Abstract
We have determined the X-ray crystal structures of the NADH-dependent alcohol dehydrogenase LlAdhA from Lactococcus lactis and its laboratory-evolved variant LlAdhARE1 at 1.9 Å and 2.5 Å resolution, respectively. LlAdhARE1, which contains three amino acid mutations (Y50F, I212T, and L264V), was engineered to increase the microbial production of isobutanol (2-methylpropan-1-ol) from isobutyraldehyde (2-methylpropanal). Structural comparison of LlAdhA and LlAdhARE1 indicates that the enhanced activity on isobutyraldehyde stems from increases in the protein’s active site size, hydrophobicity, and substrate access. Further structure-guided mutagenesis generated a quadruple mutant (Y50F/N110S/I212T/L264V), whose KM for isobutyraldehyde is ~17-fold lower and catalytic efficiency (kcat/KM) is ~160-fold higher than wild-type LlAdhA. Combining detailed structural information and directed evolution, we have achieved significant improvements in non-native alcohol dehydrogenase activity that will facilitate the production of next-generation fuels such as isobutanol from renewable resources.
Keywords: Alcohol dehydrogenase, Crystal structure, Site-saturation mutagenesis, Directed evolution, Isobutyraldehyde, Biofuel
1. Introduction
Alcohol dehydrogenases (ADH, EC 1.1.1.1) are oxidoreductases that catalyze the reversible oxidation of a wide range of alcohols to the corresponding aldehydes or ketones using nicotinamide adenine dinucleotides (NAD(P)H) as coenzymes. Ubiquitous in bacteria, yeast, plants, and mammals, ADHs can be divided into three classes based on size (Reid et al., 1994): short-chain ADHs (c.a. 250 residues), long- chain, iron-activated ADHs (c.a. 385 residues), and medium-chain, zinc-dependent ADHs (c.a. 350 residues) which also belong to the medium-chain dehydrogenase/reductase (MDR) super-family. Several MDR-ADHs have been structurally characterized, including horse liver ADH (HLADH, Eklund et al., 1976, 2008) and yeast Saccharomyces cerevisiae ADH (YADH, Leskovac et al., 2002). MDR-ADHs are either dimeric or tetrameric. Dimeric ADHs are usually found in plants and mammals, whereas tetrameric ADHs are mostly found in bacteria and yeast. The MDR-ADH catalytic mechanism was established through studies of HLADH (Ramaswamy et al., 1994; Agarwal et al., 2000) and supplemented by studies of related MDR-ADHs (Eklund and Ramaswamy, 2008; Bakera et al., 2009).
ADHs play important roles in numerous natural and engineered metabolic pathways. The latter includes the work of Liao and coworkers, who engineered Ehrlich and valine biosynthetic pathways to produce isobutanol, a next-generation biofuel, in E. coli (Atsumi et al., 2008). Liao’s isobutanol pathway diverts 2-ketoisovalerate, a valine precursor, to isobutanol by over-expression of a 2-ketoisovalerate decarboxylase and an ADH. The ADH catalyzes the final step, conversion of isobutyraldehyde to isobutanol. This pathway can be used to produce isobutanol in a variety of microorganisms including E. coli (Atsumi et al., 2008, 2009 and 2010; Cann and Liao, 2008; Shen and Liao, 2008; Connor and Liao, 2009; Savrasova et al., 2011; Baez et al., 2011), Corynebacterium glutamicum (Smith et al., 2010; Blombach et al., 2011), Bacillus subtilis (Li et al., 2011), and Clostridium cellulolyticum (Higashide et al., 2011). Atsumi and coworkers reported that the NADH-dependent AdhA from Lactococcus lactis (LlAdhA) functions in this pathway, as does an NADPH-dependent homologue, YqhD, that is native to E. coli (Atsumi et al., 2010). Although the KM of YqhD toward isobutyraldehyde is 5-fold lower than that of LlAdhA, its preference for NADPH over NADH generates an overall cofactor imbalance in the biosynthetic pathway. To relieve the cofactor imbalance and improve isobutanol production, Bastian and coworkers in this laboratory used directed evolution by sequential rounds of random mutagenesis and screening to enhance L1AdhA activity on isobutyraldehyde (Bastian et al., 2011). The resulting ADH variant, LlAdhARE1, contained three amino acid substitutions (Y50F, I212T, and L264V) that led to a ~7-fold decrease in KM and a ~30-fold increase in catalytic efficiency (kcat/KM) over wild- type LlAdhA. Inclusion of LlAdhARE1 in the isobutanol biosynthetic pathway in place of wild-type LlAdhA increased anaerobic isobutanol titer from 8.4 g/L to 13.4 g/L in a 24-hour fermentation (Bastian et al., 2011).
To better understand the activity enhancements in LlAdhARE1 and to guide further mutagenesis to decrease the KM for isobutyraldehyde, a non-native substrate of LlAdhA, we determined the crystal structures of LlAdhA and LlAdhARE1. We have used the structures to further improve LlAdhARE1 for this pathway and provide insights into how these improvements were achieved.
2. Materials and methods
2.1 General
Strains, plasmids and primers are listed in Tables S1 and S2, Supplemental Information. Biological media were purchased from Research Products International (Mt. Prospect, IL, USA), NADH from Codexis, Inc. (Redwood City, CA, USA), aldehydes from Sigma-Aldrich (St. Louis, MO, USA), oligonucleotides from Integrated DNA Technologies (San Diego, CA, USA), DNA polymerases, restriction enzymes, and T4 ligase from New England Biolabs (Ipswich, MA, USA). DNA sequencing was performed by Laragen (Los Angeles, CA, USA). Standard molecular biology methods were taken from Maniatis et al. (Sambrook et al., 1989).
2.2 Cloning, library construction, and heterologous expression
For crystallization purposes, the genes encoding LlAdhA and variant LlAdhARE1 were cloned into pET22b(+) (EMD Chemicals Group, Darmstadt, Germany) using NdeI and XhoI restriction sites as described previously (Bastian et al., 2011) and expressed in E. coli BL21(DE3). Plasmids pGVRE1 and pGV29C8 harboring variants LlAdhARE1 and LlAdhA29C8 served as templates for site-saturation mutagenesis and random mutagenesis library construction, respectively. The libraries were constructed using primers listed in Table S2, Supplemental Information, and expressed in yeast CEN.PK2 as described previously (Bastian et al., 2011). Mutant LlAhdARE1-T212I harboring only the Y50F and L264V mutations was constructed using plasmid pGVRE1 and primers RE1_T212I for and RE1_T212I_rev (Table S2, Supplemental Information).
2.3 Kinetic assay and high-throughput screening
ADH activities were detected by monitoring NADH consumption at 340 nm for isobutyraldehyde, acetaldehyde, and coniferaldehyde, and at 365 nm for 2-furaldehyde, hydroxymethylfurfural (5-HMF), cinnamaldehyde, 4-hydroxybenzaldehyde, syringaldehyde and vanillin, as described previously (Larroy et al., 2002). All variants were purified by immobilized metal affinity chromatography (IMAC) before they were assayed. High-throughput screening was conducted using yeast lysate as described previously (Bastian et al., 2011).
2.4 Thermostability measurements
To determine the half-denaturation temperature (T50) of LlAdhA and its variants, 30- µL aliquots of purified enzyme were transferred into PCR tubes. Each tube was assigned a specific incubation temperature that corresponded to a slot on the block of an Eppendorf master cycler PCR machine programmed with a gradient covering a 20°C temperature range. The measurements were conducted in duplicate. The temperature range was changed, depending on the stability of the tested enzymes, in order to cover the denaturation range. The tubes were incubated in their slots for 15 min, and the reactions were then quenched on ice. Residual activity was determined with the activity assay in a total volume of 100 µL in a plate reader (TECAN Group Ltd., Switzerland). T50 is defined as the temperature at which 50% of the initial activity is retained after 15 minutes incubation.
2.5 Protein purification, crystallization, and data collection
Wild-type LlAdhA and variants LlAdhARE1 and LlAdhARE1-T212I were expressed from pET22b(+) in E. coli BL21(DE3) and purified by IMAC as described (Bastian et al., 2011). For crystallization purposes, the IMAC-purified proteins were subjected to two sequential anion exchange chromatography runs over pre-equilibrated Q Sepharose™ columns (HiTrap™ Q HP, GE Healthcare, Piscataway, NJ, USA) using an AKTA FPLC system (GE Healthcare, Waukesha, WI, USA) after a buffer exchange to buffer A (25 mM Tris pH 7.4, and 10 mM MgCl2). The anion exchange purification method consisted of a 4-column volume equilibration step with buffer A, followed by sample injection and washout of unbound sample with buffer A for two column volumes. The proteins were eluted with a linear gradient from buffer A to 100% buffer B (25 mM Tris pH 7.4, 10 mM MgCl2, and 1 M NaCl) over 10 column volumes. Both enzymes eluted at 35% buffer B. Purified proteins (>99% purity) were then subjected to a buffer exchange to TBS buffer (50 mM Tris pH 7.4 and 150 mM NaCl) and concentrated to 12 mg/mL prior to crystallization. For determination of the oligomerization state, we performed size exclusion chromatography on a Superdex™ 200 10/300 GL column (GE Healthcare) with 20 mM Tris, pH 7.0. Prior to the gel filtration, the enzyme was purified over a HisTrap column as described above followed by a concentration step using centrifugal filter units with a 30 kDa MWCO (Millipore). The column was calibrated with gel filtration standards from Bio-Rad. Droplets (0.3 µL) of concentrated protein solutions were tested against an equal volume of 480 crystallization conditions at room temperature using the sitting drop method. The first hit appeared in 30% (v/v) polyethylene glycol 400, 0.2 M calcium acetate, 0.1 M sodium acetate pH 4.5 for LlAdhARE1 and was further refined by traditional methods employing the sitting drop method in Linbro plates (Hampton Research). The final crystallization conditions were 30% (v/v) polyethylene glycol 400, 0.2 M calcium acetate, 0.1 M sodium acetate pH 4.5 for LlAdhARE1, and 30% (v/v) polyethylene glycol 400, 0.2 M calcium acetate, 0.1 M sodium acetate pH 5.0 for LlAdhA, and both yielded crystals in two days.
Crystals were transferred into a cryoprotectant solution of mother liquor supplemented with 25% (v/v) PEG 400 prior to flash-cooling in liquid nitrogen and shipping to the Stanford Synchrotron Radiation Laboratory (SSRL). Diffraction data were collected using a Dectris Pilatus 6M detector on beamline 12-2 at SSRL at 100 K. An X-ray wavelength of 0.954 Å was used for both proteins. Diffraction datasets were integrated with XDS (Kabsch, 2010) and scaled using SCALA (Evans, 2006).
2.6 Molecular replacement and structural refinement
The crystal structure of LlAdhARE1 was determined by molecular replacement. First, a homology model for LlAdhARE1 from a variety of available homologous structures was generated using MODELLER (Šali et al., 1995). The program MOLREP (Vagin and Teplyakov, 1997) in CCP4 (Bailey, 1994) was then used to identify a candidate solution in which two monomers form a contiguous beta sheet via residues 280–285. However this solution was not amenable to refinement (rigid REFMAC Rfree = 0.52). This solution was somewhat improved (Rfree = 0.39) by PHENIX Autobuild (Adams et al., 2010). To proceed, we removed large portions (~40%) of the model, deleting most of the ill-fitting Rossmann domains. This truncated model retained an Rfree of 0.4 after applying PHENIX Refine. Given this model, a subsequent Autobuild run produced an entirely different dimer interface (via residues 97–100) with an improved Rfree of 0.34. At this point, the density map was of sufficient quality to manually rebuild missing segments of the model in Coot (Emsley and Cowtan, 2004), leading to a productive iterative cycle between manual refinement in Coot and automated refinement using PHENIX. The structure of wild-type LlAdhA was determined using the LlAdhARE1 model.
2.7 Structure analysis and modeling
The crystal structure of LlAdhA was superimposed with the crystal structures of variant LlAdhARE1, Bacillus stearothermophilus ADH (htADH, PDB ID: 1RJW, 50% identity) (Ceccarelli et al., 2004) and Pseudomonas aeruginosa ADH (PaADH, PDB ID: 1LLU, 41% identity) (Levin et al., 2004) on Cα atoms using the align tool within Pymol (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC). Root mean square deviation (rmsd) values were obtained by Pymol using the align tool. NADH coordinates were extracted from PaADH after superposition of PaADH with LlAdhA and were used to model NAD+ into LlAdhA and LlAdhARE1. Isobutanol was built into LlAdhA and LlAdhARE1 structures manually using trifluoroethyl alcohol from htADH as a starting model.
2.8 Protein Data Bank accession codes
Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 4EEX for LlAdhA and 4EEZ for LlAdhARE1.
3. Results and Discussion
3.1 Crystal structures of LlAdhA and variant LlAdhARE1
We determined the crystal structures of LlAdhA and variant LlAdhARE1 to understand the basis of the activity increase in the variant and to obtain guidance for further enzyme engineering. This work provides the first structure of an ADH from L. lactis. The crystallographic statistics for LlAdhA and LlAdhARE1 data collection and refinement are shown in Table 1. Both crystal structures were observed in space group C2221 with two molecules per asymmetric unit. Size exclusion chromatography revealed only a peak consistent with dimer formation (see Methods). The observed dimer is likely to correspond either to the two monomers found in the crystallographic asymmetric unit (~1830 Å2 buried) or to a crystallographic dimer formed via beta strand pairing of the Rossmann domain (~3450 Å2 buried). Most bacterial ADHs are tetrameric, and LlAdhA represents a rare dimeric bacterial ADH. The LlAdhARE1 structure was determined by molecular replacement, and the structure of wild-type LlAdhA was determined using LlAdhARE1 as a starting model. LlAdhA and LlAdhARE1 were refined to an Rfree of 23.0% and 20.4% at a resolution of 2.2 Å and 1.9 Å, respectively.
Table 1.
Crystallographic data collection and refinement statistics.
| Crystal Parametersa | LlAdhA PDB ID 4EEX |
LlAdhARE1 PDB ID 4EEZ |
|---|---|---|
| Space group | C2221 | C2221 |
| Unit cell dimensions | a=123.5 Å, | a=124.4 Å, |
| b=126.5 Å, | b=126.5 Å, | |
| c=94.35 Å, | c=94.2 Å, | |
| α=β=γ=90° | α=β=γ=90° | |
| Data collection statistics | ||
| Wavelength | 0.954 | 0.954 |
| Resolution (Å) | 2.2–32.2 (2.2–2.3)b | 1.9–37.8 (1.9–2.0) |
| Total reflections | 119805 | 193087 |
| Unique reflections | 34501 | 54325 |
| Rmerge (%) | 6.4 (43.1) | 5.2 (31.5) |
| Completeness (%) | 96.6 (92.8) | 97.7 (97.2) |
| I/σ I | 11.9 (2.8) | 13.8 (3.3) |
| Refinement | ||
| Rfactor /Rfreec(%) | 18.1/23.6 | 16.6/20.8 |
| Protein molecules in asymmetric unit | 2 | 2 |
| No. of residues | 680 | 681 |
| No. of water molecules | 188 | 435 |
| No. of zinc atoms | 4 | 4 |
| Average B-factor (Å2) | ||
| Protein | 32.3 | 24.4 |
| Water | 32.3 | 30.4 |
| Geometry deviation | ||
| Rmsd on bond length (Å) | 0.02 | 0.03 |
| Rmsd on bond angles (deg) | 1.81 | 1.99 |
| Ramachandran analysis (%) | ||
| Preferred regions | 97.9 | 98.1 |
| Outliers | 0.1 | 0.1 |
All data sets were collected from single crystals.
Highest-resolution shell is shown in parentheses.
Rfree is calculated using 5% of the reflections randomly excluded from refinement.
Structural alignment of LlAdhA and LlAdhARE1 resulted in a root mean square deviation (rmsd) of 0.19 Å, indicating the absence of large-scale structural changes attributable to the mutations. We do observe structural differences between asymmetric monomers within both LlAdhA and LlAdhARE1 crystal structures. For example, the rmsd between chains A and B of LlAdhARE1 is 1.36 Å. Alignment of these structures with an NADH-bound structure of the alcohol dehydrogenase from Pseudomonas aeruginosa (PaADH, PDB ID: 1LLU) shows that in both cases the tertiary structure of chain A in LlAdhA and its variant closely resembles the cofactor- bound protein (rmsd = 0.81 Å). Chain B (rmsd = 1.83 Å) shows considerable differences in the cofactor-binding domain. It is known that cofactor binding in other ADHs induces conformational changes in this region (Plapp, 2010). Together, these data suggest that the LlAdhA crystal packing permits both open and closed conformations to be observed, even in the absence of the bound reductant.
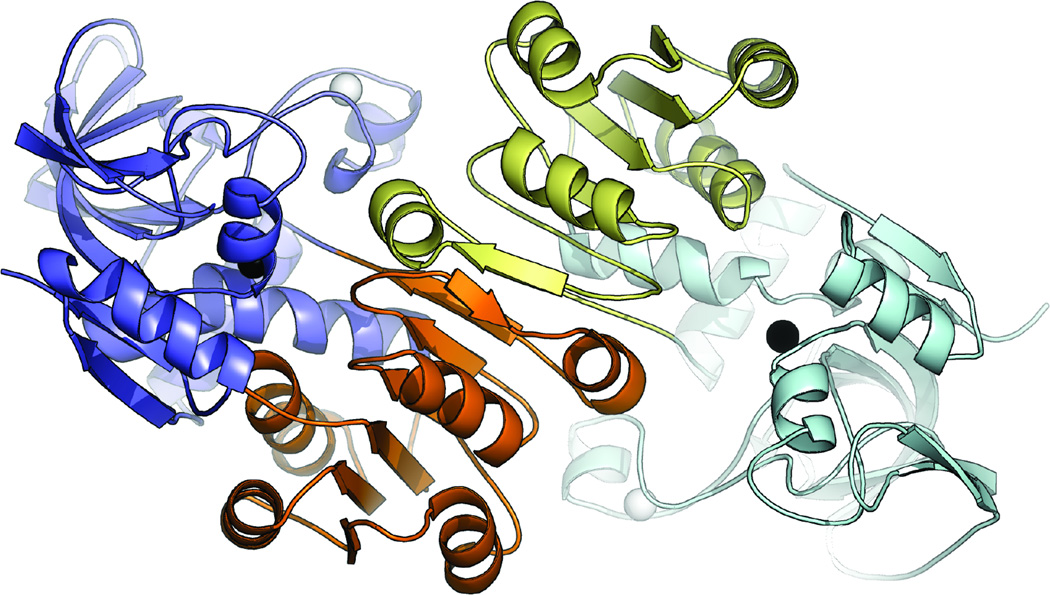
The LlAdhA monomer (Fig. 1) comprises 339 residues and contains two zinc ions, a structural zinc and a catalytic zinc. The structural zinc is tetrahedrally coordinated by four cysteine residues (Cys91, Cys94, Cys97, and Cys105). In LlAdhA, coordination of the catalytic zinc is consistent with most MDR-ADHs and involves active site amino acids Cys39, His60, and Cys147. The fourth coordination site is filled by a water molecule in the substrate/cofactor-free enzyme subunit (Aulda and Bergman, 2008).
Fig. 1.
The LlAdhA asymmetric unit dimer with catalytic domains (blue, light blue), Rossmann fold (orange, yellow), structural zinc atoms (white spheres), and catalytic zinc atoms (black spheres).
Each LlAdhA monomer contains two distinct domains: a coenzyme-binding domain, which folds into a classical α/β Rossmann fold (residues 148–287), and a catalytic domain (residues 1–147 and 288–339). These two domains are connected by an interdomain ‘hinge’ formed by a helix (residues 148–159) and a loop (residues 285- 291), and are separated by a deep cleft that houses the active site and the catalytic zinc. LlAdhA contains a conserved proton relay system composed of Thr41 and His44; this system contributes to the transfer of protons to free solvent, which serves as a driving force for catalysis (Agarwal et al., 2000; Levin et al., 2004).
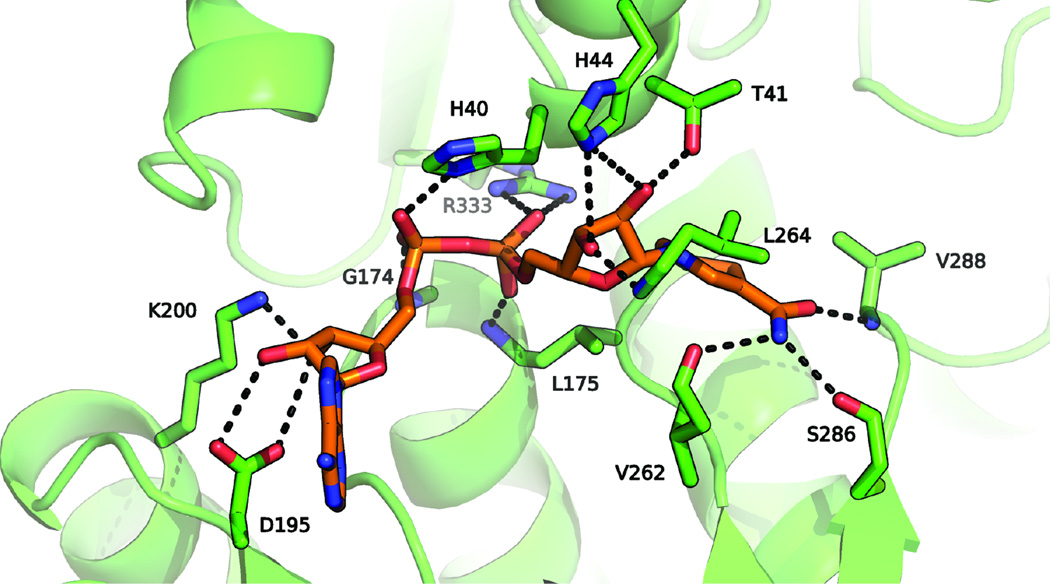
To identify molecular determinants of NADH cofactor binding, we superposed the LlAdhA and the NADH-bound PaADH structures (Levin et al., 2004). From this alignment, we infer that the cofactor interacts with LlAdhA through a number of hydrogen bonds with the protein main chain (residues Val262, Ser286, Val288, Leu264, His40, Leu175, and Gly174) as well as several amino acid side chains (residues His44, Thr41, His40, Asp195, and Arg333) (see Fig. 2). Asp195 is conserved throughout all NADH-dependent ADHs for recognition of the two NADH ribosyl hydroxyl groups. The conserved Rossmann GxGxxG motif is also observed in LlAdhA.
Fig. 2.
The cofactor-binding site of LlAdhA (green), with cofactor coordinates adopted from the superimposed NADH (orange) co-crystal structure of PaADH (PDB ID: 1LLU). Hydrogen bonds to the main chain of residues Gly174, Leu175, Val262, Leu264, Ser286, and Val288 anchor NADH to the active site. The other hydroxyl groups are bound to the protein via a series of amino acids, Thr41, His40, His44 Asp195, Lys200, and Arg333.
3.2 Protein engineering of LlAdhA to increase activity on isobutyraldehyde
LlAdhA catalyzes the final step, reduction of isobutyraldehyde, in the engineered biosynthesis of isobutanol (Atsumi et al., 2008 and 2010; Bastian et al., 2011). To improve isobutanol production by this novel pathway, we previously used directed evolution to enhance the catalytic efficiency of LlAdhA for isobutyraldehyde. Random mutagenesis and subsequent recombination of beneficial mutations yielded variant LlAdhARE1 with a ~7-fold decrease in KM and ~30-fold increase in catalytic efficiency on isobutyraldehyde (Table 2, Bastian et al., 2011). Attempts to further improve activity by random mutagenesis were unsuccessful. Using the LlAdhARE1 structure, we selected four residues based on their proximity to the substrate binding pocket, Trp86, Asn110, Leu287, and Val288, for saturation mutagenesis with the gene coding for RE1 as template. Upon screening the four individual libraries to >95% coverage we identified a variant with the additional N110S mutation. This variant, LlAdhA29C8, has a ~17-fold decrease in KM (from 12 to 0.68 mM) and a ~160-fold increase in catalytic efficiency (from 2.8 to 440 mM−1s−1) on isobutyraldehyde compared to wild-type LlAdhA (Table 2). No improved variants were isolated from the saturation mutagenesis at residues Trp86, Leu287, or Val288.
Table 2.
Kinetic parameters and thermostabilities of LlAdhA and variants, measured with isobutyraldehyde in the presence of the cofactor NADH.
| Variant | Mutations | T50 [°C] | KM [mM] | kcat [s−1] | kcat/KM [mM−1s−1] |
|---|---|---|---|---|---|
| LlAdhA | - | 54.4 ± 0.4 | 12 ± 1 | 30 ± 1 | 2.8 ± 0.2 |
| LlAdhARE1-T212I | Y50F, L264V | 55.2 ± 0.2 | 1.6 ± 0.2 | 50 ± 1 | 32 ± 4 |
| LlAdhARE1 | Y50F, I212T, L264V | 61.6 ± 0.1 | 1.70 ± 0.01 | 140 ± 1 | 82 ± 1 |
| LlAdhA29C8 | Y50F, N110S, I212T, L264V |
55.2 ± 0.2 | 0.68 ± 0.07 | 300 ± 4 | 440 ± 50 |
All enzymes were purified prior to characterization. Errors are reported as standard deviations determined from a minimum of three independent experiments. The enzyme assays were conducted in 100 mM Tris pH 7 with 1 mM DTT, 200 µM NADH, and 10 mM isobutyraldehyde. Concentrations of the purified enzymes were determined using the Bradford assay. The Michaelis-Menten constants for the substrate were measured with appropriate dilution series of isobutyraldehyde. Enzyme assays were performed at 25°C.
3.3 Structural insights into directed evolution
3.3.1 Structural basis of the increased activity of LlAdhARE1 on isobutyraldehyde
Three mutations (Y50F, I212T, and L264V) present in LlAdhARE1 are responsible for its ~30-fold higher catalytic efficiency on isobutyraldehyde. Mutation I212T is surface-exposed in the NADH-binding domain of LlAdhARE1 and distant from the active site. Because this mutation was discovered together with Y50F (Bastian et al., 2011), we constructed the variant having only the Y50F and L264V mutations, denoted LlAdhARE1-T212I, and compared its activity to that of LlAdhARE1 (Table 2); LlAdhARE1-T212I has a significantly lower kcat. I212T thus makes an important contribution to the increased activity, despite its location >20 Å from the active site.
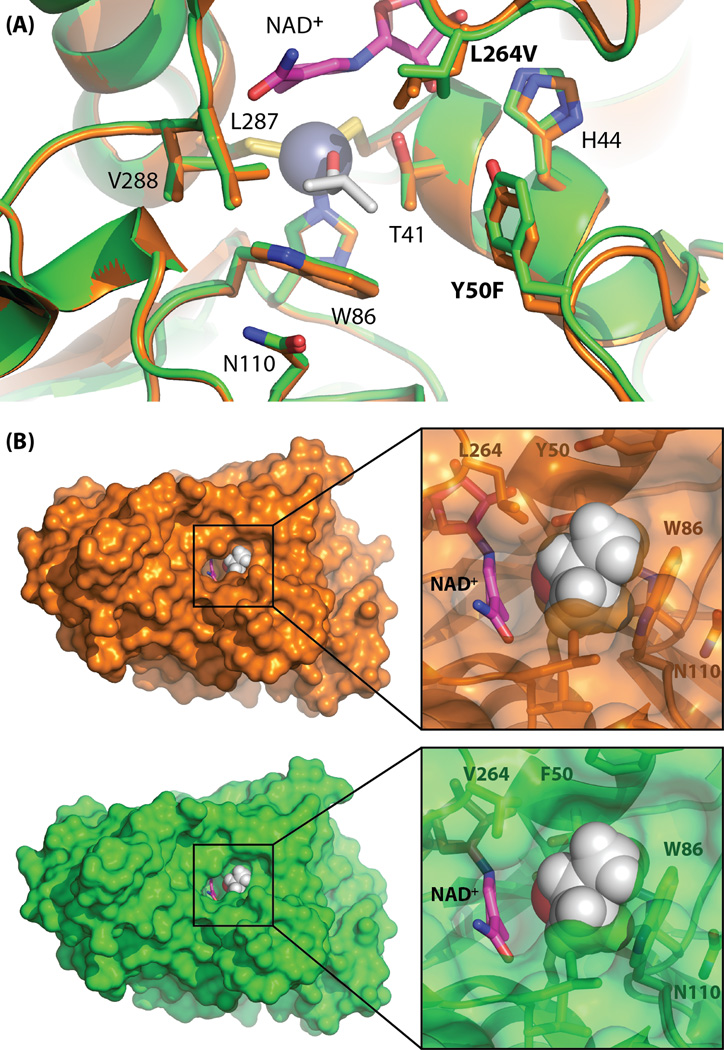
Mutations L264V and Y50F lie in loops that cap the active site and have a more direct role in regulating activity on isobutyraldehyde (Fig. 3A). Making use of the known structure of zinc-bound trifluoroethanol (see Methods), we modeled zinc-bound isobutanol in the active site of LlAdhARE1 using Pymol (Fig. 3A). From this model, it is apparent that the wild-type Leu264 side chain leaves little space for the bulky terminal methyl groups of isobutyraldehyde or isobutanol. We suspect that mutation to the smaller valine provides the space to accommodate the non-native aldehyde, which is larger than the native substrate, acetaldehyde. Deletion of the Tyr hydroxyl in Y50F, which packs against the side chain of Leu264 in wild-type LlAdhA, further enlarges the substrate-binding pocket to accommodate larger substrates. Analogous mutations that increase the size and hydrophobicity of the active site cavity have been found in the yeast homolog YADH-1 (Murali and Creaser, 1986; Creaser et al., 1990). Moreover, replacement of Trp54 in YADH-1 (analogous to Tyr50 in LlAdhA) with leucine was found to broaden substrate specificity (Weinhold et al., 1995). Thus we propose that improvements in isobutyraldehyde activity in LlAdhARE1 are due to its enlarged and increasingly hydrophobic binding pocket resulting from loss of the Tyr50 hydroxyl moiety and substitution of bulky Leu264 with valine (Fig. 3A, Table 3).
Fig. 3.
(A) The active site of LlAdhA (orange) superimposed with LlAdhARE1 (green). The catalytic zinc is shown as a blue sphere. Selected active site residues are shown in stick form. Thr41 and His44 are implicated in the proton conduction pathway from the active site to solvent. Isobutanol (white) and NAD+ (magenta) were modeled by alignment of htADH and PaADH (see Methods), respectively. (B) Surface representation of LlAdhA (orange) and LlAdhARE1 (green) active site cavities reveals an enlarged channel opening in LlAdhARE1’s that may result in improved access of larger non-native substrates to the active site.
Table 3.
Kinetic parameters of LlAdhA variants on related aldehydes.
| Aldehydes | Structure | LlAdhA | LlAdhARE1 | LlAdhA29C8 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
KM [mM] |
kcat [s−1] |
kcat/KM [mM−1s−1] |
KM [mM] |
kcat [s−1] |
kcat/KM [mM−1s−1] |
KM [mM] |
kcat [s−1] |
kcat/KM [mM−1s−1] |
||
| Isobutyraldehyde | 12 | 30 | 2.8 | 1.70 | 140 | 82 | 0.68 | 300 | 440 | |
| Acetaldehyde | 0.4 | 35 | 94 | 0.5 | 31 | 57 | 0.92 | 58 | 63 | |
| 5-HMF | 22 | 19 | 0.88 | 0.67 | 23 | 34 | 0.57 | 29 | 51 | |
| 2-Furaldehyde | 0.39 | 22 | 57 | 0.26 | 6.0 | 21 | 0.20 | 7 | 37 | |
| Cinnamaldehyde | 0.7 | 27 | 39 | 0.24 | 28 | 140 | 0.16 | 31 | 210 | |
All enzymes were purified prior to characterization. Errors are reported as standard deviations determined from a minimum of three independent experiments. The resulting standard errors are shown in Table S3, Supplemental Information. The enzyme assays were conducted in 100 mM Tris pH 7 with 1 mM DTT, 200 µM NADH, and 10 mM substrate. Concentrations of the purified enzymes were determined using the Bradford assay. The Michaelis-Menten constants for the substrate were measured with appropriate dilution series of isobutyraldehyde. Enzyme assays were performed at 25°C.
The entrance to the LlAdhA active site is formed by the side chains of four residues: Tyr50, Trp86, Leu264, and Leu287. In LlAdhARE1, shortening of the leucine side chain in L264V leads to significant opening of the entrance (Fig. 3B). It has been shown that the loop 292–299 in the horse liver enzyme HLADH, which corresponds to the loop containing Leu264 in LlAdhA, undergoes conformational changes upon cofactor binding (Plapp, 2010). Similar conformational changes are expected to be important in LlAdhA, and mutations in this loop will tune substrate binding and catalysis. We observe very subtle movements of the loop carrying Phe50 versus Tyr50 (the alpha carbon moves only 0.3 Å when residues 40–60 are aligned). As shown in Fig. 3B, the overall effect is greater substrate access that leads to the ADH catalytic zinc center. Therefore, improvements in the activity of LlAdhARE1 on isobutyraldehyde can be attributed to expansion and increased hydrophobicity of the substrate-binding pocket combined with improved substrate access to the active site.
3.3.2 Effects of N110S on isobutyraldehyde activity in LlAdhA29C8
If Leu264 and Tyr50 form one wall of the substrate pocket, the opposing wall comprises Leu287, Val288, Trp86, and Asn110. We carried out site-saturation mutagenesis at each of these sites and found one favorable mutation, N110S. We can only speculate on the effect of this mutation since the structure of LlAdhA29C8 is not available. However, it is likely that the N110S mutation perturbs the conformation of Trp86, which in turn affects the substrate binding pocket. In most bacterial ADHs the side chain of Asn110 pi-stacks with the indole ring of the highly conserved Trp86, which lies just above the catalytic zinc. The increased activity of LlAdhA29C8 may be due to a subtle, or even dramatic, reorientation of the Trp86 indole ring. For example, the N110S mutation could allow the indole ring to flip, which would significantly open the substrate channel.
Site-saturation mutagenesis of residues 86 and nearby 287 and 288 yielded no improved mutants. Residues Leu287 and Val288 are located in a loop that connects the two protein sub-domains and might be involved in the closing of the active site during cofactor binding (Colonna-Cesari et al., 1986; Hayward and Kitao, 2006). The side chain of Trp86 lies in the active site cavity; it is notable that Trp86 is conserved in almost all MDR-ADHs. Studies of Trp86 in thermophilic SsADH suggest a critical role in governing substrate specificity (Pennacchio et al., 2009). We propose that the lack of improved variants in libraries targeting Trp86, Leu287, and Val288 is due to the essential roles played by these side chains.
3.3.3 Thermostability changes during evolution
We were also interested in the stabilities of the LlAdhA variants, as stability is considered a determinant of the capacity of a lineage to evolve (Vendruscolo et al., 1997; Kirschner and Gerhart, 1998; Bornberg-Bauer and Chan, 1999; O’Loughlin et al., 2006). We therefore measured thermostabilities in terms of T50, the temperature at which 50% of the initial activity is retained after 15 minutes incubation. Decreases in (thermo)stability often accompany mutations that increase/alter protein activity during directed evolution (Shoichet et al., 1995; Tokuriki et al., 2008). The T50 of variant LlAdhARE1, however, is approximately 7°C higher than that of wild-type LlAdhA (Table 2). One underlying cause for the increase in stability is likely to be the removal of solvent-exposed hydrophobic groups. The increased thermostability associated with I212T could result from a reduced propensity for protein aggregation; this mutation removes several surface-exposed hydrophobic atoms from an edge-exposed beta strand. Similarly, the L264V mutation removes an additional solvent-exposed hydrophobic methylene as well as increasing beta-sheet propensity (Muñoz and Serrano, 1994; Street and Mayo, 1999). Introduction of the N110S mutation in LlAdhA29C8 reduces T50 back to the wild-type level (Table 2). We hypothesize that the destabilizing effect of N110S reflects changes in orientation of the active site Trp86 aromatic ring (as discussed above). LlAdhA29C8 is as stable as wild-type LlAdhA, and decreased stability cannot account for our inability to obtain further activity improvements by directed evolution (Bloom et al., 2006; Tokuriki and Tawfik, 2009).
3.3.4 Substrate specificity changes during directed evolution
The mutations selected during directed evolution reshape the substrate-binding pocket to favor isobutyraldehyde binding and catalysis. We suspected that changes in substrate specificity might reflect a general increase in ADH activity on larger substrates, as such substrate ‘promiscuity’ is often observed during directed evolution for activity on novel substrates (Roodveldt and Tawfik, 2005; Fasan et al., 2008). To investigate this, we tested the ADHs in the evolutionary lineage for activity against a panel of substrates and inhibitors, including acetaldehyde as well as other aldehydes that are biologically produced and inhibit cell growth during the fermentative processes of alcohol-producing microorganisms (Klinke et al., 2004). A summary of the kinetic parameters of LlAdhA, LlAdhARE1, and LlAdhA29C8 for different aldehydes is listed in Table 3. Neither the variants nor wild-type LlAdhA exhibited detectable activity on 4-hydroxybenzaldehyde, syringaldehyde, vanillin, or coniferaldehyde under standard assay conditions. Wild-type LlAdhA exhibited high activity on acetaldehyde, cinnamaldehyde, and 2-furaldehyde, with catalytic efficiencies of 94 mM−1s−1, 39 mM−1s−1, and 57 mM−1s−1, respectively, but showed low activity for isobutyraldehyde and hydroxymethylfurfural (5-HMF) (2.8 mM−1s−1 and 0.88 mM−1s−1, respectively). Directed evolution from LlAdhA to LlAdhA29C8 significantly decreased KM values for 5-HMF and isobutyraldehyde: the 5-HMF KM decreased ~38-fold, from 22 mM to 0.57 mM, and the isobutyraldehyde KM decreased ~17-fold, from 12 mM to 0.68 mM. Although enlarging the substrate-binding pocket facilitates access to other bulky substrates similar in size to isobutyraldehyde, the increase in kcat is specific for isobutyraldehyde (Table 3).
4. Conclusion
Two new crystal structures of ADHs from the medium-chain dehydrogenase/reductase superfamily, LlAdhA and its evolved variant LlAdhARE1, were determined at high resolution. Using structure-guided saturation mutagenesis, we were able to further engineer LlAdhA to obtain an overall ~17-fold decrease in KM and a ~160-fold increase in catalytic efficiency toward isobutyraldehyde. Structural analysis and substrate profiling showed that the increased activity is accompanied by enlargement and increased hydrophobicity of the substrate-binding pocket as well as widening of the channel that provides access to the active site for bulky aldehyde substrates. Structure-guided saturation mutagenesis of key binding pocket residues and screening for improved function can identify beneficial mutations, which can be accumulated in iterative rounds, by recombination, or upon simultaneous mutagenesis at multiple positions. These approaches can rapidly optimize alcohol dehydrogenases, which are important catalysts in engineered pathways for producing fuels and chemicals.
Supplementary Material
Highlights.
The crystal structures of L. lactis alcohol dehydrogenase LlAdhA and its variant were determined at high resolution.
The structure helped guide engineering of a new variant with lower KM and higher catalytic efficiency.
Increases in active site size, hydrophobicity, and substrate access lead to enhanced activity on isobutyraldehyde.
Acknowledgments
This research was sponsored by the Army Research Laboratory and was accomplished under Cooperative Agreement Number W911NF-09-2-0022. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Laboratory or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Government purposes notwithstanding any copyright notation herein. X.L. received support from the China Scholarship Council (CSC), E.M.B. was supported by a Ruth L. Kirschstein National Research Service Award (1F32-GM087102) from the National Institutes of Health, and C.D.S. was supported by a research fellowship (KUS-F1-028-03) from King Abdullah University of Science and Technology (KAUST). The Molecular Observatory is supported by the Gordon and Betty Moore Foundation, the Beckman Institute and the Sanofi-Aventis Bioengineering Research Program at Caltech.
Abbreviations
- ADH
alcohol dehydrogenase
- NADH
nicotinamide adenine dinucleotide
- Ll
Lactococcus lactis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
X.L., S.B., C.D.S., P.M. and F.H.A. were responsible for study concept and design, analysis and interpretation of data, and preparation of manuscript. X.L., S.B., E.B., C.D.S., T.S. and J-H.X. were responsible for acquisition of data.
Conflict of interest
F.H.A. and P.M. are co-founders and shareholders of Gevo, Inc.
Appendix A. Supplementary materials
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal PK, Webb SP, Hammes-Schiffer S. Computational studies of the mechanism for proton and hydride transfer in liver alcohol dehydrogenase. J. Am. Chem. Soc. 2000;122:4803–4812. doi: 10.1021/ja011384b. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Li Z, Liao JC. Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2009;75:6306–6311. doi: 10.1128/AEM.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2010;85:651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulda DS, Bergman T. The role of zinc for alcohol dehydrogenase structure and function. Cell. Mol. Life Sci. 2008;65:3961–3970. doi: 10.1007/s00018-008-8593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez A, Cho KM, Liao JC. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 2011;90:1681–1690. doi: 10.1007/s00253-011-3173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Bakera PJ, Brittona KL, Fishera M, Esclapezb J, Pireb C, Boneteb MJ, Ferrerb J, Ricea DW. Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily. Proc. Natl. Acad. Sci. USA. 2009;106:779–784. doi: 10.1073/pnas.0807529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MMY, Arnold FH. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 2011;13:345–352. doi: 10.1016/j.ymben.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E, Chan HS. Modeling evolutionary landscapes: mutational stability, topology, and superfunnels in sequence space. Proc. Natl. Acad. Sci. USA. 1999;96:10689–10694. doi: 10.1073/pnas.96.19.10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann AF, Liao JC. Production of 2-methyl-1-butanol in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2008;81:89–98. doi: 10.1007/s00253-008-1631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli C, Liang ZX, Strickler M, Prehna G, Goldstein BM, Klinman JP, Bahnson BJ. Crystal structure and amide H/D exchange of binary complexes of alcohol dehydrogenase from Bacillus stearothermophilus: insight into thermostability and cofactor binding. Biochemistry. 2004;43:5266–5277. doi: 10.1021/bi049736p. [DOI] [PubMed] [Google Scholar]
- Colonna-Cesari F, Perahia D, Karplus M, Eklund H, Branden C, Tapia O. Interdomain motion in liver alcohol dehydrogenase. Structural and energetic analysis of the hinge-bending mode. J. Biol. Chem. 1986;261:15273–15280. [PubMed] [Google Scholar]
- Connor MR, Liao JC. Microbial production of advanced transportation fuels in non-natural hosts. Curr. Opin. Biotechol. 2009;20:307–315. doi: 10.1016/j.copbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Creaser EH, Murali C, Britt KA. Protein engineering of alcohol dehydrogenases; effects of amino acid changes at positions 93 and 48 of yeast ADH1. Protein Eng. 1990;3:523–526. doi: 10.1093/protein/3.6.523. [DOI] [PubMed] [Google Scholar]
- Eklund H, Nordstrom B, Zeppezauer E, Soderlund G, Ohlsson I, Boiwe T, Soderberg B, Tapia O, Branden C, Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2.4 Å resolution. J. Mol. Biol. 1976;102:27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Eklund H, Ramaswamy S. Three-dimensional structures of MDR alcohol dehydrogenases. Cell. Mol. Life Sci. 2008;65:3907–3917. doi: 10.1007/s00018-008-8589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Esposito L, Sica F, Raia CA, Giordano A, Rossi M, Mazzarella L, Zagari A. Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 Å resolution. J. Mol. Biol. 2002;318:463–477. doi: 10.1016/S0022-2836(02)00088-8. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH. Evolutionary history of a specialized P450 propane monooxygenase. J. Mol. Biol. 2008;383:1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S, Kitao A. Molecular dynamics simulations of NAD+-induced domain closure in horse liver alcohol dehydrogenase. Biophys. J. 2006;91:1823–1831. doi: 10.1529/biophysj.106.085910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashide W, Li Y, Yang Y, Liao JC. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl. Environ. Microbiol. 2011;77:2727–2733. doi: 10.1128/AEM.02454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J. Evolvability. Proc. Natl. Acad. Sci. USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke HB, Thomsen AB, Ahring BK. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 2004;66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- Larroy C, Pares X, Biosca JA. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADHVII), a member of the cinnamyl alcohol dehydrogenase family. Eur. J. Biochem. 2002;269:5738–5745. doi: 10.1046/j.1432-1033.2002.03296.x. [DOI] [PubMed] [Google Scholar]
- Leskovac V, Trivic S, Pericin D. The three zinc-containing alcohol dehydrogenases from baker’s yeast, Saccharomyces cerevisiae. FEMS Yeast Res. 2002;2:481–494. doi: 10.1111/j.1567-1364.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- Levin I, Meiri G, Peretz M, Burstein Y, Frolow F. The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol. Protein Sci. 2004;13:1547–1556. doi: 10.1110/ps.03531404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wen J, Jia X. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl. Microbiol. Biotechnol. 2011;91:577–589. doi: 10.1007/s00253-011-3280-9. [DOI] [PubMed] [Google Scholar]
- Muñoz V, Serrano L. Intrinsic secondary structure propensities of the amino acids, using statistical ϕ-ψ matrices: Comparison with experimental scales. Proteins. 1994;20:301–311. doi: 10.1002/prot.340200403. [DOI] [PubMed] [Google Scholar]
- Murali C, Creaser EH. Protein engineering of alcohol dehydrogenase-1. Effects of two amino acid changes in the active site of yeast ADH-1. Protein Eng. 1986;1:55–57. doi: 10.1093/protein/1.1.55. [DOI] [PubMed] [Google Scholar]
- O’Loughlin TL, Patrick WM, Matsumura I. Natural history as a predictor of protein evolvability. Protein Eng. Des. Sel. 2006;19:439–442. doi: 10.1093/protein/gzl029. [DOI] [PubMed] [Google Scholar]
- Pennacchio A, Esposito L, Zagari A, Rossi M, Raia CA. Role of tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase. Extremophiles. 2009;13:751–761. doi: 10.1007/s00792-009-0256-0. [DOI] [PubMed] [Google Scholar]
- Plapp BV. Conformational changes and catalysis by alcohol dehydrogenase. Arch. Biochem. Biophys. 2010;493:3–12. doi: 10.1016/j.abb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Eklund H, Plapp BV. Structures of horse liver alcohol dehydrogenase complexed with NAD+ and substituted benzyl alcohols. Biochemistry. 1994;33:5230–5237. doi: 10.1021/bi00183a028. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Scholze M, Plapp BV. Binding of formamides to liver alcohol dehydrogenase. Biochemistry. 1997;36:3522–3527. doi: 10.1021/bi962491z. [DOI] [PubMed] [Google Scholar]
- Reid MF, Fewson CA. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 1994;10:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- Roodveldt C, Tawfik DS. Shared promiscuous activities and evolutionary features in various members of the amidohydrolase superfamily. Biochemistry. 2005;44:12728–12736. doi: 10.1021/bi051021e. [DOI] [PubMed] [Google Scholar]
- Šali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savrasova EA, Kivero AD, Shakulov RS, Stoynova NV. Use of the valine biosynthetic pathway to convert glucose into isobutanol. J. Ind. Microbiol. Biotechnol. 2011;38:1287–1294. doi: 10.1007/s10295-010-0907-2. [DOI] [PubMed] [Google Scholar]
- Shen CR, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 2008;10:312–320. doi: 10.1016/j.ymben.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Cho KM, Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street AG, Mayo SL. Intrinsic β-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc. Natl. Acad. Sci. USA. 1999;96:9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. Plos Comput. Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Vendruscolo M, Maritan A, Banavar JR. Stability threshold as a selection principle for protein design. Phys. Rev. Lett. 1997;78:3967–3970. [Google Scholar]
- Weinhold EG, Benner SA. Engineering yeast alcohol dehydrogenase. Replacing Trp54 by Leu broadens substrate specificity. Protein Eng. 1995;8:457–461. doi: 10.1093/protein/8.5.457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.