Abstract
Systemic lupus erythematosus (SLE) is a prototype autoimmune disease with a strong genetic involvement and ethnic differences. Susceptibility genes identified so far only explain a small portion of the genetic heritability of SLE, suggesting that many more loci are yet to be uncovered for this disease. In this study, we performed a meta-analysis of genome-wide association studies on SLE in Chinese Han populations and followed up the findings by replication in four additional Asian cohorts with a total of 5,365 cases and 10,054 corresponding controls. We identified genetic variants in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with the disease. These findings point to potential roles of cell-cycle regulation, autophagy, and DNA demethylation in SLE pathogenesis. For the region involving TET3 and that involving CDKN1B, multiple independent SNPs were identified, highlighting a phenomenon that might partially explain the missing heritability of complex diseases.
Introduction
Systemic lupus erythematosus (SLE [MIM 152700]) is an autoimmune disease with a strong genetic contribution.1 Ethnic differences have been observed in disease prevalence, clinical manifestations, and susceptibility genes for SLE, and Asians seem to have higher disease prevalence and a higher rate of lupus nephritis.2,3 Genome-wide association studies (GWASs) and related approaches have rapidly advanced our understanding of the genetics of this disease in that they have identified about 30 susceptibility loci reaching genome-wide significance for SLE.4–11 However, these loci still only explain a small portion of disease heritability. Studies with increased sample size, with meta-analysis of existing GWASs, and of non-European populations are possible approaches that could uncover some of the missing susceptibility variants for SLE and move us toward a better understanding of the disease and its intervention.
Previously, we conducted two GWASs on two different Chinese populations, one in Hong Kong and one in mainland China.4,8 We identified ETS1 (MIM 164720), WDFY4 (MIM 613316), IKZF1 (MIM 603023), RASGRP3 (MIM 609531), SLC15A4, TNIP1 (MIM 607714), and several other loci as associated with SLE in Asian populations. Interestingly, although many susceptibility genes are confirmed in multiple ethnic groups,12–16 a number of these genes were later on replicated only in Asian populations,17–20 raising the possibility of population specificity for at least some of the findings.
In this study, we performed a meta-analysis of the two existing GWASs on Chinese Han populations in Anhui and Hong Kong4,8 and followed up the findings on four additional Chinese and Thai cohorts with a total of 5,365 SLE cases and 10,054 controls matched geographically and ethnically to cases in each cohort. We identified TET3 (MIM 613555), CD80 (MIM 112203), ARID5B (MIM 608538), DRAM1 (MIM 610776), and CDKN1B (MIM 600778) as potential SLE susceptibility genes surpassing genome-wide significance (p < 5 × 10−8). We also found a number of loci with suggestive association with the disease, providing useful information for future replications. We also examined association signals in the meta-analysis data on the known loci reported from studies on European and other populations. For some of the regions, a clear population difference was indicated.
Subjects and Methods
The samples used in this study were collected from Hong Kong, Anhui province, China, and Bangkok, Thailand (Figure 1). The Hong Kong cases are SLE-affected individuals who were recruited from five hospitals in Hong Kong: Queen Mary Hospital, Tuen Mun Hospital, Queen Elizabeth Hospital, Pamela Youde Nethersole Eastern Hospital, and Princess Margaret Hospital. They are all of self-reported Chinese ethnicity and live in Hong Kong. Controls for the Hong Kong cohort are healthy blood donors from the Hong Kong Red Cross (for the Hong Kong replication panel) and individuals who participated in other GWASs conducted in the University of Hong Kong and who were genotyped on the same platform at the same time (GWAS stage). SLE-affected individuals in the Anhui replication panel 1 are all of self-reported Chinese ethnicity, live in Anhui province, central China, and have visited the Department of Rheumatology at Anhui Provincial Hospital and the First Affiliated Hospital of Anhui Medical University in Hefei, Anhui province, China.21 Controls were selected from a pool of healthy blood donors recruited from Hefei, Anhui province, to match the corresponding SLE cases in terms of age and sex. The samples for the Anhui GWAS and Anhui replication panel 2 were obtained from multiple hospitals in two geographic regions (central and southern China),8 and the corresponding controls were ethnically and geographically matched and clinically assessed to be without SLE, other autoimmune disorders, systemic disorders, or family history of autoimmune diseases. The Thai SLE cases are individuals attending King Chulalongkorn Memorial Hospital, a tertiary referral center in Bangkok, Thailand. Thai controls were recruited from unrelated voluntary healthy donors from the same ethnic background and geographic area as the Thai SLE cases. All cases involved in this study have medical records documenting fulfillment of the revised criteria of the American College of Rheumatology for diagnosis of SLE.22 The studies were approved by the respective institutional review boards at all the institutions listed above, and all subjects gave informed consent.
Figure 1.
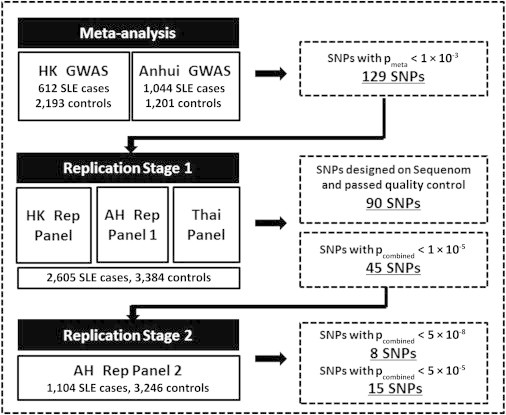
Flow Chart of the Experimental Process and SNP Selection Criteria
Genotyping
The two GWASs from Hong Kong and Anhui were conducted with the use of the Illumina 610-Quad Human Beadchip array, as previously reported.4,8 In the first stage of a two-stage replication process, 129 SNPs with a meta-analysis pmeta < 1 × 10−3 were selected for replication. A single SNP was selected for each 200 kb region unless there was clear evidence supporting independent association signals from the same locus. Of the 129 SNPs, 102 were successfully designed with the MassARRAY iPLEX Gold system (Sequenom) and were genotyped on samples from the Hong Kong replication panel, Anhui replication panel 1, and Thai replication panel. SNPs violating Hardy-Weinberg equilibrium and samples and SNPs failing to achieve a call rate of 90% or above were removed from further analysis, and 90 SNPs were analyzed on a total of 2,605 cases and 3,384 controls from replication stage 1. Forty-five SNPs reaching a pcombined value of 1 × 10−5 by joint analysis of results from replication 1 and the GWAS were selected for a second replication stage using samples from Anhui replication panel 2, which involved 1,104 cases and 3,246 controls; this second stage also used the MassARRAY iPLEX Gold system. A number of SNPs were also replicated by the TaqMan SNP genotyping method with the use of Assays-on-Demand probes and primers (Applied Biosystems). Genotyping accuracy was examined by a comparison of the results from the TaqMan SNP genotyping method and those from the Sequenom MassArray on a number of randomly chosen samples, and more than 99% concordance between the two platforms was achieved for these SNPs.
Association Analysis
After a quality-control process removing SNPs with a low call rate (<90%), low minor allele frequency (<1%), and violation of Hardy-Weinberg equilibrium (p ≤ 10−4), as well as removing samples with a low call rate (<90%) and hidden relationship detected with PLINK, 493,645 autosomal SNPs, 1,047 cases, and 1,205 controls from the Anhui cohort and 493,346 autosomal SNPs, 612 cases, and 2,193 controls from the Hong Kong cohort were analyzed. We also used PLINK to impute the GWAS data on 2,100,739 SNPs to deal with genotyping errors and missing data and to seek a complete overlap of SNPs between the two data sets; we used HapMap phase II CHB (Han Chinese in Beijing, China) and JPT (Japanese in Tokyo, Japan) samples as the reference panel and an INFO score of 0.8 as a cutoff.23 The samples from the two cohorts were assessed for population stratification with Eigenstrat24 (Figure S1, available online).
We used a weighted Z score method for the meta-analysis. To combine results across different cohorts, we oriented the alleles to the forward strand of the NCBI reference sequence of the human genome to avoid ambiguity associated with C/G and A/T SNPs. For each cohort, we converted association p values to Z scores by taking into account the direction of association relative to an arbitrary reference allele. We calculated a weighted sum of Z scores by weighing each Z score by the square root of the quotient of the effective sample size for each cohort over the sum of the effective sample size for all cohorts combined. We then converted the meta-analysis Z score to p values from a chi-square distribution. We used METAL25 to carry out the meta-analysis.
Joint association analysis, taking into account the effect of SNP differences between cohorts, was conducted with the Cochran-Mantel-Haenszel test, and homogeneity of effect size between different cohorts and different stages of the study was tested with Breslow-Day test (phet in Table 1); both tests were installed in PLINK.23 A test of independent contribution toward disease association for SNPs in a single locus was performed with logistic regression conditional on the effect of the other SNPs in the same region; this also took into account the potential differences among cohorts. Association of the newly identified SNPs was also tested conditionally on the effect of two most significant SNPs (rs9271366 and rs9275328) from the human-leukocyte-antigen region.
Table 1.
Results of Meta-analysis and Replication on the Identified Loci Associated with SLE
| SNP | Chr | Risk Allele | Gene |
p Value |
OR | phet | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| META | HK Rep | AH Rep 1 | Thai | AH Rep 2 | Combined | ||||||
| Loci with Genome-wide Significance | |||||||||||
| rs6705628 | 2p13 | C | TET3 | 2.6 × 10−8 | 0.04 | 1.0 × 10−4 | 0.11 | 1.3 × 10−5 | 6.9 × 10−17 | 0.75 | 0.43 |
| rs4852324 | 2p13 | T | TET3 | 4.8 × 10−9 | 0.33 | 1.4 × 10−5 | 0.12 | 4.4 × 10−3 | 5.7 × 10−14 | 0.79 | 0.05 |
| rs6804441 | 3q13 | A | CD80 | 1.5 × 10−5 | 0.02 | 0.02 | 0.06 | 6.1 × 10−12 | 2.5 × 10−16 | 0.79 | 0.01 |
| rs4948496 | 10q21 | C | ARID5B | 2.4 × 10−4 | 5.7 × 10−3 | 0.03 | 1.8 × 10−5 | 0.01 | 5.1 × 10−11 | 0.85 | 0.15 |
| rs12822507 | 12p13 | A | CREBL2 | 1.5 × 10−4 | 0.12 | 0.09 | 0.99 | 8.0 × 10−5 | 2.2 × 10−8 | 0.86 | 0.39 |
| rs10845606 | 12p13 | C | GPR19 | 1.1 × 10−5 | 0.16 | 1.8 × 10−7 | 0.05 | 5.8 × 10−7 | 3.8 × 10−17 | 0.79 | 0.18 |
| rs34330 | 12p13 | C | CDKN1B | 2.3 × 10−4 | 5.6 × 10−3 | 1.5 × 10−3 | 0.02 | 9.0 × 10−4 | 4.8 × 10−12 | 0.84 | 0.60 |
| rs4622329 | 12q23 | A | DRAM1 | 9.5 × 10−5 | 0.68 | 1.5 × 10−6 | 9.6 × 10−5 | 0.02 | 9.4 × 10−12 | 0.84 | 0.02 |
| Loci with Suggestive Significance | |||||||||||
| rs10911390 | 1q31 | T | APOBEC4 | 1.5 × 10−4 | 0.14 | 0.20 | 0.31 | 0.06 | 6.4 × 10−6 | 1.13 | 0.75 |
| rs1393820 | 2p13 | G | C1D | 3.9 × 10−5 | 0.26 | 0.13 | 0.09 | 0.63 | 9.1 × 10−5 | 0.82 | 0.12 |
| rs515983 | 2q22 | T | TMEM163 | 5.9 × 10−5 | 0.05 | 0.06 | 0.30 | 0.72 | 8.2 × 10−5 | 0.87 | 0.09 |
| rs11717455 | 3p22 | T | SCN10A | 2.4 × 10−5 | 0.02 | 0.86 | 0.23 | 0.07 | 6.9 × 10−6 | 0.71 | 0.10 |
| rs2023532 | 4q24 | C | UBE2D3 | 2.3 × 10−4 | 0.29 | 0.01 | 0.22 | 0.60 | 1.1 × 10−5 | 0.80 | 0.30 |
| rs2367894 | 5p13 | G | FYB | 1.3 × 10−4 | 0.50 | 0.01 | 0.65 | 0.60 | 4.0 × 10−5 | 0.84 | 0.20 |
| rs12529935 | 6q15 | C | BACH2 | 8.6 × 10−5 | 0.79 | 0.03 | 0.27 | 0.12 | 9.3 × 10−6 | 0.74 | 0.55 |
| rs6957263 | 7q22 | T | PUS7 | 1.3 × 10−4 | 0.01 | 0.11 | 0.17 | 0.09 | 2.0 × 10−5 | 0.80 | 0.03 |
| rs2252996 | 10q22 | A | SLC29A3 | 5.3 × 10−4 | 0.06 | 0.36 | 0.91 | 8.5 × 10−4 | 3.1 × 10−6 | 0.82 | 0.31 |
| rs4910907 | 11p15 | G | RRM1 | 1.6 × 10−6 | 0.44 | 0.16 | 0.22 | 0.32 | 6.5 × 10−5 | 1.07 | 0.02 |
| rs12917712 | 16p11 | T | SEZ6L2 | 8.6 × 10−6 | 0.03 | 0.03 | 0.68 | 0.84 | 7.1 × 10−6 | 1.07 | 0.19 |
| rs12461589 | 19q12 | C | PDCD5 | 2.0 × 10−5 | 0.10 | 0.67 | 0.05 | 0.25 | 7.8 × 10−5 | 0.83 | 0.04 |
| rs2303745 | 19p13.1 | A | DDA1 | 1.5 × 10−4 | 0.04 | 0.09 | 0.07 | 0.38 | 1.1 × 10−6 | 1.07 | 0.50 |
| rs13333054 | 16q24 | T | IRF8a | 6.9 × 10−4 | 3.0 × 10−4 | 0.12 | 0.12 | N/A | 4.6 × 10−5 | 1.06 | 0.02 |
| rs2785197 | 11p13 | C | CD44a | 7.7 × 10−4 | 0.75 | 3.4 × 10−3 | 0.98 | 2.6 × 10−4 | 1.9 × 10−7 | 0.81 | 0.18 |
The following abbreviations are used: chr, chromosome; META, results from the meta-analysis of the Hong Kong and Anhui GWASs; HK Rep, Hong Kong replication panel; AH Rep 1, Anhui replication panel 1; Thai, Thailand replication panel; AH Rep 2, Anhui replication panel 2; combined, p values from a combined analysis of the results from the two GWASs and the four replication panels with the use of the Cochran-Mantel-Haenszel test of disease association conditional on SNP-frequency differences among different panels; OR, odds ratio from the combined analysis; and phet, p values of the homogeneity test of ORs from the six different panels with the use of the Breslow-Day test.
Previously reported loci.
Allele-Specific Transcription Quantification
Sixty-seven healthy individuals heterozygous for rs34330 were chosen for a pyrosequencing assessment of the relative CDKN1B mRNA levels derived from the two alleles, “C” and “T.” In the meantime, DNA detection was used as a control for amplification efficiency. In brief, total RNA was extracted from peripheral-blood mononuclear cells (PBMCs) from each individual. RNA samples were treated with DNAase for the elimination of genomic DNA contamination before being reverse transcribed into cDNA with oligo-dT primer. Together with genomic DNA from the same individuals, cDNA was then amplified by PCR with transcript-specific primers. The cDNA and DNA PCR products were purified with the QIAquick PCR purification kit and were then subjected to allele-specific quantitative measuring by pyrosequencing. The sequencing primers were designed with Pyrosequencing Assay Design Software v.1.0. Reactions were performed on a Biotage PSQ96MA machine, and allele quantification was analyzed with PSQMA 2.1 software. The ratio of C:T allelic detection was performed for both DNA and cDNA. A paired Student’s t test was used for comparing the expression level from the “C” and the “T” alleles for this gene.
Analysis of Gene-Expression Data from Public Databases
To explore the expression profile of the candidate genes identified in this study in cells from SLE cases, we retrieved gene-expression data of CD4+ T lymphocytes (GSE4588 and GSE10325), B lymphocytes (GSE4588 and GSE10325), and neutrophils and/or myeloid cells (GSE27427 and GSE10325) from Gene Expression Omnibus data sets from the NCBI. Differences in gene expression between the cells from SLE-affected individuals and normal controls were analyzed by an unpaired t test. Log2 (fold change) values were also calculated from the signal-intensity ratio between cells from SLE-affected individuals and cells from controls.
Results
Meta-analysis
After a quality-control process, we performed a meta-analysis with METAL25 by using an effective sample size from the two GWASs (940 for Hong Kong and 1,110 for Anhui) as weight. Although the two Chinese populations don’t fully overlap with each other in population structure, as shown by the first two principal components analyzed with Eigenstrat24 (Figure S1), the difference was small, and a very modest genome inflation factor of 1.04 for the meta-analysis was observed. Meta-analysis results showed that variants from the major-histocompatibility-complex region, TNFSF4 (MIM 603594), STAT4 (MIM 600558), TNFAIP3 (MIM 191163), IRF5 (MIM 607218), BLK (MIM 191305), WDFY4, and ETS1 reached genome-wide significance, together with SNPs from a locus in 2p13 (Figure 2 ). After we removed SNPs in and near the known susceptibility loci (100 kb upstream and downstream of the susceptibility genes), deviation from the diagonal line at the tail remained in a quantile-quantile plot, indicating additional association signals not uncovered from the previous work (Figure S2).
Figure 2.
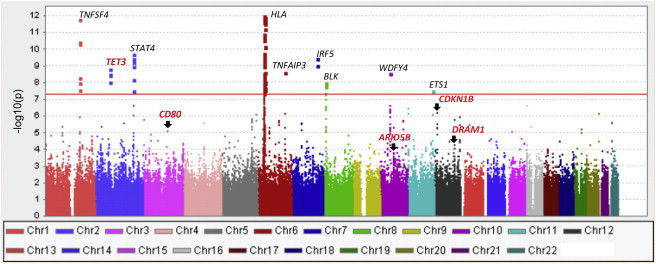
Manhattan Plot on the Meta-analysis Results of the Two SLE GWASs on Two Chinese Populations in Hong Kong and Anhui, China
Known susceptibility genes are labeled in black, and genes identified in this study are labeled in red. The y axis is the −log10(pmeta) for the autosomal SNPs from the two GWASs.
Association Signals from Meta-analysis on Previously Reported Loci
We examined meta-analysis results on previously reported loci to identify consistent cross-ethnicity associations or potential population differences in susceptibility genes. For easy observation, we grouped the results into three tables according to meta-analysis p values. Among these loci, 15 of them showed strong evidence of association (pmeta < 1 × 10−4, Table S1). Five additional loci showed suggestive evidence (1 × 10−4 < pmeta < 0.01, Table S2). However, meta-analysis showed little support for the association of several other genes reported in earlier studies on populations of European ancestry or Asians (these genes include IFIH1 [MIM 606951], PHRF1 [MIM 611780], PXK [MIM 611450], TYK2 [MIM 176941], IL10 [MIM 124092], JAZF1 [MIM 606246], PTPN22 [MIM 600716], and AFF1 [pmeta > 0.01, Table S3]) for the SNPs genotyped both within the reported gene and 100 kb upstream and downstream, suggesting potential differences between different ethnic groups, although this could also be caused by incomplete coverage of the regions or lack of study power. Interestingly, associations between SLE and WDFY4, ETS1, DDX6 (MIM 600326), SLC15A4, RASGRP3, HIP1 (MIM 601767), and ZNF689/PRR14 were only detected from studies on Asian populations (Table S4), highlighting the significance of conducting association studies on non-European populations. Further studies using imputation and fine-mapping in different ethnic groups are needed for clarifying the potential population differences in the regions that showed apparent discrepancies.
Variants Reaching Genome-wide Significance after Replication
A joint analysis of results from both GWAS and replication stages identified a number of variants associated with SLE, and some of them reached genome-wide significance.
In five loci, we identified eight SNPs reaching genome-wide significance (3.8 × 10−17 ≤ pcombined ≤ 2.2 × 10−8) (Table 1, Figures 3 and 4, and more details in Table S5). Three SNPs (rs12822507, rs10845606, and rs34330) were located at 12p13 (Figures 3A and 4A–4C), and all reached genome-wide significance and had minimum linkage disequilibrium (LD) with each other (r2 ≤ 0.17, Figure S3). Their independent contribution to disease susceptibility was also supported by logistic regression conditional on the effect of the other two SNPs (Table S6). The most significant signal was from rs10845606 (pcombined = 3.8 × 10−17, odds ratio [OR] = 0.79), a SNP located in an intron of G-protein-coupled receptor 19 (GPR19 [MIM 602927]), encoding an orphan receptor that is highly expressed in human embryonic stem cells26 but that otherwise has no known function. SNP rs12822507 (pcombined = 2.2 × 10−8, OR = 0.86) is located in an intron of cAMP-responsive element-binding protein-like 2 (CREBL2 [MIM 603476]), which encodes a protein that is well conserved during evolutionary courses and shares high sequence similarity to CREB1 on a bZip domain. CREBL2 might possess DNA-binding capability and is found to interact with CREB1.27 One of the three validated 12p13 SNPs, rs34330 (pcombined = 4.8 × 10−12, OR = 0.84), is located in the 5′ UTR of CDKN1B, and sequences near the SNP might contain promoter activity.28
Figure 3.
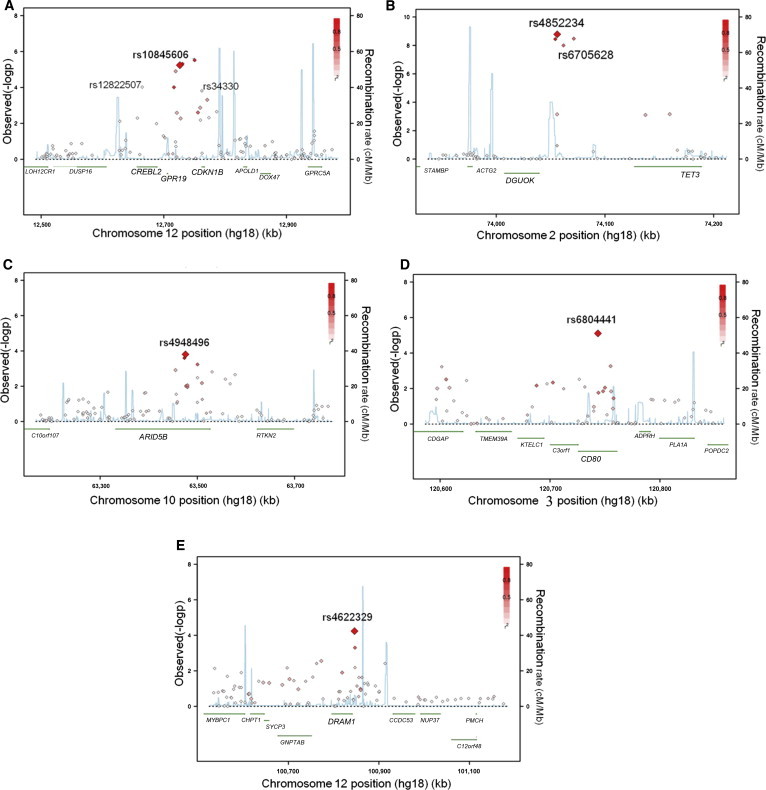
Association with SLE across the Five Regions
(A–E) Regional association plots show association from meta-analysis (−log10(pmeta)) versus chromosomal position (kb) for all the SNPs in a 500 kb region centered on the newly validated SNP. pmeta values are plotted for all SNPs, shaded white to red by the degree of LD (r2; see insets) with the validated SNP in the respective region (A, rs10845606; B, rs4852234; C, 4948496; D, rs6804441; and E, rs4622329). Local recombination rates (cM/Mb, blue lines) estimated from HapMap CHB and JPT samples are plotted against the secondary y axis, showing recombination hotspots across the region and haplotype blocks in between. Genes in the respective regions are labeled below the plots. Chromosomal regions are as follows: (A) 12p13, (B) 2p13, (C) 10q21, (D) 3q13, and (E) 12q23.
Figure 4.
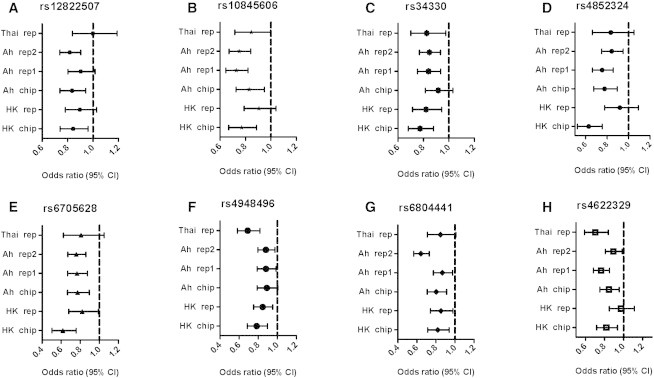
Forest Plot of Odds Ratios for the SNPs Surpassing Genome-wide Significance of 5 × 10−8 by Joint Analysis
The bars are 95% confidence intervals of odds ratios (ORs).
Two other variants found in this study, rs4852324 (pcombined = 5.7 × 10−14, OR = 0.79) and rs6705628 (pcombined = 6.9 × 10−17, OR = 0.75), are located in the upstream region of tet methylcytosine dioxygenase 3 (TET3) and downstream of deoxyguanosine kinase (DGUOK [MIM 601465]) at 2p13 (Figures 3B and 4D–4E). The two SNPs have moderate LD (r2 ≤ 0.56, Figure S6) with each other, and there was evidence of independent contribution of them toward disease association (Table S7). DGUOK encodes a protein that is responsible for phosphorylation of purine deoxyribonucleosides in the mitochondrial matrix and is highly expressed in immune cells. TET3 is most abundant in myeloid and monocytic hematopoietic cells and might play a role in malignant hematopoiesis.29 Alternatively, both SNPs are located within noncoding RNA DGUOK-AS1 (DGUOK antisense RNA 1) and might affect its expression or function.
SNP rs4948496 (pcombined = 5.1 × 10−11, OR = 0.85) is located in the fourth intron of ARID5B (AT-rich interactive domain 5B [MRF1-like]) at 10q21 (Figures 3C and 4F). More recently, rs10821944, a SNP 20 kb upstream of rs4948496, was found to be associated with rheumatoid arthritis (MIM 180300) in a Japanese population.30 The two SNPs have moderate LD with each other (r2 = 0.18), and rs10821944 showed suggestive association with SLE in our meta-analysis (pmeta = 0.0012), but there was no evidence of independent association of the effect of rs4948496 by logistic regression (p = 0.08 for rs10821944 and p = 0.0074 for rs4948496 when conditional on the effect of the other SNP). It remains to be determined whether the same or different biological mechanisms are involved in the two autoimmune diseases for this region. Another potential candidate gene in this region is rhotekin 2 (RTKN2), which encodes a lymphocytic Rho-GTPase effector that is exclusively expressed in lymphocytes and might play a role in lymphocyte development and function.31
SNP rs6804441 (pcombined = 2.5 × 10−16, OR = 0.79) is located in a 40 kb LD block harboring a single gene (CD80) at 3q13 (Figures 3D and 4G). CD80 encodes B7-1, a membrane receptor that is on antigen-presenting cells and that induces T cell proliferation and cytokine production. At 12q23, we identified SNP rs4622329 to be associated with SLE (pcombined = 9.4 × 10−12, OR = 0.84), and there are four genes (DRAM1, GNPTAB, SYCP3, and CHPT1) in this region in a LD block of 230 kb (Figures 3E and 4H). SNP rs4622329 is located about 4 kb downstream of DRAM1 (DNA-damage-regulated autophagy modulator 1), which encodes a lysosomal membrane protein that is required for the induction of autophagy and apoptosis and is a target of TP53.32
Suggestive Association Signals
In addition, we also identified 15 loci with suggestive association (1.9 × 10−7 ≤ pcombined ≤ 9.1 × 10−5) (Table 1) with SLE, providing opportunities for confirming more susceptibility genes in future studies through replications or meta-analyses. Among these 15 loci, 2 had been recently reported. SNP rs2785197 is located upstream of CD44 (MIM 107269) and has high LD (r2 = 0.98) with a SNP (rs2732552) identified in an earlier study.33 SNP rs13333054 is located downstream of IRF8 (MIM 601565), a susceptibility gene reported in a recent study on European populations. However, it has little LD (r2 = 0.10 based on Hong Kong GWAS, Figure S4) with rs2280381, the SNP that was reported in the previous study on SLE.10 We also interrogated rs2280381 in our GWAS stage, and it showed marginal association by meta-analysis (pmeta = 0.035). It is likely that the association seen for rs2280381 is dependent on rs13333054 in Asians.
Allelic Differential Expression
Of the genetic variants reaching genome-wide significance, rs34330 is located in the 5′ UTR of CDKN1B. Thus, allelic differential expression for this gene can be studied with pyrosequencing, which is a method that can detect small changes in expression between different alleles for each individual. We compared the expression levels of CDKN1B from the two alleles in PBMCs of healthy controls heterozygous on rs34330. A significant difference was detected with the risk allele (C allele), which correlated with a lower expression level of the gene (Figure S5). Differential expression of potential susceptibility genes was also examined from public databases (Table S8) and might hint at the potential functional roles of the identified susceptibility variants, although these experiments were designed for various purposes, and the results might be skewed by unknown confounding factors.
Discussion
Despite tremendous progress made in understanding genetic susceptibility of complex diseases such as SLE, the heritability explained by these variants is still very limited. For a disease like SLE, which has apparent differences in prevalence and manifestations among major ethnic groups, studies on non-European populations are an efficient approach to making discoveries without solely relying on increasing sample size. Findings in this study focus our attention on several pathways that were not uncovered in previous genetic studies and will undoubtedly broaden our understanding of the genetics of this disease and eventually help develop personalized care and risk prediction that are particularly suited to Asian populations. The independent contributors in two of the loci identified in this study also raised an important prospect that might help explain the missing heritability of complex diseases.
Among the potential candidate susceptibility genes identified in this study, CDKN1B is well studied, and its potential role in autoimmunity is supported by numerous functional studies. CDKN1B encodes p27kip1, a cyclin-dependent-kinase (CDK) inhibitor, which plays a critical role in the inhibition of cell-cycle progression, especially in T lymphocytes.34 p27kip1 is essential for the induction of tolerance, a process believed to be at the center of autoimmune diseases such as SLE, and upregulation of p27kip1 was found to correlate with induction of anergy in vitro and tolerance in vivo.35,36 p27kip1 is also involved in dendritic cell apoptosis,37 and the potential roles of the identified susceptibility genes in this study in SLE etiology are depicted in Figure 5.
Figure 5.
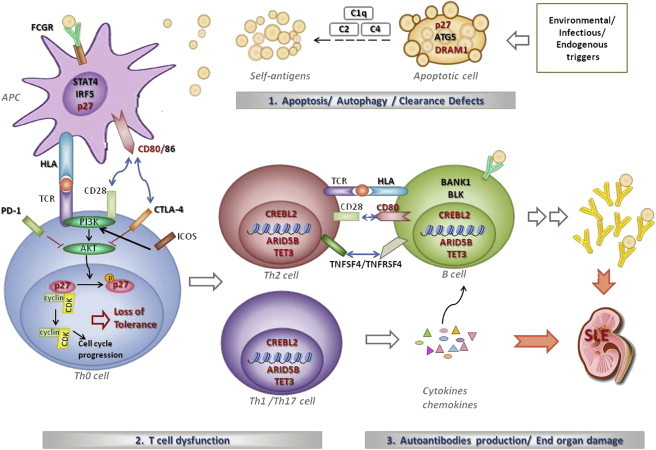
Potential Roles of the Identified Susceptibility Genes in SLE Etiology
Genes labeled in red are the ones identified in this study. Environmental triggers such as UV irradiation cause damage to cells. In affected individuals, genetic variants in DRAM1, ATG5, and CDKN1B (encoding p27kip1) might lead to defects in apoptosis or autophagy and thus increase exposure of nuclear autoantigens to the immune system. Interaction between CD28 and CD80 or CD86 plays a vital role in T cell activation. Upon T cell activation, p27kip1 is phosphorylated and the CDK-cyclin complex is released, allowing cell-cycle progression. Upregulation of p27kip1 is essential for induction of tolerance. p27kip1 also plays a role in dendritic cell apoptosis. Activated autoreactive Th2 cells interact with B cells and induce B cell differentiation and production of autoantibodies. Autoreactive Th1 and Th17 cells can exacerbate this process by secreting proinflammatory cytokines and chemokines, enhancing immune-complex deposition and end-organ damage. Susceptibility genes such as CREBL2, ARID5B, and TET3 are involved in DNA and histone modification and might regulate expression of genes in T and B lymphocytes involved in autoimmunity.
A significant difference was detected with the risk allele (C allele) of SNP rs34330, located in the 5′ UTR of CDKN1B; it was found to be correlated with a lower expression level of the gene (Figure S5). This is consistent with observed downregulation of CDKN1B in PBMCs of individuals with SLE (also see Table S8).38,39 It is noteworthy to point out that the differences are very small, and further functional studies are needed for confirming the potential role of this SNP because a larger effect might be restricted to specific cell types and/or responses to particular transactivation signals or regulators. Alternatively, this SNP might be an imperfect surrogate of another functional variant with a stronger effect.
For the 12p13 region (GPR19, CREBL2, and CDKN1B), as well as for the 2p13 region between TET3 and DGUOK, we identified multiple variants independently contributing to disease susceptibility. Given the design of this study, which focused on finding previously undescribed susceptibility regions, the exploration of independently contributing variants in a given region is by no means complete. We suspect that having multiple such variants in a single locus could be more common than expected, and this might partially explain the missing heritability of complex diseases.
Another potential susceptibility gene, TET3, is ubiquitously expressed, but it is most abundant in myeloid and monocytic hematopoietic cells and might play a role in malignant hematopoiesis.29 TET3 catalyzes conversion of 5-methylcytosine to 5-hydroxymethylcytosine and thus plays an important role in DNA demethylation,40 a process that might activate genes in T lymphocytes involved in autoreactivity. TET3 was found to be enriched specifically in the male pronucleus, and TET3-mediated DNA hydroxylation is also involved in epigenetic reprogramming of the zygotic paternal DNA after natural fertilization.41 Global hypomethylation has been observed in CD4+ T cells in individuals with SLE.42,43 It is possible that demythylation induced by TET3 might play a role in lymphocyte development and induction of tolerance.
SNP rs4948496 is located in an intron of ARID5B, which plays a role in the growth and differentiation of B-lymphocyte progenitors and possibly other lymphocytes. ARID5B is a component of a histone-demethylase complex that removes the repressive H3K9Me2 mark and activates transcription of target genes.44 Knockout of Arid5b in mice has been shown to cause transient immune abnormalities.45 Previously, polymorphisms in ARID5B were found to be associated with childhood acute lymphoblastic leukemia (ALL [MIM 613065]).46,47 SNPs found to be associated with ALL have little LD with rs4948496, indicating that different biological processes might be involved in the susceptibility of these two diseases.
CD80 encodes B7-1, a membrane receptor (on antigen-presenting cells) that induces T cell proliferation and cytokine production. Prolonged downregulation of CD80 on B cells was observed in SLE-affected individuals successfully treated with rituximab.48,49 CD80 was also found to be associated with celiac disease (MIM 212750), multiple sclerosis (MIM 126200), and primary biliary cirrhosis (MIM 109720), which all have a strong autoimmune component. Three SNPs found to be associated with these diseases were interrogated in the GWAS stage, but none of them showed association with SLE, again pointing to complexities in the biological processes conferring disease susceptibility.
Identification of genetic variants near DRAM1 might provide a missing link between genetic factors and environmental stimulators for this disease. UV exposure has long been suspected to be a trigger of lupus manifestations. It was found that induction of DNA damage by UV irradiation stabilizes TP53 and induces DRAM1 expression,50 a key molecule in autophagy. The role of autophagy in autoimmune diseases has long been suspected.51 Genetic variants near ATG5 (MIM 604261), another important player in autophagy, were previously found to be associated with SLE,7–9 and this association was also confirmed in our meta-analysis (Table S1).
Genes of interest in the regions with suggestive susceptibility include DDA1 (DET1- and DDB1-associated 1), also an important gene in the autophagy network,52 and FYN binding protein (FYB [MIM 602731]), which encodes a protein involved in LCP2-signaling cascades in T cells and in the expression of interleukin-2,53 a key cytokine that is often reduced in individuals with SLE. C1D nuclear receptor corepressor (C1D [MIM 606997]), which encodes a DNA-binding and apoptosis-inducing protein that is involved in DNA double-strand break repair inflicted by UV irradiation,54 and ubiquitin-conjugating enzyme E2D 3 (UBE2D3 [MIM 602963]), which is involved in the activation of IκB kinase and NF-κB,55 are also potential candidates for lupus susceptibility. And some of these genes were found to be differentially expressed in the immune cells of individuals with SLE (Table S8). Replication and meta-analysis focusing on these regions in the future could further improve our understanding of SLE susceptibility and the disease mechanisms.
The susceptibility genes identified in this study suggest a role for cell-cycle progression, apoptosis, autophagy, and DNA and histone modification in SLE, broadening our understanding of the genetics of this prototype autoimmune disease. However, fine mapping, targeted sequencing, and functional characterization are required for firmly establishing the role of these genes. Findings in this study also highlight potential differences in susceptibility genes between different ethnic groups, which has an impact not only on the understanding of the disease but also on risk prediction and stratified patient care based on ethnicity.
Acknowledgments
This study was supported by a generous donation from Shun Tak District Min Yuen Tong of Hong Kong. We thank Winnie Lau and her team for the collection of samples and clinical records for Hong Kong cases. This study also received support from the Research Grant Council of the Hong Kong Government (General Research Fund [GRF] grants HKU781709M, HKU 784611M, and HKU 770411M to W.Y. and Y.L.L.), the Ministry of Education of China (IRT-1046 to X.Z.), National Natural Science Foundation of China (NSFC) grants (81171505 and 30972727 to S.Y., 81071940 to Y.C., 31000528 to X.Z., 81102192 to H.F.P., and 30830089 to D.Q.Y.), the Preproject of State Key Basic Research Program 973 of China (grant 2011CB512103 to S.Y. and grant 2012CB722404 to L.D.S.), the National Research University Project of The Commission on Higher Education of Thailand (N.H. and Y.A.), the Ratchadaphiseksomphot Endowment Fund (HR1163A to N.H. and Y.A.), the National Research Council of Thailand (N.H. and Y.A.), and the Thailand Research Fund (K.S. and V.S.). We thank P.K. and A.C. for their contributions to Thai pediatric patient care and sample collections, as well as Sukkapan P. for retrieving the microarray expression data. Genotyping of samples serving as controls in this study was supported by Hong Kong GRF grants HKU 7623/08M and HKU 7747/07M. Y.Z. and J.Z. are supported by Edward the Sai Kim Hotung Pediatric Education and Research Fund.
Contributor Information
Xuejun Zhang, Email: ayzxj@vip.sina.com.
Yu Lung Lau, Email: lauylung@hkucc.hku.hk.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
BioGPS, http://biogps.org
Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
References
- 1.Arnett F.C., Shulman L.E. Studies in familial systemic lupus erythematosus. Medicine (Baltimore) 1976;55:313–322. doi: 10.1097/00005792-197607000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Mok C.C., Lau C.S. Lupus in Hong Kong Chinese. Lupus. 2003;12:717–722. doi: 10.1191/0961203303lu451xx. [DOI] [PubMed] [Google Scholar]
- 3.Yang W., Ng P., Zhao M., Hirankarn N., Lau C.S., Mok C.C., Chan T.M., Wong R.W., Lee K.W., Mok M.Y. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009;10:219–226. doi: 10.1038/gene.2009.1. [DOI] [PubMed] [Google Scholar]
- 4.Yang W., Shen N., Ye D.Q., Liu Q., Zhang Y., Qian X.X., Hirankarn N., Ying D., Pan H.F., Mok C.C., Asian Lupus Genetics Consortium Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remmers E.F., Plenge R.M., Lee A.T., Graham R.R., Hom G., Behrens T.W., de Bakker P.I., Le J.M., Lee H.S., Batliwalla F. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N. Engl. J. Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 7.Harley J.B., Alarcón-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D., Nath S.K., International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han J.W., Zheng H.F., Cui Y., Sun L.D., Ye D.Q., Hu Z., Xu J.H., Cai Z.M., Huang W., Zhao G.P. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 9.Gateva V., Sandling J.K., Hom G., Taylor K.E., Chung S.A., Sun X., Ortmann W., Kosoy R., Ferreira R.C., Nordmark G. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunninghame Graham D.S., Morris D.L., Bhangale T.R., Criswell L.A., Syvänen A.C., Rönnblom L., Behrens T.W., Graham R.R., Vyse T.J. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 2011;7:e1002341. doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nath S.K., Han S., Kim-Howard X., Kelly J.A., Viswanathan P., Gilkeson G.S., Chen W., Zhu C., McEver R.P., Kimberly R.P. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat. Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Yang W., Mok C.C., Chan T.M., Wong R.W., Mok M.Y., Lee K.W., Wong S.N., Leung A.M., Lee T.L. Two missense variants in UHRF1BP1 are independently associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2011;12:231–234. doi: 10.1038/gene.2010.66. [DOI] [PubMed] [Google Scholar]
- 13.Yang W., Zhao M., Hirankarn N., Lau C.S., Mok C.C., Chan T.M., Wong R.W., Lee K.W., Mok M.Y., Wong S.N. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum. Mol. Genet. 2009;18:2063–2070. doi: 10.1093/hmg/ddp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R., Yang W., Zhang J., Hirankarn N., Pan H.F., Mok C.C., Chan T.M., Wong R.W., Mok M.Y., Lee K.W. Association of CD247 with systemic lupus erythematosus in Asian populations. Lupus. 2012;21:75–83. doi: 10.1177/0961203311422724. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y.K., Yang W., Zhao M., Mok C.C., Chan T.M., Wong R.W., Lee K.W., Mok M.Y., Wong S.N., Ng I.O. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009;10:414–420. doi: 10.1038/gene.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen N., Fu Q., Deng Y., Qian X., Zhao J., Kaufman K.M., Wu Y.L., Yu C.Y., Tang Y., Chen J.Y. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA. 2010;107:15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Yang W., Hirankarn N., Ye D.Q., Zhang Y., Pan H.F., Mok C.C., Chan T.M., Wong R.W., Mok M.Y. ELF1 is associated with systemic lupus erythematosus in Asian populations. Hum. Mol. Genet. 2011;20:601–607. doi: 10.1093/hmg/ddq474. [DOI] [PubMed] [Google Scholar]
- 18.Zhong H., Li X.L., Li M., Hao L.X., Chen R.W., Xiang K., Qi X.B., Ma R.Z., Su B. Replicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Res. Ther. 2011;13:R186. doi: 10.1186/ar3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C.F., Liu Y.S., Cheng Y.L., Gao J.P., Pan T.M., Han J.W., Quan C., Sun L.D., Zheng H.F., Zuo X.B. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19:1181–1186. doi: 10.1177/0961203310367918. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H., Yang W., Qiu R., Li J., Xin Q., Wang X., Feng Y., Shan S., Liu Y., Gong Y., Liu Q. An intronic variant associated with systemic lupus erythematosus changes the binding affinity of Yinyang1 to downregulate WDFY4. Genes Immun. 2012;13:536–542. doi: 10.1038/gene.2012.33. [DOI] [PubMed] [Google Scholar]
- 21.Ni J.D., Yao X., Pan H.F., Li X.P., Xu J.H., Ye D.Q. Clinical and serological correlates of anti-Sm autoantibodies in Chinese patients with systemic lupus erythematosus: 1,584 cases. Rheumatol. Int. 2009;29:1323–1326. doi: 10.1007/s00296-009-0855-1. [DOI] [PubMed] [Google Scholar]
- 22.Hochberg M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Willer C.J., Li Y., Abecasis G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assou S., Le Carrour T., Tondeur S., Ström S., Gabelle A., Marty S., Nadal L., Pantesco V., Réme T., Hugnot J.P. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X., Zhang H., Yuan L., Jing H., Thacker P., Li D. CREBL2, interacting with CREB, induces adipogenesis in 3T3-L1 adipocytes. Biochem. J. 2011;439:27–38. doi: 10.1042/BJ20101475. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z., Dong Z., Han B., Yang Y., Liu Y., Zhang J.T. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 2005;33:3763–3771. doi: 10.1093/nar/gki680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langemeijer S.M., Aslanyan M.G., Jansen J.H. TET proteins in malignant hematopoiesis. Cell Cycle. 2009;8:4044–4048. doi: 10.4161/cc.8.24.10239. [DOI] [PubMed] [Google Scholar]
- 30.Okada Y., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Kawaguchi T., Stahl E.A., Kurreeman F.A., Nishida N. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 2012;44:511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 31.Collier F.M., Gregorio-King C.C., Gough T.J., Talbot C.D., Walder K., Kirkland M.A. Identification and characterization of a lymphocytic Rho-GTPase effector: rhotekin-2. Biochem. Biophys. Res. Commun. 2004;324:1360–1369. doi: 10.1016/j.bbrc.2004.09.205. [DOI] [PubMed] [Google Scholar]
- 32.Crighton D., Wilkinson S., O’Prey J., Syed N., Smith P., Harrison P.R., Gasco M., Garrone O., Crook T., Ryan K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 33.Lessard C.J., Adrianto I., Kelly J.A., Kaufman K.M., Grundahl K.M., Adler A., Williams A.H., Gallant C.J., Anaya J.M., Bae S.C., Marta E. Alarcón-Riquelme on behalf of the BIOLUPUS and GENLES Networks Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am. J. Hum. Genet. 2011;88:83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ophascharoensuk V., Fero M.L., Hughes J., Roberts J.M., Shankland S.J. The cyclin-dependent kinase inhibitor p27Kip1 safeguards against inflammatory injury. Nat. Med. 1998;4:575–580. doi: 10.1038/nm0598-575. [DOI] [PubMed] [Google Scholar]
- 35.Boussiotis V.A., Freeman G.J., Taylor P.A., Berezovskaya A., Grass I., Blazar B.R., Nadler L.M. p27kip1 functions as an anergy factor inhibiting interleukin 2 transcription and clonal expansion of alloreactive human and mouse helper T lymphocytes. Nat. Med. 2000;6:290–297. doi: 10.1038/73144. [DOI] [PubMed] [Google Scholar]
- 36.Li L., Iwamoto Y., Berezovskaya A., Boussiotis V.A. A pathway regulated by cell cycle inhibitor p27Kip1 and checkpoint inhibitor Smad3 is involved in the induction of T cell tolerance. Nat. Immunol. 2006;7:1157–1165. doi: 10.1038/ni1398. [DOI] [PubMed] [Google Scholar]
- 37.Lu C., Huang X., Zhang X., Roensch K., Cao Q., Nakayama K.I., Blazar B.R., Zeng Y., Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maas K., Chan S., Parker J., Slater A., Moore J., Olsen N., Aune T.M. Cutting edge: Molecular portrait of human autoimmune disease. J. Immunol. 2002;169:5–9. doi: 10.4049/jimmunol.169.1.5. [DOI] [PubMed] [Google Scholar]
- 39.Tang H., Tan G., Guo Q., Pang R., Zeng F. Abnormal activation of the Akt-GSK3beta signaling pathway in peripheral blood T cells from patients with systemic lupus erythematosus. Cell Cycle. 2009;8:2789–2793. doi: 10.4161/cc.8.17.9446. [DOI] [PubMed] [Google Scholar]
- 40.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 42.Patel D.R., Richardson B.C. Epigenetic mechanisms in lupus. Curr. Opin. Rheumatol. 2010;22:478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hedrich C.M., Tsokos G.C. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol. Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba A., Ohtake F., Okuno Y., Yokota K., Okada M., Imai Y., Ni M., Meyer C.A., Igarashi K., Kanno J. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 2011;13:668–675. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 45.Lahoud M.H., Ristevski S., Venter D.J., Jermiin L.S., Bertoncello I., Zavarsek S., Hasthorpe S., Drago J., de Kretser D., Hertzog P.J., Kola I. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 2001;11:1327–1334. doi: 10.1101/gr.168801. [DOI] [PubMed] [Google Scholar]
- 46.Papaemmanuil E., Hosking F.J., Vijayakrishnan J., Price A., Olver B., Sheridan E., Kinsey S.E., Lightfoot T., Roman E., Irving J.A. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treviño L.R., Yang W., French D., Hunger S.P., Carroll W.L., Devidas M., Willman C., Neale G., Downing J., Raimondi S.C. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokunaga M., Fujii K., Saito K., Nakayamada S., Tsujimura S., Nawata M., Tanaka Y. Down-regulation of CD40 and CD80 on B cells in patients with life-threatening systemic lupus erythematosus after successful treatment with rituximab. Rheumatology (Oxford) 2005;44:176–182. doi: 10.1093/rheumatology/keh443. [DOI] [PubMed] [Google Scholar]
- 49.Iwata S., Saito K., Tokunaga M., Yamaoka K., Nawata M., Yukawa S., Hanami K., Fukuyo S., Miyagawa I., Kubo S., Tanaka Y. Phenotypic changes of lymphocytes in patients with systemic lupus erythematosus who are in longterm remission after B cell depletion therapy with rituximab. J. Rheumatol. 2011;38:633–641. doi: 10.3899/jrheum.100729. [DOI] [PubMed] [Google Scholar]
- 50.Valbuena A., Castro-Obregón S., Lazo P.A. Downregulation of VRK1 by p53 in response to DNA damage is mediated by the autophagic pathway. PLoS ONE. 2011;6:e17320. doi: 10.1371/journal.pone.0017320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierdominici M., Vomero M., Barbati C., Colasanti T., Maselli A., Vacirca D., Giovannetti A., Malorni W., Ortona E. Role of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosus. FASEB J. 2012;26:1400–1412. doi: 10.1096/fj.11-194175. [DOI] [PubMed] [Google Scholar]
- 52.Behrends C., Sowa M.E., Gygi S.P., Harper J.W. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Wei B., Bismuth G., Rudd C.E. SLP-76-ADAP adaptor module regulates LFA-1 mediated costimulation and T cell motility. Proc. Natl. Acad. Sci. USA. 2009;106:12436–12441. doi: 10.1073/pnas.0900510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li G., Liu J., Abu-Asab M., Masabumi S., Maru Y. XPB induces C1D expression to counteract UV-induced apoptosis. Mol. Cancer Res. 2010;8:885–895. doi: 10.1158/1541-7786.MCR-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia Z.P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z.J. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


