Abstract
Cellulose, the most abundant biological macromolecule, is an extracellular, linear polymer of glucose molecules. It represents an essential component of plant cell walls but is also found in algae and bacteria. In bacteria, cellulose production frequently correlates with the formation of biofilms, a sessile, multicellular growth form. Cellulose synthesis and transport across the inner bacterial membrane is mediated by a complex of the multi-spanning catalytic BcsA subunit and the membrane-anchored, periplasmic BcsB protein. Here we present the crystal structure of a complex of BcsA and BcsB from Rhodobacter sphaeroides containing a translocating polysaccharide. The structure of the BcsA-B translocation intermediate reveals the architecture of the cellulose synthase, demonstrates how BcsA forms a cellulose-conducting channel, and suggests a model for the coupling of cellulose synthesis and translocation in which the nascent polysaccharide is extended by one glucose molecule at a time.
Introduction
Polysaccharides perform multiple vital functions in life, as reflected in their great variety in chemical composition and physicochemical properties. Extracellular polysaccharides, such as cellulose, chitin and hyaluronan, primarily perform structural functions and are synthesized inside the cell from nucleotide-activated donor sugars. 1–3
Cellulose is the most abundant biological polymer and consists of glucose molecules that are connected between their C1 and C4 carbons via acetal linkages.4 It is predominantly generated by vascular plants and a large number of algae, but also by some bacteria,5,6 protists7 and tunicates.8 Cellulose synthases (CESAs) are membrane-embedded glycosyltransferases, which utilize UDP-activated glucose (UDP-Glc) to processively elongate the nascent polysaccharide in a reaction that inverts the configuration at the anomeric carbon of the newly added sugar from α to β.6,9,10 Prokaryotic and eukaryotic CESAs share a similar predicted topology including eight transmembrane (TM) helices and at least one extended intracellular glycosyltransferase (GT) loop, Supplementary Fig. 1.
Bacteria, primarily Gram-negatives, produce and secrete cellulose via a protein complex consisting of at least three subunits (BcsA, -B and -C).11 The inner membrane protein BcsA is the catalytically active subunit and contains a conserved family two GT-domain between TM-helices 4 and 5 (TM4 and TM5).12 BcsB is a periplasmic protein that is anchored to the inner membrane via a single, C-terminal TM-helix. BcsA and BcsB are fused as a single polypeptide in some species, supporting the genetic observation that BcsB is essential for cellulose synthesis.13,14 BcsC, required for cellulose synthesis in vivo but not in vitro,14 is predicted to form an 18-stranded β-barrel in the outer membrane. Some bacteria contain additional, non-essential, periplasmic subunits of unknown function.11,15
While some bacteria constitutively produce cellulose,6 bacterial cellulose production is often concomitant with the formation of sessile, multi-cellular aggregates, termed biofilms.11,16 Biofilm bacteria are encased in a network of polysaccharides and proteinaceous fibers and are of particular concern to human health due to an increased tolerance to antimicrobial treatments.17 Biofilm formation is stimulated by the bacterial secondary messenger cyclic-di-GMP (cd-GMP), which bacteria sense via PilZ domains.18 Accordingly, BcsA contains a PilZ domain within its C-terminal, intracellular domain and its activity is strongly stimulated by cd-GMP.19,20
In all organisms, the cellulose polymer has to be exported across the plasma membrane. However, the mechanism by which a polymer consisting of thousands of glucose units can be translocated has been mysterious. In order to understand this process, we determined the crystal structure of the catalytically active BcsA-B complex in an intermediate state during cellulose synthesis and translocation at 3.25Å resolution (Supplementary Table).
Architecture of the BcsA-B Complex
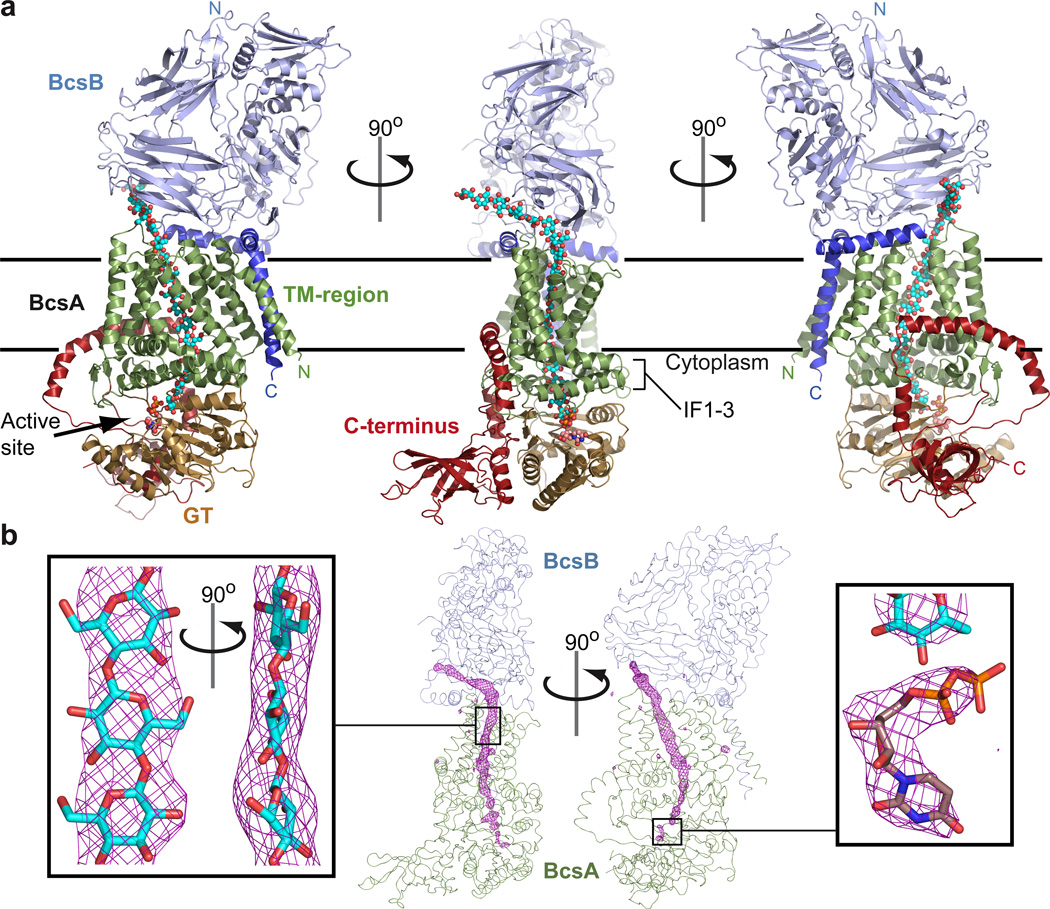
BcsA and BcsB form a 1:1 stoichiometric complex spanning approximately 150Å perpendicular and 55Å parallel to the membrane. The complex is divided into a cuboid-shaped membrane-spanning region sandwiched between large cytoplasmic and periplasmic domains, Fig 1a. The BcsA-B interface is approximately 4500Å2 including a cluster of negative and positive charges on BcsA and BcsB, respectively (Supplementary Fig. 2).
Figure 1. Architecture of the BcsA-B complex.
a, BcsA and BcsB form an elongated complex with large cytosolic and periplasmic domains. BcsA’s TM-helices are colored green, the glycosyltransferase (GT) domain sand and the C-terminal domain red. BcsB is shown in light and dark blue for its periplasmic and membrane associated regions, respectively. The N- and C-termini of both subunits are indicated and the translocating glucan and UDP are shown as cyan and violet spheres. Horizontal bars indicate the membrane boundaries. IF: Amphipathic interface helices of BcsA. b, Unbiased FoFc-difference Fourier electron density (pink mesh, contoured at 4.5σ) calculated before modeling the glucan and UDP molecules. The continuous density runs from the intracellular catalytic site to the periplasmic BcsA-B interface and accommodates 18 glucose molecules (cyan sticks). The UDP density was contoured at 3σ (right panel).
BcsA contains four N-terminal and four C-terminal TM-helices separated by a large intracellular loop (4/5-loop) that forms a GT-domain (aa 128 to 368), Fig. 1a and Supplementary Fig. 3. TM3-8 form a narrow channel for the translocating polysaccharide and BcsA’s intracellular C-terminus (aa 575 to 759) contains a 6-stranded β-barrel and a highly curved α-helical region that attaches the β-barrel to the GT-domain, Fig. 1a.
BcsB is a dome-shaped, β-strand rich, periplasmic protein. Its N-terminal region forms the tip of the dome, whereas the C-terminal TM-anchor interacts with BcsA, Fig. 1a. Two amphipathic helices further stabilize its interaction with BcsA and the periplasmic water-membrane interface, Fig. 1a.
Throughout model building, we noticed a continuous, strong positive difference Fourier electron density that could not be accounted for by BcsA or -B, Fig 1b. The ribbon-shaped electron density extended from BcsA’s GT-domain in a 45° angle toward the membrane, kinked near the putative water-lipid interface to run through the center of the TM-region, and bent sharply at the BcsA-B interface to emerge from the complex at approximately 145°, Fig. 1b. Past the BcsA-B interface, the electron density extended into the bulk solvent. The characteristic shape of the electron density, its proximity to conserved residues throughout its path (see below), its origin at the glycosyltransferase catalytic site, and extension past the BcsA-B complex strongly suggest that it represents a translocating glucan likely synthesized during or prior to crystallization.
The catalytic activity of the purified, detergent-solubilized BcsA-B complex was confirmed by incubating it with UDP-Glc and cd-GMP. The synthesized polymer, that could be completely degraded by β-1,4-glucanase (data not shown), was sedimented, purified, and analyzed by glycosyl linkage analysis (performed by the Complex Carbohydrate Research Center, University of Georgia), confirming that 94% of it consists of 4-linked glucose, Supplementary Fig. 4.
Although most likely reflecting an average of different registers of the translocating glucan, the density discerns the overall position of eighteen glucose molecules, Fig. 1b and Supplementary Fig. 5. According to the prevailing model that cellulose synthase elongates the non-reducing end of the growing polymer,21,22 the glucan was modeled with its non-reducing end (glucose-18) at the glycosyltransferase catalytic site. At the active site, we observed additional, weak electron density detached from the glucan. Based on its shape and coordination by residues that are predicted to be implicated in nucleotide binding and glycosyl transfer,23 it seemingly represents a weakly bound UDP molecule, Fig. 1b and Supplementary Fig. 5. Therefore, our structure likely represents a cellulose synthase-cellulose translocation intermediate captured after glycosyl transfer and before replacing UDP with UDP-Glc.
The Glycosyltransferase Domain
BcsA’s GT-domain contains the conserved signature “D,D,D,Q(Q/R)xRW” of three variably spaced aspartic acids and a pentapeptide consisting of a Gln that is often followed by a Gln or Arg, a variable residue and an Arg and Trp. This signature is shared with other processive β-glycosyltransferases, such as chitin and hyaluronan synthases.2,24 The GT-domain adopts a glycosyltransferase A fold (GT-A)10 consisting of a mixed, 7-stranded β-sheet that is surrounded by seven α-helices and attaches to the TM-region via three amphipathic interface helices (IF1-3), Fig. 1a, 3, and Supplementary Fig. 3. IF1 is formed by residues belonging to the 4/5-loop (aa 298 to 312) and packs against the C-termini of BcsA’s TM8 and -4 as well as the N-terminus of TM5. The second helix, IF2 (aa 373 to 395), directly precedes TM5 and includes the invariant “Q(Q/R)xRW” sequence of the signature. IF3, aa 470 to 498, follows TM6 and forms a crescent-shaped helix around the cytosolic entrance to the TM-channel (discussed below). It is connected to TM7 via a conserved loop (IF3/TM7-loop) that runs across the entrance to the GT substrate-binding site, Fig. 2b and Supplementary Fig. 3. The GT-domain protrudes from these IF-helices at approximately 45°, Fig. 1a. The inverting glycosyltransferase SpsA from Bacillus subtilis23 superimposes with BcsA’s GT-domain with 2.15Å root mean square deviation (r.m.s.d.) between C α atoms (Supplementary Fig. 6).
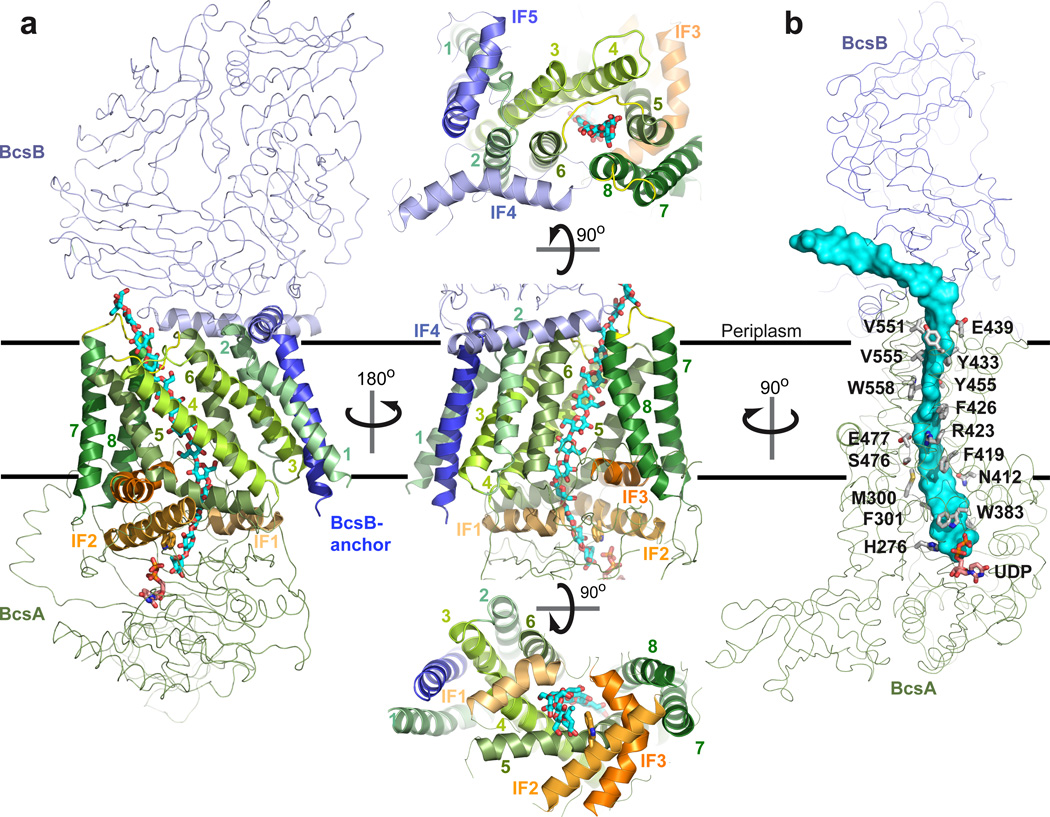
Figure 3. The membrane-integrated domain of BcsA-B.
a, The TM-region includes nine TM-helices and three cytoplasmic and two periplasmic interface helices (IF1-5). BcsA and BcsB are shown as green and blue ribbons. TM- and IF-helices are shown as cartoons and colored in shades of green and orange for BcsA’s TM1-8 and IF1-3, respectively, and shades of blue for BcsB’s IF4 and -5 and its C-terminal TM-anchor. TM3-8 of BcsA form a narrow channel. Its cytoplasmic entrance is formed by IF1-3 as well as the N-terminal half of TM5 and its periplasmic exit is between the periplasmic 5/6- and 7/8-loops of BcsA. Trp383 of the “Q(Q/R)xRW” motif is shown in sticks. b, Conserved residues of BcsA interact with the translocating glucan. BcsA and BcsB are shown as ribbons and side chains of BcsA contacting the glucan (cyan surface) are shown as sticks. Residues of BcsB contacting the polymer are not conserved and not shown.
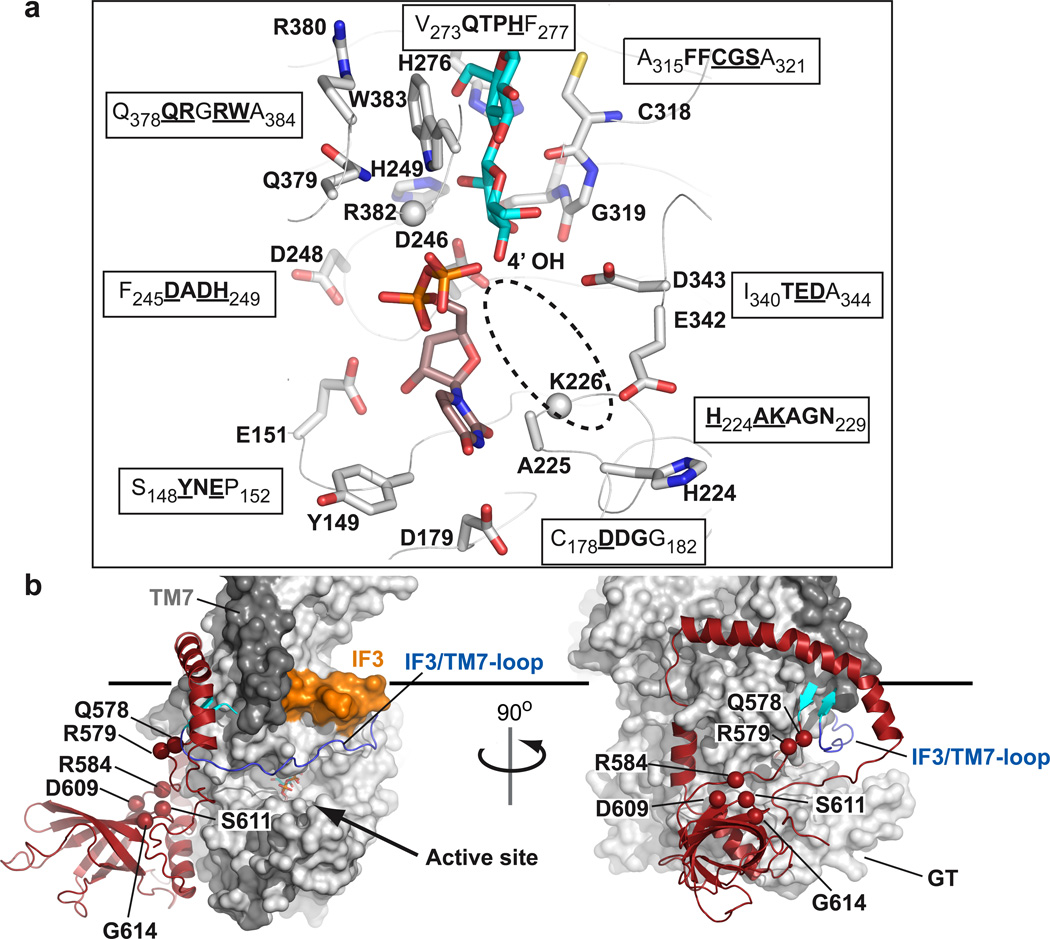
Figure 2. Organization of BcsA’s catalytic site and PilZ domain.
a, Conserved residues of BcsA coordinate UDP and the terminal disaccharide of the glucan. Side chains represented in sticks belong to the sequence motifs shown in single letter code. Conserved residues are highlighted in bold and the depicted residues are underlined. All side chains shown could be positioned unambiguously in the electron density map. No density was observed for the side chains of Lys226 and Arg382 (shown as spheres for their Cα atoms). The likely position of the donor Glc is indicated by a dashed ellipsoid. b, Surface representation of BcsA with its C-terminal domain shown as cartoon in red. The linker connecting TM8 with the β-barrel forms a 2-stranded β-sheet (shown in cyan) with the IF3/TM7-loop. TM7 is colored dark gray and IF3 orange. The Cα atoms of residues likely involved in cd-GMP binding are shown as spheres and are labeled. The horizontal bar indicates the cytoplasmic membrane boundary.
The structure of the cellulose synthase reveals the function of the signature “D,D,D,Q(Q/R)xRW” as well as other highly conserved sequence motifs. Consistent with non-processive glycosyltransferases,23,25,26 the first two aspartic acids of the signature, Asp179 and Asp246, coordinate UDP, Fig. 2a. The third “D” (Asp343), which is part of the invariant “TED” motif, likely represents the catalytic base due to its proximity to the non-reducing end of the glucan, Fig. 2a.
The “Q(Q/R)xRW” part of the signature, together with an equally conserved “FFCGS” sequence, forms a binding site for the terminal disaccharide of the glucan, the acceptor, Fig. 2a. The “Q(Q/R)xRW” sequence belonging to IF2 is a part of the cytoplasmic entry to the glucan channel, Fig. 3. The first “Q”, Gln379, hydrogen bonds to the guanidino group of the neighboring Arg380, which in turn forms a salt bridge to the invariant Glu297. Arg382, preceding the conserved Trp residue, most likely coordinates the diphosphate of UDP, however, the electron density for its side chain is weak, consistent with the apparent low occupancy of UDP. Trp383 forms van der Waals interactions with the penultimate glucose molecule at the acceptor site (glucose-17), and packs against the Gln379-Arg380 pair towards the distal site of the catalytic pocket, Fig. 2a. The FFCGS pentapeptide (aa 316–320) interacts with the opposite side of the terminal disaccharide, primarily via the backbone carbonyls of Cys318 and Gly319 with glucose-18.
The dominant negative thanatos mutation (Pro578Ser) in Arabidopsis thaliana (At) CESA327 maps to the invariant “QTPH” sequence in bacterial cellulose synthases, Supplementary Fig. 1. His276 of this motif is within hydrogen bond distance to glucose-17, Fig. 2a, and is thus likely also involved in positioning the non-reducing end with respect to the catalytic base. The motif is part of an extended β-strand and Pro275 (equivalent to Pro578 in At-CESA3) induces a “kink” in the β-strand that positions the side chain of His276 towards the glucan, which might be disrupted when Pro is replaced by another amino acid.
A cavity next to the UDP-binding pocket might accommodate the glucose donor in the UDP-Glc bound state. This pocket is flanked on one side by Ala225 and Lys226 of the invariant “AKAGN” motif and, on the other side, by Glu342 (of the conserved “TED” motif) and His224, Fig. 2a and Supplementary Fig. 7.
Activation by cyclic-di-GMP
BcsA’s activity is stimulated by cd-GMP.20 The cd-GMP-responsive PilZ-domain is localized within BcsA’s C-terminus18, right next to the GT-domain, Fig. 2b. BcsA’s C-terminus extends from the last TM-helix (TM8) via a short β-strand, which forms a 2-stranded β-sheet with the IF3/TM7-loop, before it folds into a 6-stranded β-barrel (aa 585–675). The β-barrel points away from the GT-domain at approximately 90°. Past the β-barrel, the polypeptide chain continues in an α-helical conformation on the surface of the GT-domain toward the water-lipid interface. At the interface, it forms an amphipathic helix that interacts with TM6, IF3 and TM8, Fig. 1a, before it curves away from the membrane towards the GT-domain and breaks at Glu743. After Glu743, the polypeptide continues in an extended conformation towards the β-barrel, past the substrate-binding pocket.
BcsA’s β-barrel aligns with an r.m.s.d. of 2.2Å between Cα atoms with the cd-GMP binding protein VCA004228 (Supplementary Fig. 8). The conserved residues Arg584, Asp609, Ser611 and Gly614 on the β-barrel surface as well as Gln578 and Arg579 of the TM8-β-barrel linker are likely implicated in cd-GMP binding28 (Fig. 2b and Supplementary Fig. 8). We hypothesize that cd-GMP binding to BcsA induces conformational changes that allow UDP-Glc to access the catalytic site. A likely candidate for displacement is the IF3/TM7-loop that runs across the entrance to the catalytic pocket and interacts with the TM8-β-barrel linker, Fig. 2b and Supplementary Fig. 3. Notably, replacing Thr942 with Ile in At-CESA3 confers resistance to the herbicide isoxaben.29 The corresponding, equally conserved residue in our structure is Thr506 in the IF3/TM7-loop (Supplementary Fig. 1), supporting the hypothesis that this loop is involved in regulating BcsA’s activity.
The Transmembrane Region and Polysaccharide Channel
The transmembrane region of the cellulose synthase includes eight TM-helices from BcsA (TM1-8) and one from BcsB, Fig. 3a. The TMs are framed by three cytoplasmic- and two periplasmic interface helices (IF) formed by BcsA and BcsB, respectively. In addition, the N-terminal four turns of TM5 also run parallel to the membrane before the helix bends towards the hydrocarbon core at Pro420, Fig. 3a. All TM-helices, except TM6 and -7 and BcsB’s TM-helix, tilt with respect to the membrane normal and surround a narrow channel approximately 8Å wide and 33Å long. Because the TM-helices are organized in pairs in one direction (helices 1/2, 3/6, 4/8, and 5/7 of BcsA), their overall arrangement resembles an elongated cuboid, and BcsB’s TM-anchor packs against one side of the cuboid, Fig. 3a. On the membrane periplasmic side, BcsB forms two amphipathic helices (IF4 and IF5) at the water-lipid interface. IF4 (aa 569 to 592) interacts with the periplasmic termini of BcsA’s TM2, -6 and -8 and IF5 (aa 679–693) directly precedes BcsB’s membrane anchor (aa 695–720) and contacts BcsA’s TM1 and the 1/2–loop, Fig. 3a.
The channel formed by BcsA’s TM3-8 accommodates 10 glucose units of the translocating glucan, Fig. 3. The glucan enters the channel through the cytoplasmic opening formed by IF1-3 and the N-terminal half of TM5, crosses the membrane parallel to TM5 and -6, and exits on the periplasmic side between BcsA’s 5/6- and 7/8-loops at the BcsA-B interface. Here, the glucan kinks to protrude from the complex sideways, Fig. 3b. TM6 and -8 are separated by about 9Å and 15Å (between Cα atoms) at the periplasmic and cytoplasmic sides of the membrane, respectively, and only the tips of their side chains maintain the barrier towards the lipids, Fig 3a. In the absence of a translocating glucan, TM-helices 7 and 8 might move towards the N-terminal TMs to close the channel.
All TM-helices of BcsA, with the exception of TM1 and -2, directly contact the glucan. The faces of the glucopyranose rings form van der Waals interactions with residues Met300 and Phe301 of IF1, Phe316 directly proceeding IF1, Trp383 of IF2, Phe419, Phe426, and Tyr433 of TM5, Phe441 in the 5/6-loop, and Val551, Val555, and Trp558 of TM8, Fig. 3b. The equatorial hydroxyl groups of the glucose units form hydrophilic contacts with Tyr80 of TM3, Asn118 of TM4, His276 of the GT-domain, Asn412 and Arg423 of TM5, Glu439 in the TM5/6 loop, Tyr455 of TM6, and Ser476 and Glu477 of IF3. We note that the quinoxyphen resistance mutation Ala903Val in At-CESA130 aligns with Tyr455 in TM6, which hydrogen bonds to the translocating glucan, Fig. 3b and Supplementary Fig. 1. Therefore, although the sequence alignment in this region is weak, it is likely that quinoxyphen affects the translocation rather than the synthesis of the glucan.
The Periplasmic Domain
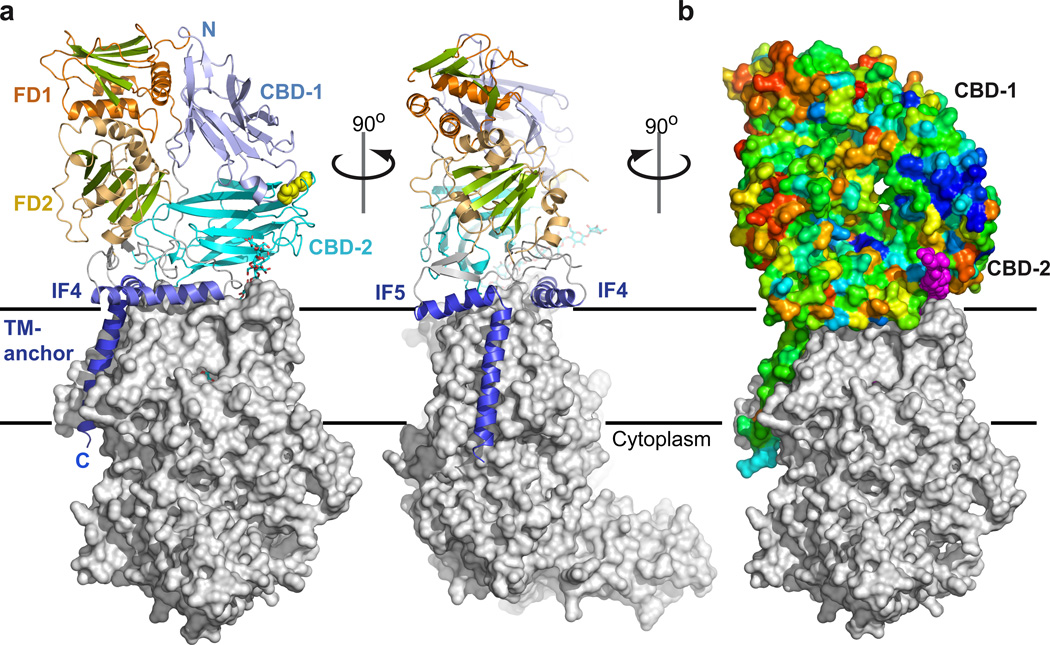
BcsB protrudes approximately 60Å into the periplasm with its N-terminal half, aa 54 to 307, forming the membrane distal part. BcsB consists primarily of β-strands that are organized into distinct domains, including two jellyroll and two flavodoxin-like folds, Fig. 4. The jellyrolls, aa 54–185 and 316–452, exhibit a striking structural similarity with carbohydrate binding modules (CBM, Supplementary Fig. 9)31 and are termed carbohydrate-binding domain (CBD) -1 and -2. Both CBDs contain eight antiparallel β-strands separated by loops 2–16 residues in length and adopt a similar fold, such that their Cα atoms superimpose with an r.m.s.d. of 2.5Å, Supplementary Fig. 9. The CBDs are positioned at a 45° angle relative to one another and interact via two extended loops, including a disulfide bond between the conserved Cys163 and Cys430, Fig 4a.
Figure 4. Organization of the periplasmic domain.
a, The periplasmic region of the cellulose synthase is primarily formed by BcsB. It consists of two periplasmic carbohydrate binding domains (CBD-1 and -2) that are connected to two α/β-domains (FD1 and FD2). The CBDs are covalently attached via a disulfide bond between the conserved Cys163 and Cys430 (yellow spheres). BcsB’s TM-anchor and IF-helices 4 and 5 are colored in dark blue. The N- and C-termini of BcsB are indicated. b, Surface representation of BcsB colored according to sequence conservation from red (variable) to deep blue (invariant). The tips of CBD-1 and CBD-2 form a patch of conserved, primarily hydrophobic residues above the periplasmic exit of the TM-channel. BcsA is shown as a grey surface and the glucan as purple spheres.
CBD-2 runs parallel to the plane of the membrane above the periplasmic exit of the TM-channel. It interacts with BcsA’s TM2 and -3, as well as with its 5/6-loop, thereby accounting for almost 30% of the complex interface. While BcsB exhibits significant sequence variability, we noticed a cluster of conserved residues including His159, Arg160, Ile161, Leu171 and Trp172 at the tip of CBD-1, close to its disulfide bond with CBD-2 and above the periplasmic exit of the glucan channel, Fig. 4b. A comparison of CBD-1 with the bacterial CBM family 35 (CBM35) in complex with a glucuronic acid disaccharide31 localizes disaccharide binding to the cluster of conserved residues in CBD-1, Supplementary Fig. 9, suggesting that this region interacts with the translocating glucan.
Both CBDs are linked to compact α/β-domains containing a central β-sheet five and six strands wide that is framed on either side by two α-helices, Fig. 4a. Although probably lacking functional similarity, both domains resemble a flavodoxin-fold32 (Supplementary Fig. 10) and are thus referred to as FD1 (aa 192–303) and FD2 (aa 457–529 together with 598–666). The repeating structural motif of BcsB therefore contains a CBD linked to a FD-domain.
Implications for Cellulose Synthesis and Translocation
The structure of the cellulose synthase-cellulose translocation intermediate provides unique insights into the mechanism of cellulose synthesis and translocation. Based on the large distance between the glycosyltransferase site and the putative water-membrane interface (approximately 25Å, Fig. 1) as well as BcsA’s activity in detergent, a reaction mechanism involving lipid-linked intermediates seems unlikely.5 The spatial restrictions in the substrate-binding pocket further suggest that the nascent glucan is extended by one33 and not by two34 glucose molecules at a time.
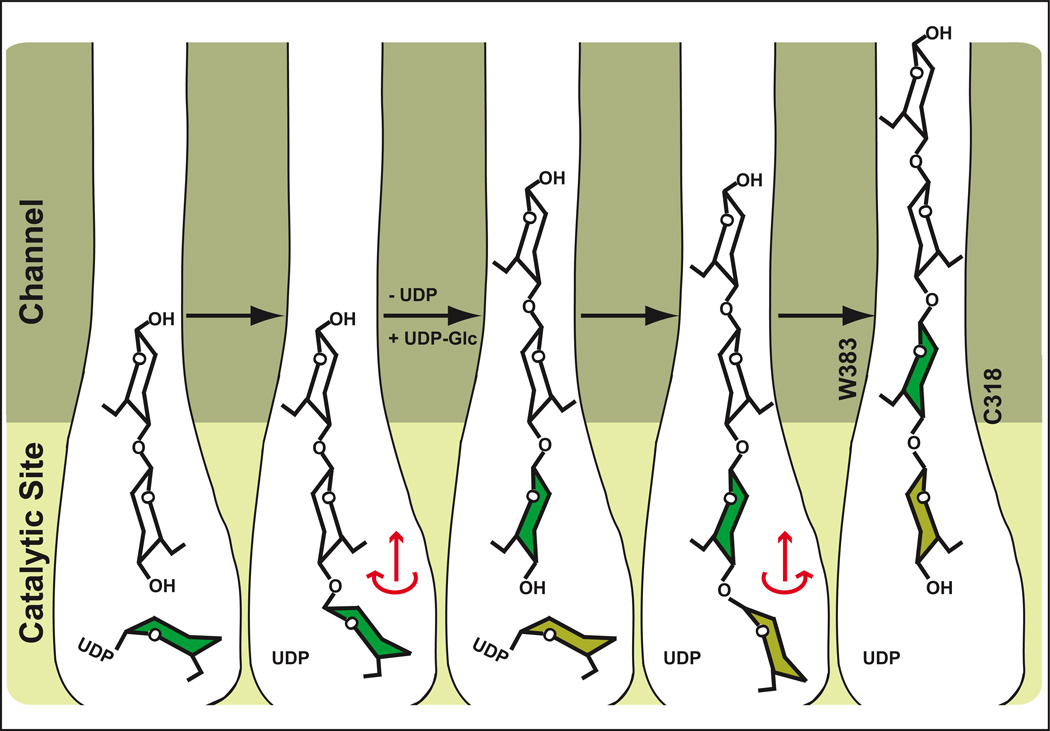
The distinct features of the glucan electron density (Fig. 1b) further imply that the polymer translocates one glucose molecule at a time. The dimensions of the TM-channel require that the glucopyranose rings have to be within the same plane in order to slide into the pore, Fig. 3b. Thus, we speculate that following glycosyl transfer the newly attached terminal glucose molecule will rotate around the acetal linkage35,36 to align with the glucan in the channel, Fig. 5. Steric interactions might dictate the rotation direction, leading to the β-1,4 glucan characteristic 180° rotation between individual glucose units and intramolecular hydrogen bonding between ring oxygens and 3’ hydroxyl groups of neighboring units.4,37 This relaxation might be sufficient to allow the polymer to slide into the channel, Fig. 5. Alternatively, the extended glucan might not translocate until UDP is replaced with UDP-Glc, necessitating translocation due to spatial restrictions at the active site. Past the channel, the induced kink of the glucan chain at the BcsA-B interface, the interaction with BcsB’s CBDs, or the aggregation with other glucans might further contribute to a unidirectional movement of the polymer.
Figure 5. Proposed model for cellulose synthesis and translocation.
Following glycosyl transfer, the newly added Glc could rotate around the acetal linkage into the plane of the polymer. The rotation direction would be determined by steric interactions and formation of the β-1,4 glucan characteristic intramolecular O3-H••O5 hydrogen bond. The glucan might translocate into the channel during this relaxation. This process would be repeated with a second UDP-Glc but the rotation direction after glycosyl transfer would be in the opposite direction due to steric constraints. Alternatively, the glucan might not translocate into the channel until UDP is replaced by UDP-Glc. Trp383 and Cys318 mark the entrance to the TM-channel (only shown in the right panel).
Cellulose, chitin and hyaluronan are likely synthesized by a conserved mechanism involving a membrane-embedded glycosyltransferase that couples polymer synthesis with its translocation across the cell membrane.3 This unique mechanism stands in contrast to the translocation of most other biological polymers where “polymer synthases” function independently of or alongside dedicated translocation machineries.38–41 Our structure now provides a basis for unraveling the details of this process.
Methods Summary
The BcsA and BcsB subunits of the Rhodobacter sphaeroides cellulose synthase complex were co-expressed and purified by metal affinity and gel filtration chromatography as described in detail in the Methods section. The BcsA-B complex was crystallized and the structure was determined after single anomalous dispersion phasing of a data set obtained from samarium(III)-chloride-soaked crystal. Additional experimental phases were obtained from selenomethionine-derivatized BcsA-B crystals. The quality of the electron density differed between peripheral and transmembrane regions of the complex. Therefore, modeling of the N-terminal region of BcsB, amino acids 54–177 and 586 to 675 of BcsA relied on the position of bulky side chains, Se-Met positions and backbone electron density. The model was built in Coot42 and includes all residues with the exception of the terminal residues 1–12 and 760–788 of BcsA and 19–53 and 721–725 of BcsB. In addition, a short loop of BcsB consisting of residues 532–543 was not visible in the electron density map.
Methods
Expression and Purification of BcsA-B
The BcsA gene was PCR amplified from Rhodobacter sphaeroides 2.4.1. genomic DNA (ATCC) and cloned using NcoI and HindIII restriction sites into an engineered pETDuet expression vector (Novagen) conferring a C-terminal dodeca-histidine tag. The BcsB gene was PCR amplified without its native, N-terminal signal sequence. The gene coding for amino acids 21 to 725 of BcsB was cloned into the pETDuet vector containing the BcsA gene, together with an N-terminal PelB signal sequence using NdeI and KpnI restriction sites. After signal sequence cleavage, this generates an additional Met-Gly dipeptide at the N-terminus of BcsB. Electro-competent Rosetta 2 cells were transformed with the pETDuetBcsA-12His-BcsB construct and plated on agar plates supplemented with 100µg/ml ampicillin and 25µg/ml chloramphenicol and grown overnight at 37°. The transformed cells were resuspended and used to inoculate 36L of ZYP-5052 auto-induction medium.43 The cells were grown shaking at 37°C for at least 4 hours after the optical density at 600nm reached 0.8. Subsequently, the cells were harvested by centrifugation, resuspended in RB1-buffer containing 20mM sodium phosphate buffer pH 7.2, 0.1M NaCl and 5mM cellobiose and then lysed in a microfluidizer. The crude membranes were harvested by centrifugation for 60min at 120,000xg in a Beckman Ti45 rotor and solubilized for 60min at 4°C in RB2-buffer containing 20mM sodium phosphate buffer pH 7.2, 0.3M NaCl, 5mM cellobiose, 5mM MgCl2, 40mM imidazole, 10% glycerol and 2% Triton X-100. The insoluble material was cleared by centrifugation for 30min at 120,000xg in a Beckman Ti45 rotor and the membrane extract was batch incubated with 10mL Ni-NTA agarose (Qiagen) for 45min at 4°C. The resin was packed in a gravity flow chromatography column, washed with 75mL WB1-buffer (RB2-buffer containing a total of 60mM imidazole and 5mM lauryldimethylamine-N-oxide, LDAO), 25mL WB2-buffer (RB2-buffer containing a total of 1M NaCl and 5mM LDAO) and the BcsA-B complex was eluted in 50mL EB-buffer containing 20mM Tris HCl pH 7.5, 0.1M NaCl, 5mM cellobiose, 5mM MgCl2, 250mM imidazole, 10% glycerol and 5mM LDAO. The eluted protein was concentrated and purified over an analytical S200 gel filtration column (GE Healthcare) equilibrated in EB-buffer containing 20mM MES pH 6.5 instead of Tris HCl and lacking MgCl2 and imidazole (GF-buffer). The eluted BcsA-B was concentrated to 50µM final concentration (using an extinction coefficient at 280nm of 161,925M−1 cm−1) in a 100kDa cut-off centrifugal filter (Millipore) and supplemented with 10mM ethylenediaminetetraacetic acid (EDTA) and 1mM uridine diphosphate-glucose (UDP-Glc) prior to crystallization.
Selenomethionine derivatized BcsA-B was prepared as described above with the exception that the cells were grown in the auto-inducing PASM-5052 minimal medium supplemented with 125µg/ml selenomethionine (Se-Met).43
The BcsA-B complex was crystallized in 30% PEG200, 0.1M MES pH 6.5 and 50mM NaCl at 4°C. Initial crystals were observed after approximately 7 days and reached their final size of about 50 by 50 by 100 microns within three weeks. The crystals were harvested and directly cryo-cooled in liquid nitrogen. Wild-type crystals were soaked with 20mM samarium(III)-chloride (Sm) or 1mM sodium ethylmercuric-thiosalicylate (EMTS) for 2 to 24 hours before harvesting.
Structure determination
X-ray diffraction data were collected at the Argonne National Laboratory beamlines SER- as well as GM/CA-CAT. The data was indexed and integrated in Mosflm44, analyzed in Pointless and scaled and reduced in space group P43212 in Scala from the CCP4 suite of programs.45 The Sm–derivatized crystals diffracted to 5Å resolution and were used for single wavelength anomalous dispersion (SAD) phasing in Shelx.46 The initial phases allowed localizing all transmembrane helices of BcsA and were further used to calculate an anomalous difference Fourier electron density map for the data set obtained from Se-Met derivatized BcsA-B. Initially, 10 Se-Met positions were identified and refined in Mlphare.45 The phases were combined with the native data set followed by density modification with phase extension to 3.25Å in DM47 using a user-generated solvent mask. Throughout the model building process, Se-Met anomalous difference Fourier electron density maps were calculated, additional Se-Met positions were refined as described above and new phases were calculated and improved by density modification in Parrot.47 The final experimental phases were calculated from thirty-six Se-Met-, four EMTS- and eight Sm positions and allowed tracing of most of the backbone of BcsA and the membrane proximal regions of BcsB. The model was built in Coot42 via an iterative process involving careful model building, combining of experimental and model phases, density modification and restraint refinement in Refmac-545 (including TLS parameters48 for the final model). The model includes a β-1,4 linked glucan containing eighteen glucose molecules and a UDP molecule, as well as two partially ordered LDAO molecules. The UDP molecule was refined with 50% occupancy due to weak electron density. The refined model has 91.8 % of its residues in the favored region of the Ramachandran diagram and includes 1.1% outliers, as determined by MolProbity. All figures were prepared in Pymol49 and the sequence conservation score for BcsB was calculated using CONSURF50.
In vitro Cellulose Synthesis
100µL of BcsA-B in GF-buffer was incubated with 5mM UDP-Glc, 20mM MgCl2 and 30µM cyclic-di-GMP at 37°C for 90min. The polymerization reaction was terminated with 40mM EDTA and the sample was centrifuged at 14,000xg at RT for 20min. The supernatant was discarded and the pellet was washed twice in 500µL deionized water, dried and subjected to linkage analysis by the Complex Carbohydrate Research Center, University of Georgia.
Supplementary Material
Acknowledgments
We are grateful to Garib Murshudov for advice on Refmac refinement and Ursula and Zygmunt Derewenda for discussions. We thank Lukas Tamm, Michael Wiener, Ann Walling, and Tom Rapoport for critical comments on the manuscript. X-ray diffraction data were collected at GM/CA- and Southeast Regional-Collaborative Access Team beamlines at the Advanced Photon Source (APS), Argonne National Laboratory. Use of the APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Contract No. DE-AC02-06CH11357 and W-31-109-Eng-38. GM/CA at APS has been funded in whole or in part with funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). The University of Georgia CCRC is supported by the Department of Energy funded Center for Plant and Microbial Complex Carbohydrates (DE-FG02-09ER-20097). JLWM is partially supported by a Peach Fellowship, University of Virginia. JZ is support by the NIH grant 1R01GM101001 and start-up funds from the University of Virginia School of Medicine.
Atomic coordinates and structure factors have been deposited at the Protein Data Bank under accession number 4HG6.
Footnotes
Contributions
J.Z. designed the experiments. J.L.W.M. and J.Z. expressed, purified, and crystallized the BcsA-B complex. J.L.W.M. and J.Z. analyzed the crystallographic data and built the model. J.S. performed in vitro cellulose synthesis assays. J.L.W.M. and J.Z. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 2.Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol B, Biochem Syst Environ Physiol. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard C, McNamara JT, Azumaya C, Patel MS, Zimmer J. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 2012;418:21–31. doi: 10.1016/j.jmb.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 4.Nishiyama Y, Langan P, Chanzy H. Crystal structure and hydrogen-bonding system in cellulose Ibeta from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc. 2002;124:9074–9082. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 5.Matthysse AG, Thomas DL, White AR. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1076–1081. doi: 10.1128/jb.177.4.1076-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bureau TE, Brown RM. In vitro synthesis of cellulose II from a cytoplasmic membrane fraction of Acetobacter xylinum. Proc Natl Acad Sci USA. 1987;84:6985–6989. doi: 10.1073/pnas.84.20.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimson MJ, Haigler CH, Blanton RL. Cellulose microfibrils, cell motility, and plasma membrane protein organization change in parallel during culmination in Dictyostelium discoideum. J Cell Sci. 1996;109:3079–3087. doi: 10.1242/jcs.109.13.3079. [DOI] [PubMed] [Google Scholar]
- 8.Kimura S, Itoh T. New cellulose synthesizing complexes (terminal complexes) involved in animal cellulose biosynthesis in the tunicate Metandrocarpa uedai. Protoplasma. 1996;194:151–163. [Google Scholar]
- 9.Aloni Y, Delmer DP, Benziman M. Achievement of high rates of in vitro synthesis of 1,4-beta-D-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci USA. 1982;79:6448–6452. doi: 10.1073/pnas.79.21.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 11.Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 12.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Standal R, Iversen TG, Coucheron DH, Fjaervik E, Blatny JM, Valla S. A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J Bacteriol. 1994;176:665–672. doi: 10.1128/jb.176.3.665-672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena IM, Kudlicka K, Okuda K, Brown RM. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol. 1994;176:5735–5752. doi: 10.1128/jb.176.18.5735-5752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, Munekata M, Tanaka I, Yao M. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci U S A. 2010;107:17957–17961. doi: 10.1073/pnas.1000601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahn CE, Selimi DA, Barak JD, Charkowski AO. The Dickeya dadantii biofilm matrix consists of cellulose nanofibres, and is an emergent property dependent upon the type III secretion system and the cellulose synthesis operon. Microbiology. 2011;157:2733–2744. doi: 10.1099/mic.0.051003-0. [DOI] [PubMed] [Google Scholar]
- 17.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 18.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 19.Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 20.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 21.Koyama M, Helbert W, Imai T, Sugiyama J, Henrissat B. Parallel-up structure evidences the molecular directionality during biosynthesis of bacterial cellulose. Proc Natl Acad Sci USA. 1997;94:9091–9095. doi: 10.1073/pnas.94.17.9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai-Kee-Him J, Chanzy H, Müller M, Putaux J-L, Imai T, Bulone V. In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J Biol Chem. 2002;277:36931–36939. doi: 10.1074/jbc.M203530200. [DOI] [PubMed] [Google Scholar]
- 23.Charnock SJ, Davies GJ. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38:6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 24.Weigel PH, Deangelis PL. Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem. 2007;282:36777–36781. doi: 10.1074/jbc.R700036200. [DOI] [PubMed] [Google Scholar]
- 25.Kozmon S, Tvaroska I. Catalytic mechanism of glycosyltransferases: hybrid quantum mechanical/molecular mechanical study of the inverting N-acetylglucosaminyltransferase I. J Am Chem Soc. 2006;128:16921–16927. doi: 10.1021/ja065944o. [DOI] [PubMed] [Google Scholar]
- 26.Kubota T, Shiba T, Sugioka S, Furukawa S, Sawaki H, Kato R, Wakatsuki S, Narimatsu H. Structural basis of carbohydrate transfer activity by human UDP-GaINAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10) J Mol Biol. 2006;359:708–727. doi: 10.1016/j.jmb.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 27.Daras G, Rigas S, Penning B, Milioni D, McCann MC, Carpita NC, Fasseas C, Hatzopoulos P. The thanatos mutation in Arabidopsis thaliana cellulose synthase 3 (AtCesA3) has a dominant-negative effect on cellulose synthesis and plant growth. New Phytologist. 184:114–126. doi: 10.1111/j.1469-8137.2009.02960.x. [DOI] [PubMed] [Google Scholar]
- 28.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies D-M, Estevez JM, Bonetta D, Urbanowicz BR, Ehrhardt DW, Somerville CR, Rose JKC, Hong M, Debolt S. Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA. 2012;109:4098–4103. doi: 10.1073/pnas.1200352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montanier C, van Bueren AL, Dumon C, Flint JE, Correia MA, Prates JA, Firbank SJ, Lewis RJ, Grondin GG, Ghinet MG, Gloster TM, Herve C, Knox JP, Talbot BG, Turkenburg JP, Kerovuo J, Brzezinski R, Fontes CMGA, Davies GJ, Boraston AB, Gilbert HJ. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc Natl Acad Sci USA. 2009;106:3065–3070. doi: 10.1073/pnas.0808972106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sancho J. Flavodoxins: sequence, folding, binding, function and beyond. Cell Mol Life Sci. 2006;63:855–864. doi: 10.1007/s00018-005-5514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delmer DP. Cellulose Biosynthesis: Exciting Times for A Difficult Field of Study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 34.Carpita NC. Update on Mechanisms of Plant Cell Wall Biosynthesis: How Plants Make Cellulose and Other (1→4)-β-D-Glycans. Plant physiology. 2011;155:171–184. doi: 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowd MK, French AD, Reilly PJ. Conformational analysis of the anomeric forms of sophorose, laminarabiose, and cellobiose using MM3. Carbohydr Res. 1992;233:15–34. doi: 10.1016/s0008-6215(00)90917-0. [DOI] [PubMed] [Google Scholar]
- 36.Momany FA, Schnupf U. DFTMD studies of β-cellobiose: conformational preference using implicit solvent. Carbohydr Res. 2011;346:619–630. doi: 10.1016/j.carres.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Brown M. Cellulose structure and biosynthesis: What is in store for the 21st century? J Pol Sci. 2004;42:487–495. [Google Scholar]
- 38.Rapoport TA. Protein transport across the endoplasmic reticulum membrane. FEBS J. 2008;275:4471–4478. doi: 10.1111/j.1742-4658.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- 39.Burton B, Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 41.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 44.Leslie AGW. The integration of macromolecular diffraction data. Acta Crystallogr D. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 45.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 47.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 49.PyMol. The PYMOL Molecular Graphics System. DeLano Scientific [Google Scholar]
- 50.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.