by ELISA that had a detection limit of ~ 0.1 ng/ml (Supplementary Fig. 15a,b). To fully capture the broad spectrum of activities from matrix remodeling proteases shared by most invasive tumors, we infused our biomarker ensemble (G1−G10) into mice bearing LS174T flank tumors (Fig. 5a,b) and detected a significant rise in urinary fluorescence. NW extravasation into the tumor parenchyma was verified by fluorescence imaging of excised tumors and analysis of tissue sections (Supplementary Fig. 16a,b). To test whether urinary responses were specifically produced via proteolysis from MMPs, a separate cohort of tumor bearing animals was treated with Marimastat for two days prior to G1−G10 administration. Pharmacological inhibition of MMPs resulted in near abrogation of urinary signals, reducing signal intensities by ~ 73% (Fig. 5b).
Figure 5. Synthetic urinary biomarkers outperform serum CEA in early cancer detection.
(a) IVIS fluorescence in vivo imaging showed urinary accumulation of synthetic biomarkers released by LS174T colorectal tumors. (b) Quantification of urinary fluorescence with (+) and without (−) Marimastat treatment (ANOVA, ** P < 0.01, * P < 0.05). (c) Growth kinetics of LS174T and HCT-15 colorectal tumors (n = 5, s.d.). (d) Circulating levels of CEA analyzed every third day post tumor implantation by ELISA (** P < 0.01, Two-way ANOVA, Tukey post test). Arrows mark NW injections. (e) Early detection of human colorectal tumors. Urinary responses were significantly elevated following NW infusions in LS174T and HCT-15 tumor-bearing animals at day 10 and 13 respectively (two-tailed paired T-test, * P < 0.05). (f) AUC comparison between ensemble urinary biomarker panel and serum CEA at day 10 (n = 10).
We next sought to directly compare synthetic urinary biomarkers to CEA for early cancer detection. Following implantation of LS174T cells, we monitored tumor growth noninvasively by quantifying serum CEA levels every 3 days by ELISA which revealed disease by day 13 when average tumor volume reached ~ 330 mm3 (** P < 0.01, Two way ANOVA, n = 5, Fig. 5c,d). In parallel, we monitored tumor growth by infusing G1−G10 at day 0 and 10 (~130 mm3). Whereas CEA was unable to detect tumor burdens < 330 mm3, ensemble urinary responses at day 10 were significantly elevated relative to pre-tumor samples, allowing the detection of tumors 2.5-times smaller than CEA (130 vs. 330 mm3, Fig. 5e). To further characterize the discriminatory sensitivity and specificity of these two approaches, serum CEA and urinary biomarker levels were subjected to ROC analysis. In contrast to the limited predictive power of CEA for early detection (AUC = 0.61), ensemble urinary fluorescence was highly discriminatory, producing a collective AUC of 0.94 (n = 10, Fig. 5f). To determine the underlying biomarkers driving the predictivity of the ensemble, the response of individual probes was quantified and plotted as ROC curves (Supplementary Fig. 17,18). Disease classification by the best-performing individual probes (G1, G2 and G3, AUC = 0.78–0.81) did not fully recapitulate the multiplexed set, but was improved in dual (G1+G2, AUC = 0.88) or triple biomarker panels (G1+G2+G3, AUC = 0.89) (Supplementary Fig. 19). This latter observation underscored the value of employing a diverse family of probes for most sensitive detection.
Having established the potential of biomarker amplification for early cancer detection, we sought cross-validation in an independent cohort of CRC-bearing mice. In light of the high variability in CEA secretion rates, we hypothesized that our ensemble library of probes could detect tumors that secrete biomarkers at low rates. We selected HCT-15 cells, a genetically distinct CRC line that secretes CEA at a rate three times lower than the median value (Supplementary Fig. 14). As anticipated by the significant reduction (~300×) in CEA production relative to LS174T tumors, growth of HCT-15 tumors could not be detected by serum analysis even up to day 29 when average tumor burden measured ~1300 mm3 (Fig. 5c,d), reaching the maximum allowable limit in this rodent model. By contrast, HCT-15 tumors were readily discriminated by NW infusion and urine analysis at day 13 (~150 mm3, Fig. 5c,e), representing, at the minimum, a > 9-fold improvement over CEA (150 versus 1300 mm3 respectively). Collectively, these results showed that synthetic urinary biomarkers have the potential to detect cancer earlier compared to conventional blood biomarkers, with particularly dramatic enhancements for tumors secreting biomarkers at low rates.
DISCUSSION
An ideal biomarker should be secreted at high levels relative to native background, remain stable or persistent in circulation until detection, be readily accessible from compositionally simple host fluids and discriminate disease with high sensitivity and specificity. In practice, these parameters are often difficult to improve or control for naturally occurring biomarkers and consequently, many promising biomarkers fail during rigorous evaluation for clinical translation. Here, we devised a system of synthetic biomarkers with the capacity to (i) amplify biomarker levels through substrate turnover by targeting aberrant protease activities, (ii) release stable, disomer enriched mass reporters designed to appear within a narrow mass window free of host molecules, (iii) trigger reporter clearance from blood into urine to reduce matrix complexity and facilitate facile extraction and (iv) simultaneously monitor libraries of candidate synthetic biomarkers in vivo to identify and validate lead biomarkers.
An enabling feature of our platform is the use of a nanoscale scaffold to direct the traffic of peptides in vivo. Although free peptides are typically cleared rapidly from circulation via urinary secretion, we showed that NW-conjugated peptides are endowed with long-circulation times to allow transport into diseased tissues across porous vasculature and appear in urine only after release from NWs by disease-associated proteases. Several reports have highlighted the potential of applying peptide substrates to patient serum samples followed by MS profiling to uncover disease-specific activity signatures4,29,30,41. However, without a delivery mechanism, in vitro serum analysis cannot sample proteases expressed on the membrane of cells residing in the disease microenvironment (e.g. MMP9 expression by liver-resident macrophages in fibrosis). Similar to blood biomarkers, secreted proteases are significantly diluted in circulation and are often challenging to detect above highly-abundant plasma proteins5, potent proteolytic cascades activated during sample collection (e.g. coagulation) and pan protease inhibitors in plasma (e.g. α2-macroglobulin). Here, we chose NWs as chaperones because iron oxide nanoparticles are safe for use in humans, but a broad range of nontoxic scaffolds including proteins and sugars (e.g. albumin and dextran respectively) would also be amenable for peptide delivery. Given the cumulative wealth of nanomaterials, targeting ligands and enhanced delivery strategies available in nanomedicine, we expect this work to be transferrable to many additional formulations to gain access to different organs, types of vasculature and tissue depths42,43.
Our library of isobaric mass tags to track the response of 10 peptides in vivo provides a level of multiplexing that is currently challenging to attain with molecular and activity-based imaging probes11–13. The vast majority of these approaches make use of modified protease substrates that emit fluorescent signals following proteolytic cleavage. Consequently, substrate multiplexing is limited by emission overlap as well as the need to emit in the near infrared window (600–900 nm) to minimize signal attenuation from tissue absorption, constraining most of these studies to single probes. Conversely, our work demonstrates the generation of a synthetic biomarker library that is 5–10× more densely multiplexed than existing state-of-the-art activity-based probes, compares favorably with commercial isobaric tags (e.g. 8-plex iTRAQ) and with additional parent peptides, is extensible to hundreds of orthogonal mass codes.
In addition to its invasiveness, a major limitation of the core biopsy for liver fibrosis is that tissue specimens are only representative of ~1/50,000th the size of an adult liver leading to sampling variation that can result in inaccurate diagnosis or staging and repeat biopsies. Here, we show how nanoparticles accumulate uniformly in the liver, penetrating without bias into regions of active fibrosis to release urinary biomarkers as integrated measures of disease burden. Our work in two models of fibrosis with different mechanisms of induction (i.e. xenobiotic vs. genetic) indicates the value of biomarker G7 for monitoring fibrosis. These results are reflective of fibrosis as a conserved tissue response to diverse chronic liver diseases (e.g. viral hepatitis, alcohol abuse, fatty liver disease, etc.) and suggest that biomarker G7 could be useful for monitoring fibrosis stemming from distinct underlying pathologies. Moving forward, an important area for future study is elucidating the biological mechanisms that are ultimately responsible for the release of individual reporters. This could be accomplished, for example, by comparing urinary signatures from animals lacking specific proteases (e.g. MMP9−/−) to their wild type counterpart or the use of clodronate liposomes to deplete liver macrophages to identify reporters that track with cellular inflammation.
A major factor preventing early detection of cancer is the tremendous dilution biomarkers experience upon release from tumor cells into systemic circulation. Recent computational estimates by Gambhir and colleagues18 revealed that solid tumors could potentially remain undetectable for 10–12 years and reach spherical diameters > 2.5 cm before biomarker levels becomes sufficiently elevated to indicate disease. Here, the advantage of our system is the ability to amplify tumor responses by leveraging enzymatic turnover (i.e. a single copy of a protease can cleave 100's of peptide substrates per hour) and the renal system's natural capacity to remove and concentrate plasma peptides into urine (i.e. from ~5 L of blood to 300 ml void volume). Our study shows that the combined effects of protease amplification and renal concentration can lead to promising results such as the detection of small tumors that CEA failed to discriminate even at the highest tumor burdens allowable in our animal models. Since many tumors do not secrete biomarkers at sufficient rates (or at all) for detection, targeting tumor proteases should allow a broader range of cancers to be discovered at an early stage because proteases are uniformly implicated during tumor invasion and metastasis. Extension of this platform to multiple types of cancers would benefit from the development of cancer-type specific tests that could be accomplished by identifying unique biomarker panels for each cancer44. Conversely, a highly-sensitive, pan-cancer test comprised of a single set of diverse probes would be useful in clinical settings when the primary tumor is already known, such as monitoring for recurrence or metastases following surgical resection of primary tumors.
The successful translation of this platform to humans would require further confirmation of our lead biomarkers in patients as well as quantifying the potential benefits of monitoring biomarker panels versus single markers. A crucial limitation of existing single biomarker assays is their relatively poor disease specificity (e.g. CEA is elevated in smokers). These assays can be improved via multiplexing (e.g. prenatal triple screening) or specifying their use in well-defined clinical contexts (e.g. PSA is now recommended for recurrence monitoring but not screening). Similarly, the ability of this platform to differentiate protease-driven diseases (e.g. inflammation vs. cancer) would benefit from multiplexing and serial measurements in high-risk populations.
Lastly, a general concern with rodent studies is the relatively small number of animals used for hypotheses testing and validation. In this study, the number of animals selected per experimental condition (n = 5–10) allowed reasonable estimation of the mean and variance based on a normal distribution. The statistical power of our study was further bolstered by the prominent effect sizes observed (e.g. large AUCs) in both diseases studied as well as concordant biomarker responses across distinct models. Nonetheless, looking forward and in light of recent expert recommendations regarding biomarker qualification37, the results of this study will require further confirmation and rigorous evaluation in humans.
In summary, we believe this study provides a framework for engineering diagnostic agents that can exploit fundamental features of human disease and physiology for noninvasive urinary monitoring. Future expansion and inclusion of additional enzymatic families (e.g. lipases, nucleases, glycosidases), organ-specific delivery strategies and broader multiplexing capabilities would provide opportunities for systems-level monitoring of disease and elucidating multi-enzymatic networks in health and disease.
METHODS
Nanomaterial synthesis
Nanoworms were synthesized according to previously published protocols.24 Peptides were synthesized at MIT (Swanson Biotechnology Center); isotopically-labeled Fmoc amino acids were purchased from Cambridge Isotopes and 3-Nα-Fmoc-Amino-3-(2-nitrophenyl)propionic acid from Advanced Chemtech. Amine-terminated NWs were first reacted with Vivotag 680 (Perkin Elmer) to enable in vivo imaging, before reacted with SIA (Pierce) to introduce sulfhydryl-reactive handles. Cysteine-peptides and PEG-SH were then mixed with NWs overnight at room temperature (95:20:1 molar ratio respectively) and excess peptides were removed by size filtration. Peptide-NW stock solutions were stored in PBS at 4°C.
In vitro protease assays
For substrate screening, Fl-peptide-NWs (2.5 μM by peptide) were mixed with recombinant MMP-2/8/9 (R&D Systems), MMP-7/14 (AnaSpec, Inc.), Thrombin, Tissue Factor, Factor Xa or Cathepsin B (Haematologic Technologies, Inc.) in a 96-well plate at 37°C in activity buffers according to manufacturer's instructions and monitored with a microplate reader (SpectroMax Gemini EM). For MS analysis, equimolar iCORE-encoded NWs (Table) were incubated with proteases for 2.5 hrs at 37°C. Cleavage fragments were purified from NWs by size filtration before UV treatment (365 nm, CL-1000 UV crosslinker, UVP). Reporters were then dried by speed vacuum centrifuge and stored at 4°C.
In vivo imaging
All animal work was approved by the committee on animal care (MIT, protocol #0408-038-11). FVB/NJ mice (Jackson Labs) were fed with 0.1% w/w DDC (Sigma) rodent chow for three weeks (Research Diets). Fibrotic and age control animals were i.v. infused with VivoTag-680 labeled reagents and visualized by IVIS imaging (Xenogen). For tumor xenografts, LS 174T cancer cells lines were maintained in 10% FBS EMEM and inoculated s.c. (5×106/flank) in NU/NU mice (Charles River) prior to imaging.
Characterization of models
For in situ zymography, fibrotic sections were covered with 90 μl solution of 0.5% w/v low melt agarose (Sigma) in MMP activation buffer (50 mM Tris, 150 NaCl, 5 mM CaCl2, 0.025% Brij 35, pH 7.5) with 10 μl of DQ-gelation (1mg/ml, Invitrogen) and Hoechst dye at 37°C. Slides were solidified at 4°C and then incubated at room temperature overnight to promote gelation proteolysis by tissue proteases. To quantify hepatic collagen, tissue from the right and left lobes (250–300 mg) were hydrolyzed in 5 ml of 6 N HCl at 110°C for 16 hours followed by hydroxyproline quantification as previously described.38 To quantify CEA, blood was collected from tumor animals into Capiject microtubes (Terumo) to isolate serum before ELISA analysis (Calbiotech). For immunoflurescence analysis, equimolar NW cocktails (5 μM/peptide) were administered in Fibrotic FVB/NJ or tumor bearing Nude mice. After perfusion, livers or tumors were fixed in 4% PFA, froze for sectioning and stained for F4/80 (AbD Serotec), MMP-9 (R&D Biosystems), CD31 (Santa Cruz Biotechnologies) and/or FITC (Genetex) before analyzed by fluorescence microscopy (Nikon Eclipse Ti).
Collection of urinary peptides
Mice were i.v. infused with 200 μl of PBS containing equimolar NW cocktails (5 μM per peptide) with EDTA-free protease inhibitor tablets (Roche) to isolate MMP activity. Marimastat was dosed at 100 mg/kg in 0.45% methylcellulose twice daily via orogastric gavage for days prior to NW infusion. Mice were placed over 96-well plates surrounded by cylindrical sleeves for urine collection. To prevent further reporter degradation, voided samples were spiked with EDTA+ complete protease inhibitors (Roche) immediately after collection. To quantify urinary fluorescence, 2 μL of each sample was incubated with magnetic beads (Dynal) coated with α-FITC antibodies (Genetex) in 50 μl binding buffer (100 nM NH4OAc, 0.01% CHAPS) for 1 hr at 37°C, washed twice with 100 mM NH4OAc and eluted twice with 15 μl of 5% acetic acid. Samples were neutralized with 2 M Tris and quantified by microplate fluorimetry. For iCORE analysis, samples were irradiated with UV for 30 min. before TCA precipitation (20% final volume) to remove proteins. Soluble fractions were applied to C18 reverse phase columns (Nest Group) and eluted via step gradients of 20% ACN increments in 0.1% formic acid. 60% ACN fractions containing Glu-fib peptides were collected and dried by vacuum centrifuge.
LC MS/MS analyses
Peptide samples were reconstituted in 5% ACN, 0.1% formic acid and analyzed at MIT or the Taplin MS facility (Harvard Medical School). At MIT, peptides were captured and eluted from a C18 nanoflow HPLC column (75 μm internal diameter Magic C18 AQ, Michrom BioResources, Inc.) at a flow rate of 300 nL/min using water-acetonitrile solvent system with 0.1% formic acid. ESI mass spectrometry was carried out on a QSTAR Elite Q-TOF mass spectrometer (AB Sciex). At Harvard, samples are reconstituted in 2.5% ACN, 0.1% formic acid. Samples are injected using a Famos autosampler (LC Packings) into an Agilent 1100 HPLC prior to mass analysis on a LTQ-Orbitrap (Thermo Electron). To account for discrepancies in urine volumes and concentration, peak intensities of individual reporters were scaled relative to its respective total iCORE ion current before normalization against control samples to account for technical and age-related variations.
Statistical Analyses
Pearson's correlation coefficients between different protease profiles were calculated with MatLAB. ANOVA analyses were calculated with GraphPad 5.0 (Prism). For ROC analysis, risk score functions were first estimated by logistic regression on individual biomarkers followed by ROC curve analyses of single or biomarker combinations (SigmaPlot).
Supplementary Material
Acknowledgements
We thank R. Cook, N. Schiller and M. Brown (Swanson Biotechnology Center (SBC), MIT) for peptide synthesis and tissue sectioning, C. Whittaker (SBC, MIT) for bioinformatic insight, R. Tomaino (Harvard Taplin MS facility) for MS analysis, F. Giammo and D. Kim (MIT) for initial probe work and urine purification, S. Carr (Broad Institute of MIT/Harvard) for MS expertise and M. Sailor (UCSD), J. Park (UCSD) and S. Friedman (Mount Sinai School of Medicine) for insightful discussions. This work was funded by the NIH (BRP: R01 CA124427), Kathy and Curt Marble Cancer Research Fund to S.N.B. and grants U19 AI066313, 1R21 DK075857 to D.S. G.A.K. is supported by the Ruth L. Kirschstein NRSA (F32CA159496-01). S.N.B. is a Howard Hughes Institute Investigator.
Footnotes
Author contributions G.A.K., G.v.M. and S.N.B conceived the study and designed the experiments. G.v.M. and S.M. performed in vitro substrate screen and initial pharmacokinetic studies; G.A.K. developed the mass encoding scheme; G.A.K., G.M. and O.A. performed the fibrosis experiments; Y.P., D.Y.S., S.B.L. and D.S. developed and provided expertise for fibrosis progression and reversal protocols, and performed fibrosis quantification assays; G.A.K. and A.D.W. performed cancer experiments; G.A.K. and I.A.P collected in vivo MS data; G.A.K., G.v.M. and S.N.B. wrote the manuscript.
References
- [1].Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- [2].Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- [3].Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [4].Findeisen P, Neumaier M. Functional protease profiling for diagnosis of malignant disease. Proteomics Clin Appl. 2012;6:60–78. doi: 10.1002/prca.201100058. [DOI] [PubMed] [Google Scholar]
- [5].Surinova S, et al. On the development of plasma protein biomarkers. J Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- [6].Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- [7].Moon P-G, You S, Lee J-E, Hwang D, Baek M-C. Urinary exosomes and proteomics. Mass Spectrom Rev. 2011;30:1185–1202. doi: 10.1002/mas.20319. [DOI] [PubMed] [Google Scholar]
- [8].Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lutz AM, Willmann JK, Cochran FV, Ray P, Gambhir SS. Cancer screening: a mathematical model relating secreted blood biomarker levels to tumor sizes. PLoS Med. 2008;5:e170. doi: 10.1371/journal.pmed.0050170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haun JB, et al. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3:71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Edgington LE, Verdoes M, Bogyo M. Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Curr Opin Chem Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat Rev Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hilderbrand SA, Weissleder R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol. 2010;14:71–79. doi: 10.1016/j.cbpa.2009.09.029. [DOI] [PubMed] [Google Scholar]
- [14].Braet F, Wisse E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp Hepatol. 2002;1:1. doi: 10.1186/1476-5926-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].López-Otín C, Bond JS. Proteases: multifunctional enzymes in life and disease. J Biol Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med. 2011;3:109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bremer C, Tung CH, Weissleder R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nat Med. 2001;7:743–748. doi: 10.1038/89126. [DOI] [PubMed] [Google Scholar]
- [20].Kridel SJ, et al. A unique substrate binding mode discriminates membrane type-1 matrix metalloproteinase from other matrix metalloproteinases. J Biol Chem. 2002;277:23788–23793. doi: 10.1074/jbc.M111574200. [DOI] [PubMed] [Google Scholar]
- [21].Lutolf MP, et al. Repair of bone defects using synthetic mimetics of collagenous extracellular matrices. Nat Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- [22].Mahmood U, Weissleder R. Near-infrared optical imaging of proteases in cancer. Mol Cancer Ther. 2003;2:489–496. [PubMed] [Google Scholar]
- [23].Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- [24].Park J-H, et al. Systematic surface engineering of magnetic nanoworms for in vivo tumor targeting. Small. 2009;5:694–700. doi: 10.1002/smll.200801789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fickert P, et al. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am J Pathol. 2007;171:525–536. doi: 10.2353/ajpath.2007.061133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Morris TA, et al. Urine and plasma levels of fibrinopeptide b in patients with deep vein thrombosis and pulmonary embolism. Thromb Res. 2003;110:159–165. doi: 10.1016/s0049-3848(03)00334-7. [DOI] [PubMed] [Google Scholar]
- [27].Choi HS, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Park J-H, et al. Magnetic iron oxide nanoworms for tumor targeting and imaging. Adv Mater. 2008;20:1630–1635. doi: 10.1002/adma.200800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Villanueva J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Villanueva J, et al. A sequence-specific exopeptidase activity test (sseat) for “functional” biomarker discovery. Mol Cell Proteomics. 2008;7:509–518. doi: 10.1074/mcp.M700397-MCP200. [DOI] [PubMed] [Google Scholar]
- [31].Brown BB, Wagner DS, Geysen HM. A single-bead decode strategy using electrospray ionization mass spectrometry and a new photolabile linker: 3-amino-3-(2-nitrophenyl)propionic acid. Mol Divers. 1995;1:4–12. doi: 10.1007/BF01715804. [DOI] [PubMed] [Google Scholar]
- [32].Ross PL, et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents.ralph. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- [33].Thompson A, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- [34].Rockey DC, et al. Liver biopsy. Hepatology. 2009;49:1017–1044. doi: 10.1002/hep.22742. [DOI] [PubMed] [Google Scholar]
- [35].Popov Y, Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology. 2009;50:1294–1306. doi: 10.1002/hep.23123. [DOI] [PubMed] [Google Scholar]
- [36].Bedossa P, Dargère D, Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology. 2003;38(6):1449–1457. doi: 10.1016/j.hep.2003.09.022. [DOI] [PubMed] [Google Scholar]
- [37].Mischak H, et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- [38].Popov Y, Patsenker E, Fickert P, Trauner M, Schuppan D. Mdr2 (abcb4)−/− mice spontaneously develop severe biliary fibrosis via massive dysregulation of pro- and antifibrogenic genes. J Hepatol. 2005;43:1045–1054. doi: 10.1016/j.jhep.2005.06.025. [DOI] [PubMed] [Google Scholar]
- [39].Etzioni R, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- [40].D'Souza AL, et al. A strategy for blood biomarker amplification and localization using ultrasound. Proc Natl Acad Sci U S A. 2009;106:17152–17157. doi: 10.1073/pnas.0903437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dekker LJM, et al. Differential expression of protease activity in serum samples of prostate carcinoma patients with metastases. Proteomics. 2010;10:2348–2358. doi: 10.1002/pmic.200900682. [DOI] [PubMed] [Google Scholar]
- [42].Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sugahara KN, et al. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kulasingam V, Pavlou MP, Diamandis EP. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat Rev Cancer. 2010;10:371–378. doi: 10.1038/nrc2831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.