Abstract
Compelling evidence is now available that gentamicin and geneticin (G418) can induce mammalian ribosome to suppress disease-causing nonsense mutations and partially restore the expression of functional proteins. However, toxicity and relative lack of efficacy at subtoxic doses limit the use of gentamicin for suppression therapy. Although G418 exhibits strongest activity, it is very cytotoxic even at low doses. We describe here the first systematic development of the novel aminoglycoside (S)-11 exhibiting similar in vitro and ex vivo activity to that of G418, while its cell toxicity is significantly lower than those of gentamicin and G418. Using a series of biochemical assays, we provide proof of principle that antibacterial activity and toxicity of aminoglycosides can be dissected from their suppression activity. The data further indicate that the increased specificity towards cytoplasmic ribosome correlates with the increased activity, and that the decreased specificity towards mitochondrial ribosome confers to the lowered cytotoxicity.
Keywords: Aminoglycosides, Genetic Diseases, Nonsense Mutations, Translational Readthrough, Cystic Fibrosis
INTRODUCTION
Nonsense mutations are in-frame premature termination codons (PTCs) that convert a sense codon of mRNA to UAA, UAG or UGA stop codon and lead to the production of truncated, nonfunctional proteins.1 PTCs are responsible for more than 1,800 inherited human diseases, including cystic fibrosis (CF), Duchenne muscular dystrophy (DMD), Usher syndrome (USH), Hurler syndrome (HS) and numerous types of cancer.2 For many of those diseases there is presently no effective treatment and the only treatment widely used is symptomatic.
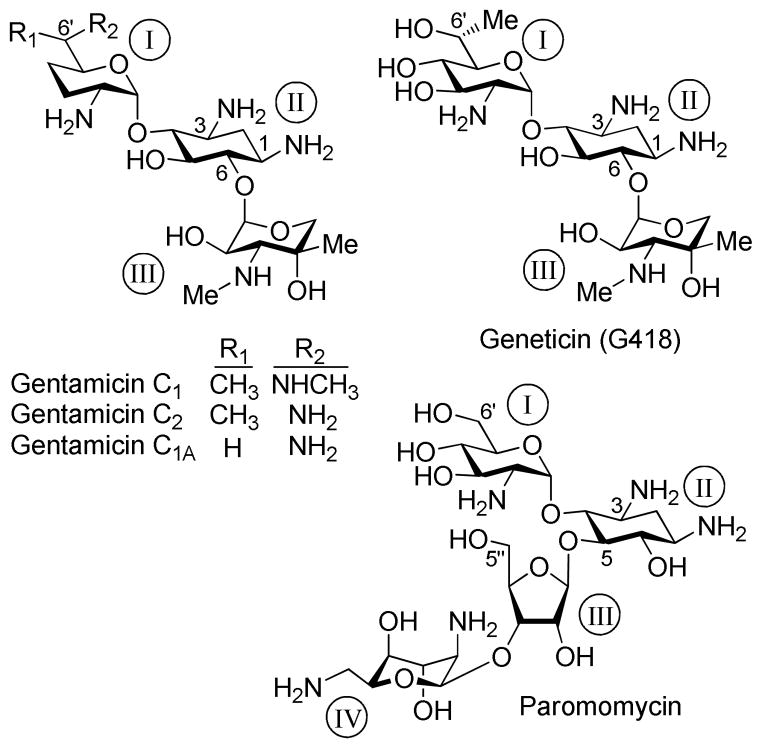
One potential approach to treatment considers the use of small molecule drugs to selectively suppress the normal proofreading function at PTCs, but not at normal termination codons. 1 This leads to a favorable competition of near-cognate aminoacyl-tRNAs with the release factor and to the insertion of a near-cognate amino acid at PTCs, allowing continued translation to full-length proteins. This approach, also called “translational readthrough” or “suppression therapy”, was first validated by using aminoglycoside (AG) antibiotics. Numerous in vitro and in vivo experiments including clinical trials have demonstrated the ability of selected structures of AGs (namely gentamicin, paromomycin and G418, Fig. 1) to induce readthrough at PTCs and partially restore functional proteins.1, 3, 4 However, severe side-effects of AGs, including high human toxicity, along with the reduced readthrough efficiency at subtoxic doses, have limited their clinical benefit for suppression therapy.5
Figure 1.
Chemical structures of standard aminoglycosides including gentamicin, paromomycin and G418 that were investigated in this study.
AGs selectively bind to the decoding A site on the 16S subunit of bacterial rRNA, and kill bacteria by disturbing the fidelity of the decoding process.6 Although prokaryotic selectivity is critical to their utility as antibiotics, they are not perfectly selective for the bacterial ribosome; they also bind to the eukaryotic A site7 resulting in PTC readthrough.3 Gentamicin and paromomycin are three orders of magnitude more selective to the prokaryotic versus the eukaryotic ribosome.8 For suppression therapy, this necessitates their use in high quantity, which in turn causes deleterious toxic side-effects, and hence, largely limits their utility.
A noteworthy exception is G418. In addition to its strong antibacterial activity, it also exhibits the highest readthrough activity among all AGs tested to date.9, 10 G418 is however very cytotoxic to mammalian cells. 11 It has not been clear whether its high cytotoxicity is due to higher specificity to the mammalian ribosome or to some other feature.12 Although the mitochondrial protein synthesis machinery is very similar to the prokaryotic machinery and the AGs-induced cytotoxicity may, at least in part, be connected to drug-mediated dysfunction of the mitochondrial ribosome,13 the direct impact of synthetic AGs to the human mitochondrial ribosome has not yet been studied into details.14, 15 The molecular mechanism of AGs-induced toxicity to mammalian cells is still mostly obscure. Clearly, a systematic search for new structures with improved PTC suppression activity and lower toxicity, along with a deeper understanding on structure-activity-toxicity relationship, are required to extrapolate the approach to the point where it can actually help patients suffering from genetic diseases caused by nonsense mutations.
Towards these ends, we hypothesized that by separating the structural elements of AGs that induce readthrough from those that affect toxicity, we might obtain potent AG-derivatives with improved readthrough activity and reduced toxicity. By systematically fine-tuning the structure-activity-toxicity relationship, we recently reported a series of structures, 1-8 (Fig. 2), exhibiting significantly reduced toxicity and higher PTC suppression activity than either gentamicin or paromomycin.16–19 Protein translation inhibition studies along with antibacterial tests indicated that 1-8 have increased selectivity in their action towards eukaryotic cells than towards prokaryotic cells in comparison to gentamicin and paromomycin. However, none of those leads were able to outreach G418’s peak suppression potency, nor its elevated eukaryotic specificity.
Figure 2.
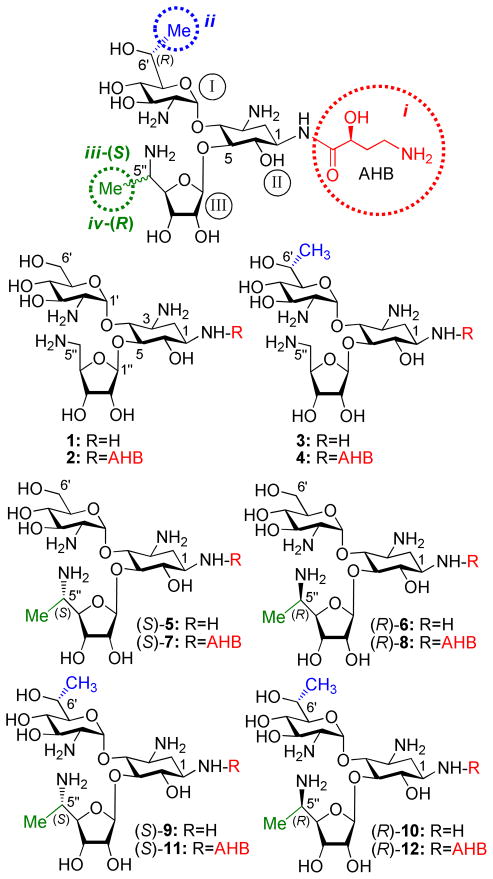
Structures of the synthetic aminoglycosides 1-12 that were investigated in this study. The identity of each pharmacophore and its attachment site are highlighted: (S)-4-amino-2- hydroxybutanoyl (AHB, i, red), (R)-6′-Me (ii, blue), (S)-5″-Me (iii, green) and (R)-5″-Me (iv, green).
The observed increased selectivity of action of 1-8 towards eukaryotic versus prokaryotic ribosome along with their reduced toxicity drew our attention and prompted us to ask several fundamental questions: what structural and mechanistic features are responsible for the observed selectivity increase and toxicity decrease of these synthetic derivatives? Can a general molecular principle for their structure-activity-toxicity relationship be devised? Using this principle, can a synthetic variant with similar or higher PTC suppression activity and lowered toxicity than those of G418 be generated?
To address these questions, here we report on the design, synthesis and evaluation of a new set of structures, 9-12 (Scheme 1) that perform better than G418 by the above criteria while exhibiting lower toxicity. Furthermore, by using a series of comparative readthrough, protein translation inhibition, antibacterial and toxicity assays between standard and the entire set of designer aminoglycosides 1-12, we demonstrate that the increased specificity towards human cytoplasmic ribosome correlates with the increased PTC suppression activity, and that the decreased specificity towards mitochondrial ribosome confers, at least in part, to the lowered cell toxicity. These observations provide proof of principle that AG-induced inhibition of cytoplasmic ribosome is a key determinant for PTC suppression activity, and that the inhibition of mitochondrial ribosome is key to AG-induced cell toxicity. These results are therefore beneficial for further research on the development of AG-based drug for the treatment of genetic diseases caused by nonsense mutations.
Scheme 1.
Reagents and conditions: (a) BF3·Et2O, CH2Cl2, 4 Ǻ MS, −20° C; (b) MeNH2-EtOH, rt; (c) Me3P, NaOH, THF, rt.
RESULTS AND DISCUSSION
Design hypothesis and synthesis
Our previously reported lead compounds 1-816–19 (Fig. 2) preserve the pseudo-trisaccharide scaffold 1 as a main recognition element for the rRNA. The extended side-chain structural elements of each structure, including the (S)-4-amino-2-hydroxybutanoyl (AHB) group at N-1 position (N1-AHB, pharmacophore-i), the (R)-6′-Me (pharmacophore-ii), and the (S)-5″-Me (pharmacophore-iii) or (R)-5″-Me (pharmacophore-iv), are four pharmacophores that we identified as key functionalities allowing an efficient discrimination between the eukaryotic and prokaryotic ribosomes, with preference towards the eukaryotic target.17–19 Whereas the first-generation lead, compound 1, exhibited significantly reduced cytotoxicity in comparison to gentamicin and paromomycin20, and promoted dose-dependent suppression of nonsense mutations of the PCDH15 gene, one of the underlying causes of type 1 Usher syndrome (USH1) 20; its suppression potency was significantly lower relative to that of gentamicin and paromomycin. Installation of each of the four pharmacophores on compound 1 generated the structures 2, 3, (S)-5 and (R)-6, respectively, exhibiting substantially greater suppression activity than those of gentamicin and the parent compound 1. In attempts to further improve the suppression efficiency and reduce the toxicity of the developed leads, we then tested the combination of two different pharmacophores on scaffold 1. We combined N1-AHB with (R)-6′-Me to generate 4,18 and the combination of N1-AHB with either (S)-5″-Me or (R)-5″-Me gave (S)-7 and (R)-8, respectively.19 Comparative PTC suppression and translation inhibition tests clearly indicated that the compounds with two pharmacophores, 4, (S)-7 and (R)-8, exhibit substantially increased readthrough activity and eukaryotic specificity than those of their parent structures with only one pharmacophore, 3, (S)-5 and (R)-6, respectively. The observed gradual increase in readthrough efficiency with an increased number of the pharmacophores on scaffold 1 also indicated that these pharmacophores can operate in an additive manner.
Encouraged from the observed data with compounds 1-8, especially from the additive impact of different pharmacophores on the readthrough activity, we sought to explore the effect of further combinations of these pharmacophores on scaffold 1. We anticipated that the remained four possible combinations of either two or three pharmacophores to generate a new set of structures 9-12 could allow us, for the first time, to surpass the peak readthrough activity of the natural antibiotic G418. To test this hypothesis, we combined (R)-6′-Me with either (S)-5″-Me or (R)-5′-Me to yield (S)-9 and (R)-10, respectively. Addition of N1-AHB to the latter two structures gave (S)-11 and (R)-12, and thus completed all twelve possible combinations of these pharmacophores.
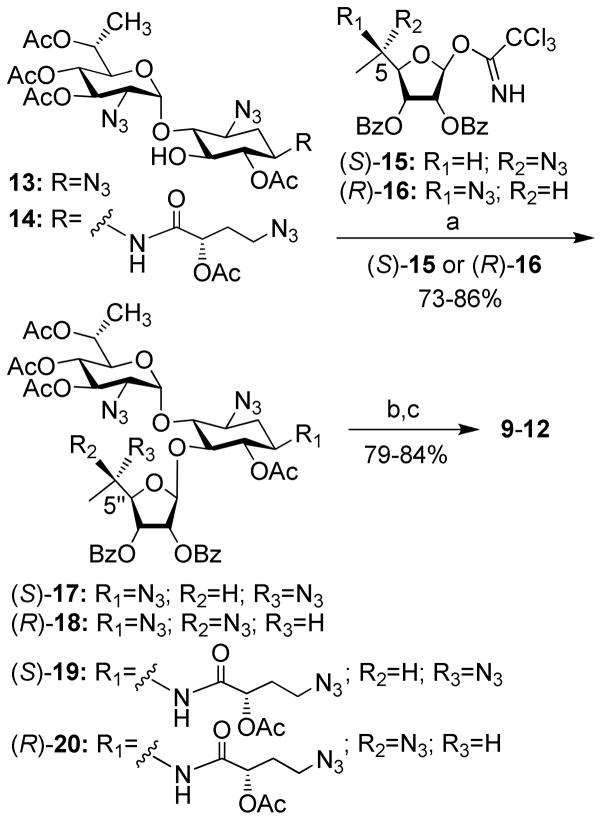
The synthesis of compounds 9-12 was accomplished from the corresponding selectively protected acceptors 13 and 14,18 and the donors (S)-15 and (R)-1619, previously reported by us, by using essentially the same chemical transformations as illustrated in Scheme 1. Lewis acid (BF3·Et2O) promoted glycosylation furnished the protected pseudo-trisaccharides 17-20 in 73–86% isolated yields, exclusively as β-anomers at the newly generated glycosidic linkage. Two sequential deprotection steps: treatment with methylamine to remove all the ester protection, and the Staudinger reaction (Me3P, THF/NaOH) to convert azides to corresponding amines, then afforded the target derivatives 9-12 in 79–84% isolated yields for two steps. The structures of all new compounds (9-12) were confirmed by a combination of various 1D and 2D NMR techniques, along with mass spectral analysis (see the Supporting Information).
Comparative in vitro PTC suppression tests to evaluate the additive effect impact of different pharmacophores in 9-12
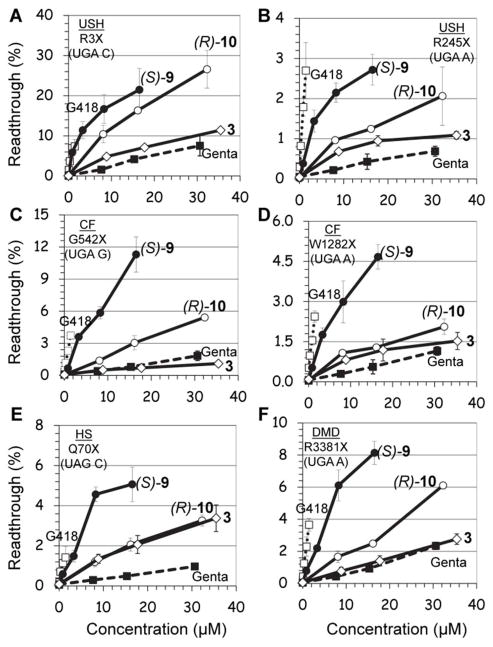
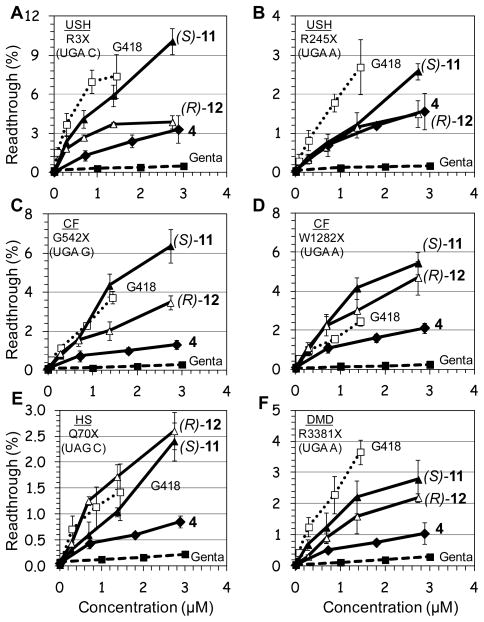
Previous studies have shown that the efficiency of aminoglycosidesinduced readthrough is highly dependent on: (i) the identity of stop codon (UGA > UAG > UAA), (ii) the identity of the first nucleotide immediately downstream from the stop codon (C > U > A ≥ G) and (iii) the local sequence context around the stop codon 9, 21. Therefore, for broader understanding on structure-activity relationship of the designed structures, we used a series of constructs containing different sequence contexts around premature stop codons derived from the PCDH15, CFTR, dystrophin and IDUA genes that underlie USH1, CF, DMD and HS, respectively. The prevalent nonsense mutations of these diseases that we chose were: R3X and R245X for USH120, G542X and W1282X for CF22, 23, R3381X for DMD24, and Q70X for HS25. Briefly, DNA fragments containing the nonsense mutation or the corresponding wild type codon, in their natural context, were cloned in frame between the Renilla and the Firefly luciferase genes of the p2luc vector as described previously by us17. The resulting six nonsense mutation-carrying plasmids, were transcribed and translated in the presence of varying concentrations of the tested compound, and the stop codon suppression efficiency was calculated as previously reported17, 26. Inhibition of translation was monitored as the ratio of Renilla luciferase activity with and without the presence of aminoglycosides26. In all the constructs tested, the highest concentrations of G418, 4, (S)-9, (S)-11 and (R)-12 resulted in approximately 50% reduction in overall translation, whereas the effects of 3, (R)-10, and gentamicin on translation were significantly milder resulting in approximately 20–40% reduction in overall translation. Initially, we tested the effect of combination of two pharmacophores [two chiral methyl groups, (R)-6′-Me with either (S)-5″-Me or (R)-5″-Me] by evaluating the comparative readthrough potential of (S)-9 and (R)-10 versus that of compound 3, which consists of only one pharmacophore [(R)-6′-Me, pharmacophoreii), and the observed data are shown in Fig. 3.
Figure 3. In vitro.
stop codon suppression levels induced by compounds (S)-9 (●), (R)-10 (○), 3 (◇), G418 (…□…) and gentamicin (--■--) in a series of nonsense mutation context constructs representing various genetic diseases (shown in parenthesis): (A) R3X (USH1), (B) R245X (USH1), (C) G542X (CF), (D) W1282X (CF), (E) Q70X (HS), and (F) R3381X (DMD). Readthrough activity was measured as previously described by us17. The results are the average of at least three independent experiments.
As seen from the data in Fig. 3, in all the mutations tested, installation of (S)-5″-methyl group (compound (S)-9) on compound 3 dramatically increases its in vitro readthrough activity, whereas that of the (R)-5″-methyl group (compound (R)-10) is comparatively small. In addition, in all mutations tested, the readthrough activity of (S)-9 was significantly greater than that of the clinical drug gentamicin. The observed stereochemical preference of (S)-5″-Me group in compound (S)-9 over that of (R)-5″-Me group in (R)-10 on readthrough activity is in accordance to our earlier observations with the compounds (S)-5 and (R)-6. 19 In the latter study, very similar preference of (S)-5 over that of (R)-6 were observed when the activities of these two were compared to that of their parent compound 1.
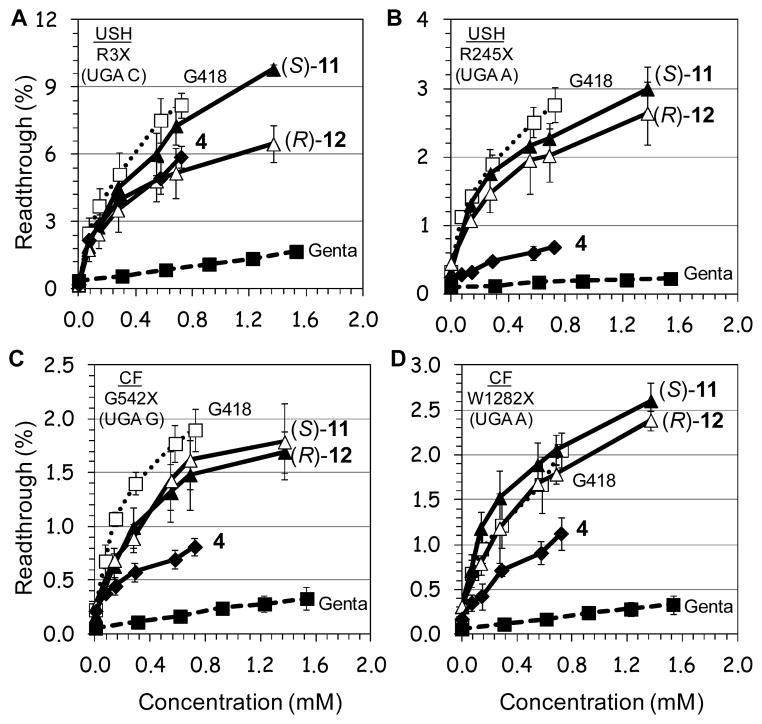
Even though we were encouraged from the data observed with the compound (S)-9, its readthrough potency was still significantly lower from that of G418 (note that G418 is strongly active already at very concentrations, Fig. 3). Therefore, we decided to explore the possibility of further potency enhancement. In particular, we were intrigued if the same potency enhancement, due to addition of the (S)-5″-methyl group, would apply in compound 4 as well. It is noteworthy that compound 4 contains two pharmacophores, (R)-6′-Me and N1-AHB, and was considered the best readthrough inducers among our entire designer AGs until the current work. To evaluate the impact of the stereochemistry at C5″-position, we constructed and tested both C5″-diastereomers (S)-11 and (R)-12. Comparative in vitro suppression tests of the pseudo-trisaccharides 4, (S)-11, (R)-12, gentamicin and G418 were performed under the same experimental conditions as above and the observed data are shown in Fig. 4.
Figure 4. In vitro.
stop codon suppression levels induced by compounds (S)-11 (▲), (R)-12 (△), 4 (◆), G418 (…□…) and gentamicin (--■--) in a series of nonsense mutation context constructs representing various genetic diseases (shown in parenthesis): (A) R3X (USH1), (B) R245X (USH1), (C) G542X (CF), (D) W1282X (CF), (E) Q70X (HS), and (F) R3381X (DMD). Readthrough activity was measured as previously described by us17. The results are the average of at least three independent experiments.
From the data in Fig. 4, it can be seen that all the tested compounds induced readthrough in a dosedependent manner. However, the efficacy of readthrough is substantially different between different constructs and compounds tested, with no obvious dependence of readthrough effectiveness on the introduced type of modification on aminoglycoside. The UGA C tetracodon sequence (R3X) showed the best translational readthrough than UGA A and UGA G, with the UAG C tetracodon least efficient, in agreement with earlier observations 9, 17, 21. Nevertheless, in all mutations tested (except Q70X, Fig. 4E) compound (S)-11 induced the highest level of readthrough among the synthetic derivatives, and the observed efficacy followed the order (S)-11 ≥ (R)-12 > 4. Thus, the impact of three pharmacophores in (S)-11 and (R)-12 is significantly greater than that of two pharmacophores (pharmacophores i + ii) in compound 4. Both (S)-11 and (R)-12 were drastically more active than gentamicin. The most impressive observation, however, was that of six different mutations tested, in three of them, including W1282X, G542X and Q70X, (S)-11 and/or (R)-12 exhibited similar or greater activity than that of G418. To our knowledge, this is the first demonstration that a synthetic derivative exhibits similar or greater stop codon readthrough activity than that of G418.
Comparative PTC suppression tests in mammalian cell line
To further evaluate the readthrough potential of compounds (S)-11 and (R)-12, their activity was assayed in cultured mammalian cells using four different dual luciferase reporter plasmids harboring the PCDH15-R3X and PCDH15-R245X nonsense mutation of USH1, and the CFTR-WG542X and CFTR-W1282X nonsense mutation of CF. These reporter constructs were the same as we used in the in vitro study, and have distinct advantage to control for differences in mRNA levels between normal and nonsensecontaining sequences over those of single reporter or direct protein analysis, as previously noted21, 26. The constructs were transfected into a human embryonic kidney cell line (HEK-293) and incubated with varying concentrations of (S)-11, (R)-12, 4, gentamicin and G418 (Fig. 5). In order to ensure suitable cell viability for each of the tested compounds at the concentrations tested, we determined cell toxicity for each compound by measuring the half-maximal-lethal concentration value (LC50 values) in HEK-293 cells (Table 1).
Figure 5. Ex vivo.
stop codon suppression levels induced by (S)-11 (▲), (R)-12 (△), 4 (◆), G418 (…□…) and gentamicin (--■--) in a series of nonsense mutation context constructs representing various genetic diseases (shown in parenthesis): A) R3X (USH1), B) R245X (USH1), C) G542X (CF), and D) W1282X (CF). The results are averages of at least three independent experiments.
Table 1.
Comparative cell toxicity, antibacterial activity and inhibition of protein translation in eukaryotic, prokaryotic and mitochondrial systems of gentamicin, G418 and synthetic compounds 1-12a
| Pharmacophore
|
Aminoglycoside | Cell toxicity LC50 (mM)b | Antibacterial activity MIC (μM)c | Translation Inhibition
|
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| i | ii | iii | iv | Eukaryotic IC50Euk (μM)d | Prokaryotic IC50Prok (μM)d | Mitochondria IC50Mit(μM)e | |||||
| x | Gentamicin | 2.5 ± 0.3 | 6 | 62.0 ± 9.0 | 0.03 ± 0.00 | 26 ± 2 | 2,214 | ||||
| Paromomycin | 4.1 ± 0.5 | 22 | 57.0 ± 4.0 | 0.05 ± 0.01 | - | 1,118 | |||||
| x | G418 | 1.3 ± 0.1 | 9 | 2.0 ± 0.3 | 0.01 ± 0.00 | 13 ± 1 | 225 | ||||
|
| |||||||||||
| 1 | 21.9 ± 4.5 | 790 | 31.0 ± 4.0 | 0.5 ± 0.1 | 1785 ± 133 | 68 | |||||
| x | 2 | 5.5 ± 0.6 | 588 | 24.0 ± 1.0 | 0.2 ± 0.0 | 492 ± 12 | 151 | ||||
| x | 3 | 22.2 ± 1.1 | 680 | 17.0 ± 0.6 | 1.1 ± 0.1 | 965 ± 155 | 15 | ||||
| x | x | 4 | 5.8 ± 0.7 | 556 | 2.8 ± 0.3 | 1.0 ± 0.1 | 404 ± 26 | 2.9 | |||
| x | (S)-5 | 23.5 ± 0.6 | 2659 | 16.0 ± 1.0 | 2.0 ± 0.2 | 1408 ± 58 | 7.9 | ||||
| x | (R)-6 | 19.8 ± 0.4 | 4989 | 28.0 ± 1.1 | 2.1 ± 0.5 | 710 ± 68 | 13 | ||||
| x | x | (S)-7 | 10.1 ± 0.8 | 1067 | 5.2 ± 0.7 | 2.3 ± 0.2 | 251 ± 21 | 2.3 | |||
| x | x | (R)-8 | 13.9 ± 1.3 | 1057 | 4.6 ± 0.6 | 0.8 ± 0.1 | 152 ± 12 | 5.7 | |||
| x | x | (S)-9 | 5.4 ± 0.5 | 768 | 1.5 ± 0.1 | 1.1 ± 0.2 | 1493 ± 28 | 1.3 | |||
| x | x | (R)-10 | 16.5 ± 3.1 | 1536 | 8.0 ± 0.3 | 1.9 ± 0.2 | 681 ± 30 | 4.3 | |||
| x | x | x | (S)-11 | 5.1 ± 0.3 | 384 | 0.7 ± 0.1 | 1.8 ± 0.3 | 506 ± 50 | 0.4 | ||
| x | x | x | (R)-12 | 5.4 ± 0.3 | 384 | 0.9 ± 0.1 | 1.8 ± 0.2 | 400 ± 38 | 0.5 | ||
In all biological tests, all tested AGs were in their sulfate salt forms and the concentrations reported refer to that of the free amine form of each AG. The presence of pharmacophores (i–iv) in each compound is noted by “x”. All assays were performed in duplicate and analogous results were obtained in at least three independent experiments.
Cell toxicity was measured in human embryonic kidney (HEK-293) cells, and calculated as a ratio between the numbers of living cells in cultures grown in the presence of the tested compound, versus cultures grown without compound.
The minimal inhibitory concentration (MIC) values were measured in E. coli R477/100 and determined by using the double-microdilution method.
Prokaryotic and eukaryotic translation inhibition was quantified in coupled transcription/translation assays by using active luciferase detection as previously described by us 8.
For the IC50Mit value measurements, the freshly isolated mitochondria (2 mg protein/mL) from HeLa cells (Qproteome Mitochondria Isolation Kit, Qiagen, CA) were incubated in 93 μL of a protein synthesis medium in the presence of emetine (5 μM, an inhibitor of 80S ribosome), [35S]methionine (150 μCi, Izotop) and a varied concentrations of AG. The incorporation of [35S]methionine into mitochondrial protein was determined by a filter paper disk assay as described in Experimental Section. The LC50, IC50Euk, IC50Pro and IC50Mit values were estimated from fitting concentration response curves to the data of at least three independent experiments, using GraFit5 software. The IC50Euk/IC50Pro ratio for (S)-11 and (R)-12 is highlighted.
In all the mutations tested, the observed efficacy of aminoglycoside-induced readthrough was in the order G418 ≥ (S)-11 ≥ (R)-12 > 4 > gentamicin (Fig. 5). This trend for (S)-11 and (R)-12 was similar to that observed for the suppression of the same stop mutations in vitro (Fig. 4), even though the gap of potency difference between the (S)-11 and (R)-12 was smaller than the one observed for the suppression of the same mutations in cell-free extracts. While these data may point to a different cell permeability of (S)-11 and (R)-12, due to the different stereochemistry at the 5″-methyl group, more experiments are needed to understand this issue satisfactorily. Nevertheless, the observed similar cell toxicity of the compounds (S)-11, (R)-12 and 4 in HEK-293 cells (Table 1), along with substantially elevated suppression activities of (S)-11 and (R)-12 over that of 4, both in vitro and ex vivo in cultured cells, indicate that compounds (S)-11 and (R)-12 may represent a superior choice than compound 4 in suppression therapy.
Compounds 9-12 inhibit prokaryotic protein translation with significantly lower potency and exhibit markedly reduced bactericidal activity and cell toxicity than those of gentamicin and G418
In our previous studies,18 we have shown that compounds 3 and 4 are about 30-fold weaker inhibitors of prokaryotic translation than gentamicin and exhibit almost no bactericidal activity against both Gram-negative and Gram-positive bacteria. To assess whether the compounds 9-12 retain similar properties, we conducted comparative translation inhibition of compounds 3, 4, 9-12, G418 and gentamicin in a prokaryotic system, by using an in vitro luciferase assay (Table 1). The measured halfmaximal inhibitory concentration (IC50Pro) values show that the efficacy with which 9-12 inhibit the prokaryotic ribosome is significantly lower (high IC50 values) than that of gentamicin and G418. These data are in accordance with the observed antibacterial data of this set of compounds (Table 1 and Table S1; note that for clarity part of the data appear in Supporting Information). Thus, while gentamicin and G418 exhibit excellent antibacterial activities against both Gram-negative Escherichia coli (R477-100) (Table 1) and Gram-positive Baccilus subtilis (ATCC-6633) (Table S1 in Supporting Information), compounds 9-12 lack significant antibacterial activity. The observed data with 9-12 is similar to that observed for 3 and 4, along with for the entire set of 1-8, and further support the previously reported correlation in aminoglycosides between prokaryotic antitranslational activity (IC50Pro) and MIC values: decreased inhibition of prokaryotic translation is associated with the decrease in antibacterial activity27.
We have previously reported that compounds 1-8 exhibit significantly reduced cytotoxicity than that of gentamicin, as measured in a variety of kidney-derived cells.17–19 To assess whether our novel compounds 9-12 retain similar properties, we determined comparative cell toxicity of 3, 4, 9-12, G418 and gentamicin in HEK-293 cells (Table 1). All the tested compounds exhibited lower toxicity than that of gentamicin. Among all aminoglycosides tested, the aminoglycoside antibiotic, G418, which is known as one of the most cytotoxic aminoglycoside11, exhibited the lowest LC50 value (1.3 mM). Very similar trend to that in HEK-293 cells (Table 1) was observed when such comparative cell toxicity was determined in human foreskin fibroblasts (HFF) cells (Table S1 in Supporting Information).
Comparison of the observed cell toxicity data in Table 1 with the readthrough activity data in Figs. 3–5, demonstrates that compound 3 that contains only one pharmacophore, (R)-6′-Me (pharmacophore-ii), exhibits similar to better suppression activity than that of gentamicin, while its cytotoxicity is about 10-fold lower than that of gentamicin. Introduction of second pharmacophore on 3, either the N1-AHB (pharmacophore-i) to give 4 (3 → 4), or the (S)-5″-Me (pharmacophore-ii) to give (S)-9 (3 → 9), results in about 4-fold increase in cytotoxicity of the resulted structures (LC50 values of 5.8 and 5.4 mM for 4 and (S)-9, and 22.2 mM for 3), with a concomitant drastic increase in the observed stop codon suppression activity ((S)-9 ≥ 4 > 3). Additional increase in the number of pharmacophores to three, in compounds (S)-11 and (R)-12 (4 → (S)-11 and 4 → (R)-12), does not affect on the cytotoxicity (LC50 values of 5.1 and 5.4 mM, for (S)-11 and (R)-12, versus 5.8 for the compound 4), while it greatly increases the observed stop codon suppression activity of (S)-11 and (R)-12 compared to that of the compound 4 (Figs. 4 and 5, and Table 1). Thus, the combined structureactivity- toxicity data clearly indicate that, whereas the gradual increase in the number of pharmacophores is accompanied with a concomitant significant increase in the suppression activity, the cell toxicity trend does not really follow with this concept.
The increased specificity of AGs towards cytoplasmic ribosome correlates with the increased PTC suppression activity
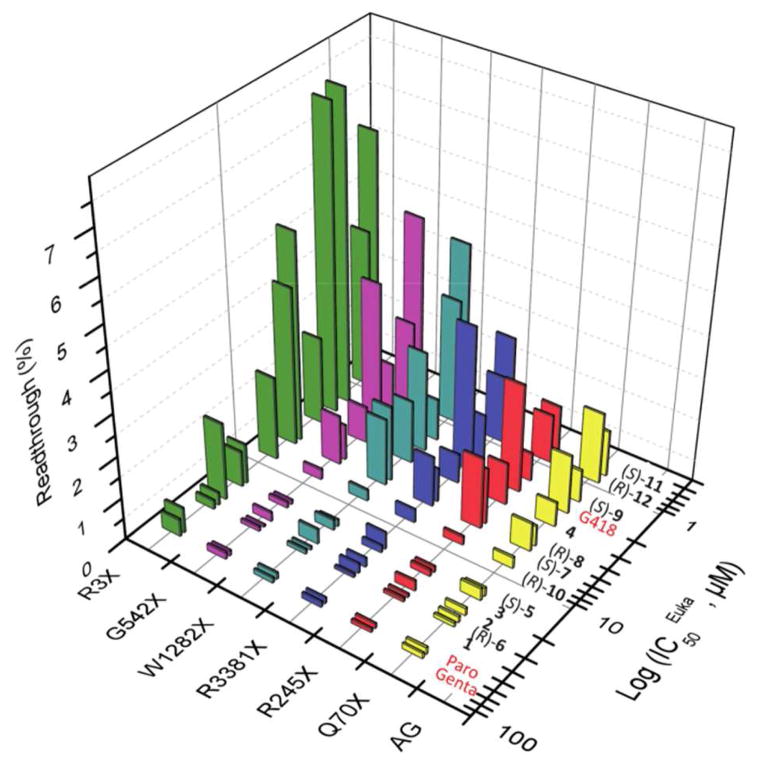
The impact of the number of pharmacophores on the elevated readthrough activities of the designer structures is further supported from the observed eukaryotic antitranslational data (Table 1). The efficacy with which (S)-11 (IC50Euk = 0.7 μM) inhibits eukaryotic translation is greater than that of (S)-9 (IC50Euk = 1.5 μM) and 3 (IC50Euk = 17 μM), a similar trend to that observed for readthrough activity: (S)-11 (contains 3 pharmacophores) > (S)-9 (contains 2 pharmacophores) > 3 (contains 1 pharmacophore) (Figs. 3 and 4). Likewise, (R)-5″-diasteromers that were in general less active than the corresponding (S)-5″-diasteromers, also were less specific to the eukaryotic ribosome: (R)-12 (IC50Euk = 0.9 μM) and (R)-10 (IC50Euk = 8.0 μM) are 1.2-fold and 5.3-fold less specific than (S)-11 and (S)-9, respectively. Finally, by plotting the observed IC50Euk values against the in vitro readthrough activity data of all the standard and synthetic AGs tested, a close correlation was observed: increased inhibition of the cytoplasmic protein synthesis is associated with the increased readthrough activity (Fig. 6 and Table S1). Since the readthrough activity is dose dependent and is also affected by the various above-mentioned factors, the data in Fig. 6 was collected at a single concentration, 1.4 μM, in which all the compounds were tested on all six different nonsense constructs used in this study.
Figure 6.
A semi-logarithmic plot of the in vitro readthrough activity at 1.4 μM concentrations versus the eukaryotic inhibition of translation (IC50Euk values) for gentamicin, paromomycin, G418 and 1-12, in a series of PTC constructs representing the genetic diseases: USH (R3X and R245X), CF (W1282X and G542X), DMD (R3381X) and HS (Q70X). For the data points see Table S2 in the Supporting Information.
As seen from the data in Fig. 6, among the synthetic derivatives, compounds (S)-11, (R)-12 and (S)-9 (IC50Euk values of 0.7, 0.9 and 1.5 μM, respectively), which exhibited particularly strong inhibition of the eukaryotic ribosome, were generally strongest readthrough inducers in all different constructs tested. Following three compounds, including 4, (R)-8 and (S)-7, with IC50Euk values of 2.8, 4.6, and 5.2 μM, respectively, induced readthrough with a lesser potency. Further decrease in eukaryotic inhibition by (R)-10, (S)-5, 3, 2, (R)-6 and 1 (IC50Euk values of 8, 16, 17, 24, 28, and 31 μM, respectively) was associated with a decrease in the observed readthrough activity. Thus, the impact of three pharmacophores in (S)-11 and (R)-12, on the inhibition of eukaryotic ribosome and the subsequent readthrough activity, is significantly bigger than those by two pharmacophores in (S)-9, 4, (R)-8, (S)-7, and (R)-10, with preference of (S)-5″-Me over that of (R)-5″-Me. Interestingly, among different combinations of two pharmacophores, the best combination was (R)-6′-Me with (S)-5″-Me in the compound (S)-9, while that of (R)-6′-Me with (R)-5″-Me in compound (R)-10, was the least effective. Very similar trend with respect to (S)-5″-Me and (R)-6′-Me, was also observed in the compounds with only one pharmacophore, and was in the order (S)-5 > 3 > 2 > (R)-6. In aggregate, the observed gradual increase in readthrough efficacy with an increase in the number of pharmacophores on the ligand indicates that these pharmacophores can operate in an additive manner, and that the structural features of the ligand play an important role in the proper recognition of mammalian rRNA.
Interestingly, very similar correlation/trend to that of synthetic AGs was also observed for the standard antibiotics gentamicin, paromomycin and G418, even though these three antibiotics are structurally significantly different (Fig. 1). G418 and gentamicin belong to a class of 4, 6-disubstitued 2-deoxystreptamine (2-DOS, ring II) AGs, while paromomycin is 4, 5-disubstituted 2-DOS-containing AG. Furthermore, while both G418 and paromomycin contain a hydroxyl group at 6′ position of the ribosamine ring (ring I), gentamicin has an amino group at the same position. Nevertheless, gentamicin and paromomycin that were very weak inhibitors of the eukaryotic ribosome (IC50Euk values of 62 and 57 μM, respectively), also exhibited very poor readthrough activity. And, G418 (IC50Euk = 2 μM) that was 30-fold more potent inhibitor than both gentamicin and paromomycin, also was very strong readthrough inducer. Thus the observed combined data with standard and synthetic AGs broadly demonstrate that a general feature of AGs with PTC suppression properties has the ability to inhibit eukaryotic cytoplasmic protein synthesis, and that the increased specificity towards cytoplasmic ribosome correlates with the increased PTC suppression activity.
Finally, we note that the observed correlation between the PTC suppression activity and inhibition of cytoplasmic ribosome in Fig. 6 encompasses compounds that inhibit translation through the same mechanism, namely by interference with the fidelity of decoding. This because, we have previously reported8 that the compound NB33 (two pieces of paromamine, ring I and ring II of paromomycin, are connected by methylene bridge at 3′ position) is a strong inhibitor of the eukaryotic translation (IC50Euk=2.4 μM), but lacks readthrough activity. This lack of readthrough by NB33 was explained by its different mechanism of translation inhibition: NB33 binds to the H. sapiens 18S cytoplasmic A site rRNA and selectively stabilizes a “non-decoding” conformational state, therefore, it does not interfere with the decoding process and subsequently lacks readthrough activity.8 Collectively, the observed data suggests that the compounds studied here might induce readthrough by selectively stabilizing a “decoding” conformation of the rRNA. Recent 3D-structure of G418 complexed to the protozoal 16S cytoplasmic A site construct28 supports this suggestion: G418 selectively stabilizes a “decoding” conformation, mainly because the pseudo pair contact between the 6′-OH of G418 and the N2-H of guanine at position 1408. Although the similar pseudo pair interactions are also expected for the entire 1-12 possessing 6′-OH, the secondary structure of H. sapiens 18S and protozoal 16S A sites are yet significantly different29, and the molecular mechanism of readthrough should await until the crystal structure of an eukaryotic ribosome30 in complex with a readthrough-inducing AG is available.
The newly designed structures (S)-11 and (R)-12 are more selective towards the eukaryotic than prokaryotic ribosome and thus show “reversed” selectivity in comparison to standard AG antibiotics
The comparative translation inhibition data in Table 1 show that gentamicin and paromomycin are 2,214-fold and 1,118-fold more selective towards the prokaryotic versus the eukaryotic ribosome. However, the difference in selectivity by G418 is only 225-fold. This order of magnitude drop in selectivity is mainly due to G418’s efficient inhibition of eukaryotic translation (IC50Euk=2 μM). Thus, one could consider this as the main reason for both extraordinary features of G418: its high cytotoxicity and its very strong readthrough activity. The results in Table 1, however, suggest that while the elevated inhibition of eukaryotic translation does indeed promote its strong readthrough activity, the inhibition of eukaryotic translation is not the only toxic event of G418. Indeed, three out of the four new structures, including (S)-9, (S)-11 and (R)-12, are similar or greater inhibitors of eukaryotic translation, while at the same time being significantly less toxic than G418. The notable decrease in the IC50Euk/IC50Pro ratio of compounds 1-12 relative to the reference AGs further demonstrates that the systematic development of a comprehensive pharmacophore and its installation on scaffold 1 could gradually increase the specificity of the developed lead to the cytoplasmic ribosome while decreasing its specificity to the prokaryotic ribosome; up till the synthesis of (S)-11 and (R)-12 derivatives, wherein all the pharmacophores are implemented, which instead exhibit “reversed” selectivity towards the eukaryotic versus the prokaryotic ribosome. To our knowledge, (S)-11 and (R)-12 represent the first examples of synthetic AGs that show such high eukaryotic versus prokaryotic selectivity, while in parallel exhibit strong PTC suppression activity.
Decreased specificity of the entire set of designer AGs 1-12 towards mitochondrial ribosome confers to the lowered cytotoxicity
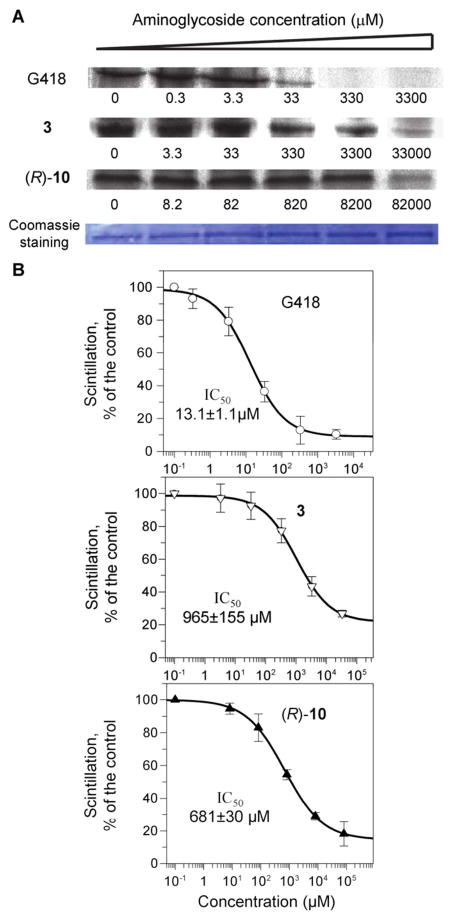
As mentioned above in the introduction section, high similarity between the bacterial and mitochondrial ribosome A sites may be connected at least partially to the AGs-induced cytotoxicity 13. Therefore, the observed continuous inability of our previous leads, 1-8, along with the current 9-12, to show significant antibacterial activity, in conjunction with their decreased prokaryotic ribosome specificity (Table 1), suggested that by reducing the specificity of 1-12 to the prokaryotic ribosome we might reduce their action on the mitochondrial ribosome and subsequently reduce their toxicity on humans. The observed significantly reduced cytotoxicity of compounds 1-12 in comparison to those of the standard AGs (Table 1) supported this indication. In addition, it has recently been shown that the high toxicity of oxazolidinone class of antibiotics is associated with their ability to strongly inhibit mitochondrial protein synthesis14. These antibiotics’ mode of action is similar to AGs as they bind to the bacterial ribosome and inhibit protein translation. Based on these observations, and our attempts to further assess the veracity of the hypothesis in connection to AGs, we examined the direct impact of AGs on mitochondrial protein synthesis. Since an in vitro luciferase assay analogous to the bacterial and cytoplasmic systems is not available for mitoribosomes, we used a radioactive assay14, 31 in which the translation levels of endogenous polypeptides were measured in intact mitochondria isolated from HeLa cells (Fig. 7, Table 1 and Fig. S1).
Figure 7.
Dose-response effects of aminoglycosides on mitochondrial protein synthesis. Isolated mitochondria from HeLa cells were incubated with varied concentrations of G418, 3 and (R)-10 as indicated. (A) After lyses, equal amounts of proteins (comassie staining) were fractionated on SDS-PAGE and [35S]methionine labeled COX1 protein levels were determined by densitometry. (B) Semi-logarithmic plots of scintillation counting as a function of aminoglycoside concentration. Radioactivity was measured on the lysed mixture after acid-precipitation as described in Experimental Section. The 50% inhibitory concentrations (IC50Mit) were determined by Grafit5 software[3].
As a representative example, the data in Fig. 7 shows the comparative effects of the standard AG, G418, and the designer structures 3 and (R)-10 on mitochondrial protein synthesis. Dose-dependent inhibition of mitochondrial protein synthesis was observed, and the observed data was consistent whether the protein levels were quantified by densitometry (COX1 protein levels on SDS-PAGE, panel-A in Fig. 7) or by direct scintillation counting of the radiolabeled proteins after acidprecipitation (panel-B in Fig. 7, for more details see Experimental Section). Furthermore, to ensure the accuracy of the observed data, we used chloramphenicol as a control in all our experiments. The measured half-maximal inhibitory value (IC50Mito) of chloramphenicol in our study (IC50Mito = 7.4 ± 0.8 μM, Fig. S1 in Supporting Information) was essentially the same to that reported (IC50Mito = 9.8 ± 0.8 μM) in the original procedure.14, 31
The measured IC50Mito values (Table 1) show that all the tested compounds inhibit translation in mitochondria. The antibiotic G418 is the most potent inhibitor of the mitoribosome (IC50Mito = 13.1 μM) followed by gentamicin (IC50Mito = 25.8 μM), and that the entire synthetic library 1-12 exhibit a roughly 12- to 140-fold reduced inhibition relative to that of G418.
We note that, by assessment of the relative hair cell toxicity potential in ex vivo cultures of cochlear explants, we have previously reported that compound 2, in addition to its significantly reduced cell toxicity (Table 1), also exhibits substantially reduced ototoxicity potential, relative to those of gentamicin17. Here we show that compound 2 (IC50Mito = 492 μM) exhibits 19-fold reduced inhibition of mitoribosome relative to that of gentamicin (Table 1). This data therefore suggests that the inhibition of mitoribosome by compound 2, in addition to its cell toxicity is probably, at least in part, also responsible for its reduced ototoxicity potential. Interestingly, as shown in Table 1, the new structures 9-12 exhibit very similar or lower inhibition potency for mitoribosome relative to that of compound 2, suggesting on the probability of their reduced ototoxicity potential. This suggestion is in agreement with the recently reported data on the AG apramycin15 in cochlear explants and in the in vivo guinea pig model of ototoxicity; apramycin caused little hair cell damage and hearing lose, while in parallel, it exhibited significantly reduced inhibition of mitochondrial protein synthesis than gentamicin. Nevertheless, it is clear that further structure-toxicity studies are required to understand whether the direct correlation between the cytotoxicity and ototoxicity of AGs can be made. These studies are currently underway and will be reported in due course.
SUMMARY AND CONCLUSION
The results of this study show that the newly developed AGs, 9-12, exhibit both appreciably higher PTC suppression efficiency and lower cytotoxicity than those of the clinical drug gentamicin. Furthermore, the data also show that compound (S)-11 exhibits similar activity to that of G418, while its cell toxicity is significantly lower than those of gentamicin and G418. Based on these findings, compound (S)-11 can be considered the best AG for the potential use in suppression therapy.
The most compelling evidence for the superior PTC suppression efficiency of compound (S)-11 over that of its parent compound 4 and gentamicin was demonstrated in vitro on six different DNA fragments derived from the mutant CFTR, PCDH15, IDUA and dystrophin genes carrying nonsense mutations and representing the underlying causes for the genetic diseases CF, USH1, HS and DMD, respectively (Fig. 4). Analogous advantage of (S)-11 was also demonstrated ex vivo in cultured cell lines (Fig. 5). Importantly, the comparative in vitro and ex vivo PTC suppression study also demonstrated the ability of (S)-11 to show comparable activity to that of G418. This was demonstrated in vitro in three different constructs, and was further verified in four different constructs ex vivo. Consistent with our previous findings, the data observed here has also established that the joint presence of three different pharmacophores in (S)-11 (Fig. 2), including N1-AHB, (pharmacophore-i), (R)-6′-Me (pharmacophore-ii) and (S)-5″-Me (pharmacophore-iii), contributes to its particularly enhanced readthrough efficiency. Here we show that (S)-11 inhibits eukaryotic protein translation by 88-fold and ~3-fold stronger than gentamicin and G418, suggesting that the enhanced readthrough activity of (S)-11 is probably due to its correspondingly enhanced interaction with the cytoplasmic ribosome (Table 1). The observed correlation between the IC50Euk values and the in vitro readthrough activity data, broadly showing that the increased inhibition of the cytoplasmic protein synthesis is associated with the increased readthrough activity (Fig. 6), supports this suggestion.
The observed data on the eukaryotic translation inhibition (Table 1) in conjunction with the cytotoxicity data (Table 1) indicated that the strong inhibition of eukaryotic translation is not the only toxic event of G418, and that other effect(s) of G418 on the mammalian cells is critical to its extraordinarily high cytotoxicity. Here we show that G418 is the most potent inhibitor of the human mitochondrial ribosome; compound (S)-11 is 38-fold less inhibitory, and the entire synthetic library 1-12 exhibit a roughly 12- to 140-fold reduced inhibition of mitochondrial ribosome relative to that of G418. The combined data thus suggest that the strong inhibition of both cytoplasmic and mitochondrial ribosomes confers on G418 its exceptionally high cytotoxicity, and that the reduced cytotoxicity of the entire 1-12 set of derivatives is mainly achieved through their reduced inhibition of the mitoribosome.
In conclusion, the results presented here provide proof of principle that, by using structure-based design, antibacterial activity and toxicity of AGs can be dissected from their PTC suppression activity. The data further indicate that the increased specificity towards cytoplasmic ribosome correlates with the increased PTC suppression activity, and that the decreased specificity towards mitochondrial ribosome confers to the lowered cytotoxicity. We have recently demonstrated the ability of some of our developed lead compounds to partially restore protein function in various clinically relevant cellular and animal models of genetic diseases caused by nonsense mutations: compound 2 in cellular and animal models of CF32, and cellular models of Rett syndrome33, 34; compounds 1 and 2 in cellular in vivo models of USH117, 35, 36; compound 4 in cellular and animal models of HS37. These observations, together with the relatively low toxicity and high degree of potency of the new generation structures 9-12 in targeting all six different nonsense constructs underlying USH1, CF, DMD and HS, support the feasibility of testing these novel AGs in treating these diseases in animal and human subjects. Finally, this study provides a new strategy for the development of novel AG-based structures by means of optimizing drug-induced suppression efficacy and toxicity; further progress in this direction may offer promise for the treatment of many genetic diseases caused by nonsense mutations.
EXPERIMENTAL SECTION
General methods
1D and 2D NMR spectra were routinely recorded on a Bruker Avance™ 500 spectrometer. Mass spectra analysis were obtained either on a Bruker Daltonix Apex 3 mass spectrometer under electron spray ionization (ESI), or by a TSQ-70B mass spectrometer (Finnigan Mat). Reactions were monitored by TLC on Silica Gel 60 F254 (0.25 mm, Merck), and spots were visualized by charring with a yellow solution containing (NH4)Mo7O24·4H2O (120 g) and (NH4)2Ce(NO3)6 (5 g) in 10% H2SO4 (800 mL). Column chromatography was performed on a Silica Gel 60 (70–230 mesh). All reactions were carried out under an argon atmosphere with anhydrous solvents, unless otherwise noted. All chemicals and biochemicals, unless otherwise stated, were obtained from commercial sources. G418 (geneticin), paromomycin and gentamicin were purchased from Sigma. In all biological tests, all the tested aminoglycosides were in their sulfate salt forms [Mw (gr/mol) of the sulfate salts were as follow: gentamicin – 653.2; G418 – 692.7; comp. 1 – 563.0; comp. 2 – 652.8; comp. 3 – 564.3; comp. 4 - 695.5; comp. (S)-5 – 577.7; comp. (R)-6 – 615.7; comp. (S)-7 – 719.5; comp. (R)-8 – 726.2; comp. (S)-9 – 605.9; comp. (R)-10 – 619.6; comp. (S)-11 – 730.2; comp. (R)-12 – 730.5]. Purity of the new compounds 9-12 was determined by using HPLC-ESI-MS analysis, which indicated 99.54% ((S)-9), 99.21% ((R)-10), 95.21% ((S)-11) and 97.30% ((R)-10) purity (see Supporting Information).
6′-(R)-Methyl-5-O-(5-azido-5,6-dideoxy-2,3-O-dibenzoyl-α-L-talofuranosyl)-3′,4′,6′,6-tetra- O-acetyl-2′,1,3-triazido paromamine, (S)-17
Anhydrous CH2Cl2 (15mL) was added to a powdered, flame-dried 4 Å molecular sieves (2.0 g), followed by the addition acceptor 1318 (0.9 g, 0.0015 mol) and donor (S)-1519 (2.0 g, 0.0037 mol). The reaction mixture was stirred for 10 min at room temperature and was then cooled to −20°C. A catalytic amount of BF3-Et2O (0.1 ml) was added and the mixture was stirred at −15°C and the reaction progress was monitored by TLC, which indicated the completion after 120 min. The reaction mixture was diluted with ethyl acetate and washed with saturated NaHCO3 and brine. The combined organic layer was dried over MgSO4, evaporated and subjected to column chromatography (EtOAc/Hexane) to obtain the titled compound (S)-17 (1.1 g) in 75% yield.1HNMR (500 MHz, CDCl3): “Ring I” δH 1.27 (d, 3H, J = 6.0 Hz, CH3), 3.58 (dd, 1H, J1 = 5.5, J2 = 10.5 Hz, H-2′), 4.45 (d, 1H, J = 10.7 Hz, H-5′), 4.96–5.02 (m, 2H, H-4′ and H-6′), 5.42 (t, 1H, J = 9.6 Hz, H-3′), 5.95 (d, 1H, J = 3.7 Hz, H-1′); “Ring II” δH 1.51 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 2.41 (td, 1H, J1 = 4.5, J2 = 12.5 Hz, H-2eq), 3.55 (m, 2H, H-1 and H-3), 3.76 (t, 1H, J = 9.4 Hz, H-4), 3.88 (t, 1H, J = 9.0 Hz, H-5), 5.03 (t, 1H, J = 9.1 Hz, H-6); “Ring III” δH 1.27 (d, 3H, J = 5.6 Hz, CH3), 3.76 (m, 1H, H-5″), 4.35 (dd, 1H, J1 = 6.9, J2 = 10.9 Hz, H-4″), 5.45 (t, 1H, J = 5.5 Hz, H-3″), 5.62 (m, 2H, H-2″ and H-1″). The additional peaks in the spectrum were identified as follows: δH 2.08 (s, 3H, OAc), 2.09 (s, 6H, OAc), 2.38 (s, 3H, OAc), 7.37 (t, 2H, J = 7.8 Hz, Ar), 7.41 (t, 2H, J = 7.8 Hz, Ar), 7.53–7.60 (m, 2H, Ar), 7.89 (d, 2H, J = 8.0 Hz, Ar), 7.93 (d, 2H, J = 8.2 Hz, Ar). 13CNMR (125 MHz, CDCl3): δC 13.3 (C-7′), 15.4 (C-6″), 20.6 (2C, OAc), 20.9 ( OAc), 21.1 (OAc), 32.1 (C-2), 58.4, 58.8, 59.5, 61.7, 68.5, 69.0, 70.1, 70.8. 71.8, 73.6, 74.6, 77.3, 79.6, 84.4, 96.0 (C-1′), 107.6 (C-1″), 128.4 (Ar), 128.5 (Ar), 128.6 (Ar), 128.7 (Ar), 129.6 (Ar), 129.7 (Ar), 133.5 (Ar), 133.6 (Ar), 164.9 (C=O), 165.3 (C=O), 169.7 (C=O), 169.9 (C=O), 170.1 (C=O), 170.2 (C=O). MALDI TOFMS calculated for C41H46N12O16 Na ([M+Na]+) m/e: 985.3; measured m/e: 985.4.
6′-(R)-Methyl-5-O-(5-azido-5,6-dideoxy-2,3-O-dibenzoyl-β-D-allofuranosyl)-3′,4′,6′,6-tetra- O-acetyl- 2′,1,3-triazido paromamine, (R)-18
Anhydrous CH2Cl2 (15mL) was added to a powdered, flame-dried 4 Å molecular sieves (2.0 g), followed by the addition acceptor 1318 (1.0 g, 0.0017 mol) and donor (R)-1619 (2.2 g, 0.004 mol). The reaction mixture was stirred for 10 min at room temperature and was then cooled to −20°C. A catalytic amount of BF3-Et2O (0.1 ml) was added and the mixture was stirred at −15 °C and the reaction progress was monitored by TLC, which indicated the completion after 120 min. The reaction mixture was diluted with ethyl acetate and washed with saturated NaHCO3 and brine. The combined organic layer was dried over MgSO4, evaporated and subjected to column chromatography (EtOAc/Hexane) to obtain the titled compound (R)-18 (1.2 g) in 75% yield. 1HNMR (500 MHz, CDCl3): “Ring I” δH 1.28 (d, 3H, J = 6.7 Hz, CH3), 3.46 (dd, 1H, J1 = 4.5, J2 = 10.4 Hz, H-2′), 4.47 (d, 1H, J = 10.7 Hz, H-5′), 4.96–5.02 (m, 2H, H-4′ and H-6′), 5.44 (t, 1H, J = 9.6 Hz, H-3′), 5.93 (d, 1H, J = 3.3 Hz, H-1′); “Ring II” δH 1.50 (ddd, 1H, J1=J2=J3=12.5 Hz, H-2ax), 2.41 (td, 1H, J1=4.5 and J2= 12.5 Hz, H-2eq), 3.56 (m, 2H, H-1 and H-3), 3.76 (t, 1H, J= 10.0 Hz, H-4), 3.92 (t, 1H, J = 9.5 Hz, H-5), 5.04 (t, 1H, J = 9.6 Hz, H-6); “Ring III” δH 1.42 (d, 3H, J = 6.9 Hz, CH3), 3.78 (m, 1H, H-5″), 4.40 (t, 1H, J = 4.6 Hz, H-4″), 5.50 (t, 1H, J = 5.0 Hz, H-3″), 5.59 (t, 1H, J = 3.7 Hz, H-2″), 5.64 (s, 1H, H-1″). The additional peaks in the spectrum were identified as follows: δH 2.09 (s, 9H, OAc), 2.33 (s, 3H, OAc), 7.37–7.41 (m, 4H, Ar), 7.56 (m, 2H, Ar), 7.92 (d, 4H, J= 8.0 Hz Ar). 13CNMR (125 MHz, CDCl3): δH 13.3 (C-7′), 15.0 (C-6″), 20.6 (OAc), 20.7 (OAc), 20.8 (OAc), 21.2 (OAc), 32.1 (C-2), 58.1, 58.2, 58.8, 61.5, 68.9, 70.2, 70.6, 71.4, 73.8, 74.6, 77.0, 77.1, 79.4, 83.9, 96.1 (C-1′), 107.0 (C-1″), 128.4 (2C, Ar), 128.7 (2C, Ar), 129.6 (2C, Ar), 133.5 (Ar), 133.6 (Ar), 164.9 (C=O), 165.4 (C=O), 169.8 (C=O), 169.9 (2C, C=O), 170.1 (C=O). MALDI TOFMS calculated for C41H46N12O16Na ([M+Na]+) m/e: 985.3; measured m/e: 985.4.
6′-(R)-Methyl-5-O-(5-azido-5,6-dideoxy-2,3-O-dibenzoyl-α-L-talofuranosyl)-3′,4′,6′,6-tetra- O-acetyl-2′,3-diazido-1-N-[(S)-4-azido-2-O-acetyl-butanoyl]paromamine, (S)-19
Anhydrous CH2Cl2 (15mL) was added to a powdered, flame-dried 4 Å molecular sieves (2.0 g), followed by the addition acceptor 1418 (1.0 g, 0.0014 mol) and donor (S)-1519 (2.5 g, 0.0046 mol). The reaction mixture was stirred for 10 min at room temperature and was then cooled to −20°C. A catalytic amount of BF3-Et2O (0.1 ml) was added and the mixture was stirred at −15 °C and the reaction progress was monitored by TLC, which indicated the completion after 60 min. The reaction mixture was diluted with ethyl acetate and washed with saturated NaHCO3 and brine. The combined organic layer was dried over MgSO4, evaporated and subjected to column chromatography (EtOAc/Hexane) to obtain the titled compound (S)-19 (1.1 g) in 73% yield. 1HNMR (500 MHz, CDCl3): 1HNMR (500 MHz, CDCl3): “Ring I” δH 1.27 (d, 3H, J = 5.2 Hz, CH3), 3.54 (dd, 1H, J1 = 4.3, J2 = 10.5 Hz, H-2′), 4.45 (dd, 1H, J1 = 1.8, J2 = 10.6 Hz, H-5′), 4.96–5.02 (m, 2H, H-4′ and H-6′), 5.43 (t, 1H, J = 9.4 Hz, H-3′), 5.94 (d, 1H, J = 3.7 Hz, H-1′); “Ring II” δH 1.44 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 2.52 (td, 1H, J1 = 4.5, J2 = 12.5 Hz, H-2eq), 3.60 (m, 1H, H-3), 3.66 (t, 1H, J = 4.5 Hz, H-4), 3.99 (t, 1H, J = 6.4 Hz, H-5), 4.05 (m, 1H, H-1), 4.94 (t, 1H, J = 9.2 Hz, H-6); “Ring III” δH 1.32 (d, 3H, J = 6.9 Hz, CH3), 3.72 (m, 1H, H-5″), 4.32 (dd, 1H, J1 = 5.85, J2 = 8.0 Hz, H-4″), 5.55 (dd, 1H, J1 = 4.7, J2 = 7.4 Hz, H-3″), 5.65 (m, 2H, H-2″ and H-1″). The additional peaks in the spectrum were identified as follows: δH 2.04–2.10 (m, 2H, H-8 and H-8), 2.11 (m, 9H, OAc), 2.22 (s, 3H, OAc), 2.30 (s, 3H, OAc), 3.37 (t, 2H, J = 6.8 Hz, H-9 and H-9), 5.20 (t, 1H, J = 4.85 Hz, H-7), 6.70 (d, 1H, J = 7.5 Hz, NH), 7.35 (t, 2H, J = 7.6 Hz, Ar), 7.43 (t, 2H, J = 7.8 Hz, Ar), 7.53–7.61 (m, 2H, Ar), 7.86 (dd, 2H, J1 = 1.1, J2 = 8.2 Hz, Ar), 7.95 (dd, 2H, J1 = 1.2, J2 = 8.2 Hz, Ar). 13CNMR (125 MHz, CDCl3): δC 13.5 (C-7′), 15.5 (C-6″), 20.6 (3C, OAc), 20.9 (OAc), 21.1 (OAc), 30.4, 32.2 (C-1), 47.0, 48.4, 58.6, 58.7, 61.6, 68.6, 69.0, 70.3, 70.8 (2C), 71.4, 73.1, 74.7, 77.5, 79.8, 83.6, 96.3 (C-1′), 107.4 (C-1″), 128.4 (Ar), 128.5 ( Ar), 128.7 (2C, Ar), 129.6 (Ar), 129.7 (Ar), 133.5 (Ar), 133.6 (Ar), 165.0 (C=O), 165.2 (C=O), 168.8 (C=O), 169.7 (2C, C=O), 169.9 (C=O), 170.0 (C=O), 172.4 (C=O). MALDI TOFMS calculated for C47H55N13O19 Na ([M+Na]+) m/e: 1128.4; measured m/e: 1128.2.
6′-(R)-Methyl-5-O-(5-azido-5,6-dideoxy-2,3-O-dibenzoyl-β-D-allofuranosyl)-3′,4′,6′,6-tetra- O-acetyl-2′,3-diazido-1-N-[(S)-4-azido-2-O-acetyl-butanoyl]paromamine, (R)-20
Anhydrous CH2Cl2 (15mL) was added to a powdered, flame-dried 4 Å molecular sieves (2.0 g), followed by the addition acceptor 1418 (1.0 g, 0.0014 mol) and donor (R)-1619 (2.5 g, 0.0046 mol). The reaction mixture was stirred for 10 min at room temperature and was then cooled to −20°C. A catalytic amount of BF3-Et2O (0.1 ml) was added and the mixture was stirred at −15 °C and the reaction progress was monitored by TLC, which indicated the completion after 90 min. The reaction mixture was diluted with ethyl acetate and washed with saturated NaHCO3 and brine. The combined organic layer was dried over MgSO4, evaporated and subjected to column chromatography (EtOAc/Hexane) to obtain the titled compound (R)-20 (1.15 g) in 76% yield. 1HNMR (500 MHz, CDCl3): 1HNMR (500 MHz, CDCl3): 1HNMR (500 MHz, CDCl3): “Ring I” δH 1.28 (d, 3H, J = 6.6 Hz, CH3), 3.43 (dd, 1H, J1 = 4.3, J2 = 10.6 Hz, H-2′), 4.49 (dd, 1H, J1 = 2.2, J2 = 10.7 Hz, H-5′), 4.96–5.02 (m, 2H, H-4′ and H-6′), 5.45 (t, 1H, J = 10.6 Hz, H-3′), 5.92 (d, 1H, J = 3.7 Hz, H-1′); “Ring II” δH 1.42 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 2.52 (td, 1H, J1 = 4.5, J2 = 12.5 Hz, H-2eq), 3.64 (m, 1H, H-3), 3.76 (t, 1H, J = 4.5 Hz, H-4), 4.05 (m, 2H, H-1 and H-5), 4.93 (t, 1H, J = 10.0 Hz, H-6); “Ring III” δH 1.39 (d, 3H, J = 6.4 Hz, CH3), 3.85 (m, 1H, H-5″), 4.36 (dd, 1H, J1 = 4.3, J2 = 6.3 Hz, H-4″), 5.63 (m, 2H, H-2″ and H-3″), 5.67 (s, 1H, H-1″). The additional peaks in the spectrum were identified as follows: δH 2.04–2.10 (m, 2H, H-8 and H-8), 2.08 (s, 3H, OAc), 2.09 (s, 3H, OAc), 2.10 (s, 3H, OAc), 2.21 (s, 3H, OAc), 2.25 (s, 3H, OAc), 3.37 (t, 2H, J = 6.7 Hz, H-9 and H-9), 5.18 (t, 1H, J = 5.0 Hz, H-7), 6.66 (d, 1H, J = 7.5 Hz, NH), 7.38–7.42 (m, 4H, Ar), 7.53–7.59 (m, 2H, Ar), 7.89–7.92 (m, 4H, Ar). 13CNMR (125 MHz, CDCl3): δC 13.5 (C-7′), 15.2 (C-6″), 20.6 (3C, OAc), 20.8 (OAc), 21.1 (OAc), 30.4, 32.4 (C-1), 47.0, 48.4, 58.1, 58.7, 61.4, 68.6, 69.0, 70.3, 70.5, 70.8, 70.9, 73.4, 74.8, 77.2, 79.6, 83.3, 96.3 (C-1′), 106.9 (C-1″), 128.4 (2C, Ar), 128.7 (2C, Ar), 129.5 (Ar), 129.6 (Ar), 133.5 (2C, Ar), 164.9 (C=O), 165.2 (C=O), 168.8 (C=O), 169.7 (2C, C=O), 169.9 (C=O), 170.0 (C=O), 172.3 (C=O). MALDI TOFMS calculated for C47H55N13O19 Na ([M+Na]+) m/e: 1128.4; measured m/e: 1128.4.
6′-(R)-Methyl-5-O-(5-amino-5,6-dideoxy-α-L-talofuranosyl)-paromamine, (S)-9
The glycosylation product (S)-17 (1.0 g, 0.001 mol) was treated with a solution of MeNH2 (33% solution in EtOH, 50 mL) and the reaction progress was monitored by TLC (EtOAc/MeOH 85:15), which indicated completion after 8 hours. The reaction mixture was evaporated to dryness and the residue was dissolved in a mixture of THF (5 mL) and aqueous NaOH (1 mM, 5.0 mL). The mixture was stirred at room temperature for 10 minutes, after which PMe3 (1 M solution in THF, 5.0 mL, 5.0 mmol) was added. The reaction progress was monitored by TLC [CH2Cl2/MeOH/H2O/MeNH2 (33% solution in EtOH) 10:15:6:15], which indicated completion after 1 hour. The product was purified by column chromatography on a short column of silica gel. The column was washed with the following solvents: THF (800 mL), CH2Cl2 (800 mL), EtOH (200 mL), and MeOH (400 mL). The product was then eluted with a mixture of 20% MeNH2 (33% solution in EtOH) in 80% MeOH. Fractions containing the product were combined and evaporated to dryness. The residue was re-dissolved in a small volume of water and evaporated again (2–3 repeats) to afford the free amine form of 3. The analytically pure product was obtained by passing the above product through a short column of Amberlite CG50 (NH4+ form). The column was first washed with a mixture of MeOH/H2O (3:2), then the product was eluted with a mixture of MeOH/H2O/NH4OH (80:10:10) to afford compound (S)-9 (0.400 g, 79% yield). For the storage and biological tests, compound was converted to its sulfate salt form: the free base was dissolved in water, the pH was adjusted around 7.0 with H2SO4 (0.1 N) and lyophilized. 1HNMR (500 MHz, CD3OD): 1HNMR (500 MHz, CD3OD): “Ring I” δH 1.21 (d, 3H, J = 5.8 Hz, CH3), 2.61 (dd, 1H, J1 = 3.5, J2 = 10.0 Hz, H-2′), 3.22 (t, 1H, J = 10.0 Hz, H-4′), 3.51 (t, 1H, J = 8.9 Hz, H-3′), 3.81 (dd, 1H, J1 = 3.0, J2 = 10.0 Hz, H-5′), 4.12 (m, 1H, H-6′), 5.20 (d, 1H, J = 3.3 Hz, H-1′); “Ring II” δH 1.18 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 1.98 (td, 1H, J1 = 4.5, J2 = 12.5 Hz, H-2eq), 2.63 (m, 1H, H-1), 2.79 (m, 1H, H-3), 3.19 (t, 1H, J = 9.7 Hz, H-6), 3.38 (t, 1H, J = 9.3 Hz, H-4), 3.48 (t, 1H, J = 9.2 Hz, H-5); “Ring III” δH 1.18 (d, 3H, J = 6.3 Hz, CH3), 2.95 (m, 1H, H-5″), 3.57 (t, 1H, J =6.4 Hz, H-4″), 4.03 (t, 1H, J = 5.6 Hz, H-3″), 4.07 (m, 1H, H-2″), 5.25 (d, 1H, J = 2.5 Hz, H-1″). 13CNMR (125 MHz, CD3OD): δC 16.9 (C-7′), 19.3 (C-6″), 37.5 (C-1), 50.6, 52.3, 52.6, 57.8, 67.8, 72.2, 73.6, 75.5, 76.2, 76.7, 78.6, 84.6, 87.3, 88.6, 101.9 (C-1′), 109.6 (C-1″). MALDI TOFMS calculated for C19H39N4O10 ([M+H]+) m/e: 483.3; measured m/e: 483.2.
6′-(R)-Methyl-5-O-(5-amino-5,6-dideoxy-β-D-allofuranosyl)-paromamine, (R)-10
The glycosylation product (R)-18 (1.0 g, 0.001 mol) was treated with a solution of MeNH2 (33% solution in EtOH, 50 mL) and the reaction progress was monitored by TLC (EtOAc/MeOH 85:15), which indicated completion after 8 hours. The reaction mixture was evaporated to dryness and the residue was dissolved in a mixture of THF (5 mL) and aqueous NaOH (1 mM, 5.0 mL). The mixture was stirred at room temperature for 10 minutes, after which PMe3 (1 M solution in THF, 5.0 mL, 5.0 mmol) was added. The reaction progress was monitored by TLC [CH2Cl2/MeOH/H2O/MeNH2 (33% solution in EtOH) 10:15:6:15], which indicated completion after 1 hour. The product was purified by column chromatography on a short column of silica gel. The column was washed with the following solvents: THF (800 mL), CH2Cl2 (800 mL), EtOH (200 mL), and MeOH (400 mL). The product was then eluted with a mixture of 20% MeNH2 (33% solution in EtOH) in 80% MeOH. Fractions containing the product were combined and evaporated to dryness. The residue was re-dissolved in a small volume of water and evaporated again (2–3 repeats) to afford the free amine form of 4. The analytically pure product was obtained by passing the above product through a short column of Amberlite CG50 (NH4+ form). The column was first washed with a mixture of MeOH/H2O (3:2), then the product was eluted with a mixture of MeOH/H2O/NH4OH (80:10:10) to afford compound (R)-10 (0.398 g, 79% yield). For the storage and biological tests, compound was converted to its sulfate salt form: the free base was dissolved in water, the pH was adjusted around 7.0 with H2SO4 (0.1 N) and lyophilized. 1HNMR (500 MHz, CD3OD): “Ring I” δH 1.22 (d, 3H, J = 5.8 Hz, CH3), 2.61 (dd, 1H, J1 = 2.5, J2 = 9.6 Hz, H-2′), 3.22 (t, 1H, J = 9.8 Hz, H-4′), 3.50 (t, 1H, J = 9.9 Hz, H-3′), 3.83 (dd, 1H, J1 = 3.0, J2 = 10.1 Hz, H-5′), 4.12 (m, 1H, H-6′), 5.20 (d, 1H, J = 3.3 Hz, H-1′); “Ring II” δH 1.21 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 1.98 (td, 1H, J1 = 4.5, J2 = 12.5 Hz, H-2eq), 2.65 (m, 1H, H-1), 2.78 (m, 1H, H-3), 3.18 (t, 1H, J = 9.3 Hz, H-6), 3.38 (t, 1H, J = 9.1 Hz, H-4), 3.46 (t, 1H, J = 9.2 Hz, H-5); “Ring III” δH 1.17 (d, 3H, J = 6.4 Hz, CH3), 3.10 (m, 1H, H-5″), 3.71 (t, 1H, J =5.0 Hz, H-4″), 4.06 (t, 1H, J = 5.6 Hz, H-2″), 4.16 (t, 1H, J = 3.0 Hz, H-3″), 5.20 (d, 1H, J = 3.0 Hz, H-1″). 13CNMR (125 MHz, CD3OD): δC 16.6 (C-7′), 18.7 (C-6″), 37.6 (C-1), 49.5, 52.2, 52.5, 57.8, 67.8, 70.8, 73.6, 75.4, 76.1, 76.7, 78.4, 84.7, 87.5, 88.0, 101.9 (C-1′), 109.6 (C-1″). MALDI TOFMS calculated for C19H39N4O10 ([M+H]+) m/e: 483.3; measured m/e: 483.2.
6′-(R)-Methyl-5-O-(5-amino-5,6-dideoxy-α-L-talofuranosyl)-1-N-[(S)-4-amino-2-hydroxybutanoyl] paromamine, (S)-11
The glycosylation product (S)-19 (1.05 g, 0.001 mol) was treated with a solution of MeNH2 (33% solution in EtOH, 50 mL) and the reaction progress was monitored by TLC (EtOAc/MeOH 85:15), which indicated completion after 8 hours. The reaction mixture was evaporated to dryness and the residue was dissolved in a mixture of THF (5 mL) and aqueous NaOH (1 mM, 5.0 mL). The mixture was stirred at room temperature for 10 minutes, after which PMe3 (1 M solution in THF, 5.0 mL, 5.0 mmol) was added. The reaction progress was monitored by TLC [CH2Cl2/MeOH/H2O/MeNH2 (33% solution in EtOH) 10:15:6:15], which indicated completion after 1 hour. The product was purified by column chromatography on a short column of silica gel. The column was washed with the following solvents: THF (800 mL), CH2Cl2 (800 mL), EtOH (200 mL), and MeOH (400 mL). The product was then eluted with a mixture of 20% MeNH2 (33% solution in EtOH) in 80% MeOH. Fractions containing the product were combined and evaporated to dryness. The residue was re-dissolved in a small volume of water and evaporated again (2–3 repeats) to afford the free amine form of 5. The analytically pure product was obtained by passing the above product through a short column of Amberlite CG50 (NH4+ form). The column was first washed with a mixture of MeOH/H2O (3:2), then the product was eluted with a mixture of MeOH/H2O/NH4OH (80:10:10) to afford compound (S)-11 (0.480 g, 86% yield). For the storage and biological tests, compound was converted to its sulfate salt form: the free base was dissolved in water, the pH was adjusted around 7.0 with H2SO4 (0.1 N) and lyophilized. 1HNMR (500 MHz, CD3OD) “Ring I” δH 1.21 (d, 3H, J = 6.0 Hz, CH3), 2.63 (dd, 1H, J1 = 3.5, J2 = 10.0 Hz, H-2′), 3.23 (t, 1H, J = 8.9 Hz, H-4′), 3.52 (t, 1H, J = 9.9 Hz, H-3′), 3.82 (dd, 1H, J1 = 3.0, J2 = 10.0 Hz, H-5′), 4.13 (m, 1H, H-6′), 5.22 (d, 1H, J = 3.3 Hz, H-1′); “Ring II” δH 1.34 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 1.99 (td, 1H, J1 = 4.5 and J2 = 12.5 Hz, H-2eq), 2.85 (m, 1H, H-3), 3.40 (t, 1H, J = 8.8 Hz, H-4), 3.50–3.59 (m, 2H, H-5 and H-6), 3.83 (m, 1H, H-1); “Ring III” δH 1.17 (d, 3H, J = 6.6 Hz, CH3), 2.94 (m, 1H, H-5″), 3.56 (t, 1H, J = 7.1 Hz, H-4″), 4.01 (t, 1H, J = 5.7 Hz, H-3″), 4.09 (dd, 1H, J1= 2.7 and J2 = 5.4 Hz, H-2″), 5.26 (d, 1H, J = 2.5 Hz, H-1″). The additional peaks in the spectrum were identified as follows: δH 1.82 (m, 1H, H-8), 1.95 (m, 1H, H-8), 2.83 (t, 2H, J = 5.7 Hz, H-9 and H-9), 4.13 (dd, 1H, J1= 4.2 and J2 = 7.6 Hz, H-7). 13CNMR (125 MHz, CD3OD): δC 16.6 (C-7′), 19.2 (C-6″), 35.9, 37.8, 39.0, 50.8, 50.9, 52.3, 57.8, 67.8, 71.7, 72.4, 73.6, 75.5, 75.6, 76.3, 76.8, 84.8, 86.7, 88.6, 101.9 (C-1′), 110.0 (C-1″), 177.1 (C=O). MALDI TOFMS calculated for C23H45N5O12Na ([M+Na]+) m/e: 606.3; measured m/e: 606.6.
6′-(R)-Methyl-5-O-(5-amino-5,6-dideoxy-β-D-allofuranosyl)-1-N-[(S)-4-amino-2-hydroxybutanoyl] paromamine, (R)-12
The glycosylation product (R)-20 (1.12 g, 0.001 mol) was treated with a solution of MeNH2 (33% solution in EtOH, 50 mL) and the reaction progress was monitored by TLC (EtOAc/MeOH 85:15), which indicated completion after 8 hours. The reaction mixture was evaporated to dryness and the residue was dissolved in a mixture of THF (5 mL) and aqueous NaOH (1 mM, 5.0 mL). The mixture was stirred at room temperature for 10 minutes, after which PMe3 (1 M solution in THF, 5.0 mL, 5.0 mmol) was added. The reaction progress was monitored by TLC [CH2Cl2/MeOH/H2O/MeNH2 (33% solution in EtOH) 10:15:6:15], which indicated completion after 1 hour. The product was purified by column chromatography on a short column of silica gel. The column was washed with the following solvents: THF (800 mL), CH2Cl2 (800 mL), EtOH (200 mL), and MeOH (400 mL). The product was then eluted with a mixture of 20% MeNH2 (33% solution in EtOH) in 80% MeOH. Fractions containing the product were combined and evaporated to dryness. The residue was re-dissolved in a small volume of water and evaporated again (2–3 repeats) to afford the free amine form of 6. The analytically pure product was obtained by passing the above product through a short column of Amberlite CG50 (NH4+ form). The column was first washed with a mixture of MeOH/H2O (3:2), then the product was eluted with a mixture of MeOH/H2O/NH4OH (80:10:10) to afford compound (R)-12 (0.500 g, 84% yield). For the storage and biological tests, compound was converted to its sulfate salt form: the free base was dissolved in water, the pH was adjusted around 7.0 with H2SO4 (0.1 N) and lyophilized. 1HNMR (500 MHz, CD3OD) “Ring I” δH 1.22 (d, 3H, J = 6.3 Hz, CH3), 2.63 (dd, 1H, J1 = 3.8, J2 = 10.0 Hz, H-2′), 3.22 (t, 1H, J = 9.8 Hz, H-4′), 3.52 (dd, 1H, J1 = 8.6, J2=10.3 Hz, H-3′), 3.83 (dd, 1H, J1 = 3.1, J2 = 10.2 Hz, H-5′), 4.13 (m, 1H, H-6′), 5.23 (d, 1H, J = 3.7 Hz, H-1′); “Ring II” δH 1.34 (ddd, 1H, J1=J2=J3= 12.5 Hz, H-2ax), 1.99 (td, 1H, J1 = 4.5 and J2 = 12.5 Hz, H-2eq), 2.85 (m, 1H, H-3), 3.39 (t, 1H, J = 8.8 Hz, H-4), 3.49–3.56 (m, 2H, H-5 and H-6), 3.82 (m, 1H, H-1); “Ring III” δH 1.16 (d, 3H, J = 6.7 Hz, CH3), 3.08 (m, 1H, H-5″), 3.69 (t, 1H, J = 5.5 Hz, H-4″), 4.07 (dd, 1H, J1 = 2.1, J2 = 5.2 Hz, H-2″), 4.14 (t, 1H, J = 5.7 Hz, H-3″), 5.21 (d, 1H, J = 3.7 Hz, H-1″). The additional peaks in the spectrum were identified as follows: δH 1.82 (m, 1H, H-8), 1.95 (m, 1H, H-8), 2.84 (t, 2H, J = 7.2 Hz, H-9 and H-9), 4.13 (dd, 1H, J1 = 3.9, J2 = 7.5 Hz, H-7). 13CNMR (125 MHz, CD3OD): δH 16.6 (C-7′), 18.8 (C-6″), 36.0, 37.7, 38.9, 49.6, 50.8, 52.3, 57.8, 67.8, 71.0, 71.7, 73.6, 75.5 (2C), 76.2, 76.7, 85.0, 86.9, 87.9, 101.9 (C-1′), 110.0 (C-1″), 177.1 (C=O). MALDI TOFMS calculated for C23H45N5O12Na ([M+Na]+) m/e: 606.3; measured m/e: 606.6.
Dual Luciferase Readthrough Assays
DNA fragments derived from PCDH15, CFTR, Dystrophin, and IDUA cDNAs, including the tested nonsense mutation or the corresponding wild type codon and four to six upstream and downstream flanking codons, were created and inserted into the polylinker of the p2luc plasmid, as previously described by us17. The polylinkers inserted to p2luc vector were as follow:
Usher Syndrome (PCDH15):
p.R3X mut/wt:5′-CAGAAGATGTTTT/CGACAGTTTTATCTCTGGACA-3′
p.R245 Xmut/wt 5′ AAAATCTGAATGAGAGGT/CGAACCACCACCACCACCCTC-3′
Cystic Fibrosis (CFTR):
p.G542X mut/wt: 5′-TCGACCAATATAGTTCTTT/GGAGAAGGTGGAATCGAGCT-3′
p.W1282X mut/wt: 5′-TCGACAACTTTGCAACAGTGA/GAGGAAAGCCTTTGAGCT-3′
Duchenne Muscular Dystrophy (Dystrophin):
p.R3381X mut/wt: 5′-TCGACAAAAAACAAATTTTGA/CACCAAAAGGTATGAGCT-3′
Hurler Syndrome (IDUA):
p.Q70X mut/wt: 5′-TCGACCCTCAGCTGGGACT/CAGCAGCTCAACCTCGAGCT-3′
For in vitro readthrough assays, the obtained plasmids in the presence of the tested AGs, were transcribed and translated using the TNT reticulocyte lysate quick coupled transcription/translation system (Promega™). Luciferase activity was determined 90 min post incubation at 30°C, using Dual luciferase reporter assay system (Promega™). Readthrough activity was calculated as previously described26 (Fig. 3 and 4).
For ex vivo readthrough assays, constructs harboring R3X, R245X, G542X and W1282X mutations were inserted to HEK-293 (human embryonic kidney) cells using Lipofectamine 2000 (Invitrogen), while the addition of the tested compounds was performed 6 hours post transfection. The cells were harvested following 16 hours incubation with AG using passive lysis buffer (Promega™). Readthrough activity was calculated as previously described26 (Fig. 5).
Antibacterial and Cell Toxicity Assays
Comparative antibacterial activities were determined in two representative strains of Gram-negative (E. coli R477-100, Table 1) and Gram-positive (B. subtilis ATCC-6633, Table S1) bacteria, by measuring the MIC values using the double-microdilution method according to the National Committee for Clinical Laboratory Standards (NCCLS)38 with two different starting concentration of 384 μg/mL and 6,144 μg/mL of the tested compound. All the experiments were performed in triplicates and analogous results were obtained in three different experiments.
For the cytotoxicity assays, HEK-293 (Table 1) and HFF (Table S1) cells were grown in 96-well plates (5,000 cells/well) in DMEM medium containing 10% FBS and 1% glutamine (90 μL; Biological Industries) at 37 °C and 5% CO2. Following overnight incubation, different concentrations of the tested AGs were added (10 μL per well) and the cells were incubated for further 48 hr. A cell proliferation assay (XTT based colorimetric assay, Biological Industries) was performed by using 3 hr incubation protocol, accordin,g to the manufacturer’s instructions. O.D. (optical density) was measured using an Elisa plate reader. Cell viability was calculated as the ratio between the numbers of living cells in cultures grown in the presence of the tested compounds, versus cultures grown under the identical protocol but without the tested compound. The half-maximal lethal concentration (LC50) values were obtained from fitting concentration-response curves to the data of at least three independent experiments, using GraFit 5 software39.
Eukaryotic and Prokaryotic Protein Synthesis Assays
Prokaryotic in vitro translation inhibition by different aminoglycosides was quantified in coupled transcription/translation assays27 using E. coli S30 extract for circular DNA with the pBESTluc plasmid (Promega), according to the manufacturer’s protocol. Variable concentrations of tested AG were incubated along with translation reaction (10 μl) at 37°C for 60min, ice cooled for 5 min, and diluted with a dilution reagent (tris-phosphate buffer (25 mM, pH 7.8), DTT (2 mM), 1,2-diaminocyclohexanetetraacetate (2 mM), glycerol (10%), Triton X-100 (1%) and BSA (1 mg mL−1) into 96-well plates.
Eukaryotic in vitro translation inhibition was quantified by using TNT® T7 Quick Coupled transcription/translation system with the T7 control DNA plasmid (Promega), according to the manufacturer protocol. Variable concentrations of the tested AG were incubated along with translation reaction (10μl) at 30 °C for 60 min, ice cooled for 5 min, diluted with similar dilution reagent as described above, and transferred into 96-well plates. In both prokaryotic and eukaryotic systems luminescence was measured immediately after the addition of Luciferase Assay Reagent (50 μL; Promega), and light emission was recorded with an FLx800 Fluorescence Microplate Reader (Biotek). Half-maximal inhibition concentration (IC50) values were obtained from concentration-response fitting curves of at least three independent experiments using Grafit5 software 39.
Mitochondrial Protein Synthesis Inhibition Assays
Mitochondria were isolated from HeLa cells by using Qproteome mitochondria isolation kit (Qiagen, CA), according to the manufacture’s protocol with slight modifications. Briefly, HeLa cells were grown in 10 cm dishes in Dulbecco’s modified Eagles medium (DMEM, Sigma) supplemented with 10% fetal bovine serum, without the addition of penicillin/streptomycin (Sigma), till cells reached 80% confluence. Approximately 2×107 cells were trypsinized (Biological Industries) and centrifuged at 500 g for 10 min. The cellular pellet was washed with PBS twice, and centrifuged again under the same conditions. The pellet was resuspended in 1 mL lysis buffer, incubated on ice for 10 min, and centrifugated at 1000 g for 10 min at 4°C. The pellet was resuspended in 1.5 mL ice-cold disruption buffer and centrifuging at 1000 × g for 10 min at 4°C. The lysate was transferred into a microcentrifuge tube following centrifugation at 6000 × g for 10 min at 4°C. Finally, high mitochondrial purity was obtained according to manufacturer’s protocol; the mitochondrial pellet was resuspeded in MSE buffer (220 mM mannitol, 70 mM sucrose, 5 mM MOPS, 2 mM EGTA, pH 7.0), and protein concentration was determined by the method of Bradford using bovine serum albumin as standard40.
Mitochondrial protein synthesis was measured in a 93 μL total volume containing KCl (90 mM), MgSO4 (4 mM), KH2PO4 (2.5 mM), MOPS (25 mM), pH 7.0, a mixture of the 19 L-amino acids without L-methionine (AppliChem) to a final concentration of 0.1 mM each, glutamate (20 mM), malate (0.5 mM), ADP (2 mM), BSA (1 mg/ml), emetine (5μM, an inhibitor of 80S ribosome, Sigma) and mitochondrial protein (2 mg/ml) in the presence of different concentrations of AGs (G418: 0, 0.3, 3.3, 33, 330, 3,300 μM; gentamicin and compounds 1-4: 0, 3.3, 33, 330, 3,300, 33,000 μM; compounds 5-12: 0, 8.2, 82, 820, 8,200, 88,000 μM). Pre- incubation was carried out at 37°C for 60 min followed by addition of [35S]methionine (150 μCi) and additional incubation for 45 min. Then the reaction mixture was centrifuged at 12,000 g for 15 min at 4°C, washed with MSE buffer, and centrifuged again under the same conditions. The mitochondrial pellet was resuspended in 30 μL of lysis buffer (2% sodium dodecyl sulfate, 2 mM EDTA, 0.2% (v/v) 2-mercaptoethanol, 0.05 M Tris (pH 6.8), and 10% (v/v) glycerol) and heated for 2 min at 90°C. The resulted mixture was then either stored in a freezer, or used immediately for electrophoresis or scintillation measurements31.
Autoradiography
Radioactivity was measured by acid-precipitation of the labeled proteins: the lysed mixture from the above (15 μL) was added with trichloroacetic acid (15%), methionine (1 mM) and BSA (50 μg/mL) to a total volume of 1.9 mL. The resulted mixture was incubated on ice for 60 min. The precipitated proteins were harvested onto filter paper disks (Whatman 3mm, 2.3cm) using a Tomtec harvester, washed twice with 2 mL of 5% trichloroacetic acid and the filters were dried at 60°C for 30 min. The filters were then inserted into the scintillation vials containing 5 mL of scintillation solution: toluene (1L), triton x 100 (0.5 L), 2,2′-p-Phenylene-bis(5-phenyloxazole) (0.3 gr) and 2,5-diphenyloxazole (3 gr), followed by counting on a scintillation counter.31 The half-maximal inhibitory concentration (IC50Mit) values of mitochondrial protein synthesis were determined by Grafit5 software39 (Fig. 7 and Fig. S1). To verify the suitability of the entire protocol, we used chloramphenicol (Sigma) as a control for each experiment as a routine test. The IC50Mit value we observed for chloramphenicol was in the range of 7.4 ± 0.9 μM, which is very similar to that observed (9.8 ± 0.5 μM) in the previous report14. Resolution of labeled mitochondrial proteins was also carried out by electrophoresis on 15% acrylamide gel. After electrophoresis the gel was fixed, stained by comassie blue, dried and visualized by autoradiography (Fig. 7A).
Supplementary Material
Acknowledgments
This work was supported by the NIH/NIGMS grant (1 R01 GM094792-01 A1) and GIF research grant (G-1048-95.5/2009). We thank John F. Atkins (University of Utah) for providing the p2luc plasmid and Gadi Schuster (Technion) for helping us in mitochondrial protein synthesis assays. We thank Marianna Hainrichson of our group for her assistance at the initial steps of mitochondrial protein synthesis assays. J.K. acknowledges the Schulich Postdoctoral Fellowship; D.A-G. acknowledges the Miriam and Aaron Gutwirth Memorial Fellowship; V.B. acknowledges the Ministry of Science and Technology, Israel (Kamea Program).
Abbreviations Used
- AG
Aminoglycoside
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- DMD
Duchenne muscular dystrophy
- USH
Usher syndrome
- HS
Hurler syndrome
- PTC
Premature termination codon
- PCDH
Protocadherin
- HFF
human foreskin fibroblasts
- LC50
half-maximal lethal concentration
Footnotes
Conflict of Interest: TB has proprietary and financial interests in the treatment of genetic diseases with novel aminoglycosides (PCT application # WO 2007113841 A2 20071011).
Supporting Information Available: Comparative cell toxicity (HFF cells) and antibacterial activity (B. subtilis) data, tabular representation of the data points of correlation in Figure 7, semi logarithmic plots representing dose-response effect on the inhibition of intact mitochondrial protein translation, purity determination and 1H and 13C NMR spectra of compounds 9-12. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Keeling KM, Bedwell DM. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip Rev RNA. 2011;2(6):837–852. doi: 10.1002/wrna.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Human Mutat. 2008;29(8):1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 3.Burke JF, Mogg AE. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 1985;13(17):6265–6272. doi: 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zingman LV, Park S, Olson TM, Alekseev AE, Terzic A. Aminoglycosideinduced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin Pharmacol Ther. 2007;81(1):99–103. doi: 10.1038/sj.clpt.6100012. [DOI] [PubMed] [Google Scholar]
- 5.Karpati G, Lochmuller H. When running a stop sign may be a good thing. Ann Neurol. 2001;49(6):693–694. [PubMed] [Google Scholar]
- 6.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292(5518):897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 7.Bottger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2(4):318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo J, Hainrichson M, Nudelman I, Shallom-Shezifi D, Barbieri CM, Pilch DS, Westhof E, Baasov T. Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites. ChemBioChem. 2007;8(14):1700–1709. doi: 10.1002/cbic.200700271. [DOI] [PubMed] [Google Scholar]
- 9.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. Rna. 2000;6(7):1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floquet C, Rousset JP, Bidou L. Readthrough of premature termination codons in the adenomatous polyposis coli gene restores its biological activity in human cancer cells. PLoS One. 2011;6(8):e24125. doi: 10.1371/journal.pone.0024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernikov VG, Terekhov SM, Krokhina TB, Shishkin SS, Smirnova TD, Kalashnikova EA, Adnoral NV, Rebrov LB, Denisov-Nikol’skii YI, Bykov VA. Comparison of cytotoxicity of aminoglycoside antibiotics using a panel cellular biotest system. Bull Exp Biol Med. 2003;135(1):103–105. doi: 10.1023/a:1023474719042. [DOI] [PubMed] [Google Scholar]
- 12.Hainrichson M, Nudelman I, Baasov T. Designer aminoglycosides: the race to develop improved antibiotics and compounds for the treatment of human genetic diseases. Org Biomol Chem. 2008;6(2):227–239. doi: 10.1039/b712690p. [DOI] [PubMed] [Google Scholar]
- 13.Hobbie SN, Bruell CM, Akshay S, Kalapala SK, Shcherbakov D, Bottger EC. Mitochondrial deafness alleles confer misreading of the genetic code. Proc Natl Acad Sci of the U S A. 2008;105(9):3244–3249. doi: 10.1073/pnas.0707265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother. 2006;50(6):2042–2049. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matt T, Ng CL, Lang K, Sha SH, Akbergenov R, Shcherbakov D, Meyer M, Duscha S, Xie J, Dubbaka SR, Perez-Fernandez D, Vasella A, Ramakrishnan V, Schacht J, Bottger EC. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci U S A. 2012;109(27):10984–10989. doi: 10.1073/pnas.1204073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nudelman I, Rebibo-Sabbah A, Shallom-Shezifi D, Hainrichson M, Stahl I, Ben-Yosef T, Baasov T. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg Med Chem Lett. 2006;16:6310–6315. doi: 10.1016/j.bmcl.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Nudelman I, Rebibo-Sabbah A, Cherniavsky M, Belakhov V, Hainrichson M, Chen F, Schacht J, Pilch DS, Ben-Yosef T, Baasov T. Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of diseasecausing premature stop mutations. J Med Chem. 2009;52(9):2836–2845. doi: 10.1021/jm801640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nudelman I, Glikin D, Smolkin B, Hainrichson M, Belakhov V, Baasov T. Repairing faulty genes by aminoglycosides: development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg Med Chem. 2010;18(11):3735–3746. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 19.Kandasamy J, Atia-Glikin D, Belakhov V, Baasov T. Repairing faulty genes by aminoglycosides: Identification of new pharmacophore with enhanced suppression of diseases-causing nonsense mutations. Med Chem Comm. 2011;2:165–171. [Google Scholar]
- 20.Rebibo-Sabbah A, Nudelman I, Ahmed ZM, Baasov T, Ben-Yosef T. In vitro and ex vivo suppression by aminoglycosides of PCDH15 nonsense mutations underlying type 1 Usher syndrome. Hum Genet. 2007;122(3–4):373–381. doi: 10.1007/s00439-007-0410-7. [DOI] [PubMed] [Google Scholar]
- 21.Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, Atkins JF. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol. 2000;48(2):164–169. [PubMed] [Google Scholar]
- 22.Correlation between genotype and phenotype in patients with cystic fibrosis. The Cystic Fibrosis Genotype-Phenotype Consortium. N Engl J Med. 1993;329(18):1308–1313. doi: 10.1056/NEJM199310283291804. [DOI] [PubMed] [Google Scholar]
- 23.Shoshani T, Kerem E, Szeinberg A, Augarten A, Yahav Y, Cohen D, Rivlin J, Tal A, Kerem B. Similar levels of mRNA from the W1282X and the delta F508 cystic fibrosis alleles, in nasal epithelial cells. J Clin Invest. 1994;93(4):1502–1507. doi: 10.1172/JCI117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuffery-Giraud S, Chambert S, Demaille J, Claustres M. Point mutations in the dystrophin gene: evidence for frequent use of cryptic splice sites as a result of splicing defects. Hum Mutat. 1999;14(5):359–368. doi: 10.1002/(SICI)1098-1004(199911)14:5<359::AID-HUMU1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 25.Keeling KM, Brooks DA, Hopwood JJ, Li P, Thompson JN, Bedwell DM. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10(3):291–299. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- 26.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dualluciferase reporter system for studying recoding signals. RNA. 1998;4(4):479–486. [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg WA, Priestley ES, Sears PS, Alper PB, Rosenbohm C, Hendrix M, Hung SC, Wong CH. Design and synthesis of new aminoglycoside antibiotics containing neamine as an optimal core structure: correlation of antibiotic activity with in vitro inhibition of translation. J Am Chem Soc. 1999;121:6527– 6541. [Google Scholar]
- 28.Kondo J. A Structural Basis for the Antibiotic Resistance Conferred by an A1408G Mutation in 16S rRNA and for the Antiprotozoal Activity of Aminoglycosides. Angew Chem Int Ed Engl. 2011;50:465–468. doi: 10.1002/anie.201106084. [DOI] [PubMed] [Google Scholar]
- 29.Hobbie SN, Kalapala SK, Akshay S, Bruell C, Schmidt S, Dabow S, Vasella A, Sander P, Bottger EC. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 2007;35(18):6086–6093. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3. 0 A resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 31.McKee EE, Grier BL, Thompson GS, McCourt JD. Isolation and incubation conditions to study heart mitochondrial protein synthesis. Am J Physiol. 1990;258(3 Pt 1):E492–502. doi: 10.1152/ajpendo.1990.258.3.E492. [DOI] [PubMed] [Google Scholar]
- 32.Rowe SM, Sloane P, Tang LP, Backer K, Mazur M, Buckley-Lanier J, Nudelman I, Belakhov V, Bebok Z, Schwiebert E, Baasov T, Bedwell DM. Suppression of CFTR premature termination codons and rescue of CFTR protein and function by the synthetic aminoglycoside NB54. J Mol Med. 2011;89(11):1149–1161. doi: 10.1007/s00109-011-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brendel C, Belakhov V, Werner H, Wegener E, Gartner J, Nudelman I, Baasov T, Huppke P. Readthrough of nonsense mutations in Rett syndrome: evaluation of novel aminoglycosides and generation of a new mouse model. J Mol Med. 2011;89(4):389–98. doi: 10.1007/s00109-010-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vecsler M, Ben Zeev B, Nudelman I, Anikster Y, Simon AJ, Amariglio N, Rechavi G, Baasov T, Gak E. Ex vivo treatment with a novel synthetic aminoglycoside NB54 in primary fibroblasts from Rett syndrome patients suppresses MECP2 nonsense mutations. PLoS One. 2010;6(6):e20733. doi: 10.1371/journal.pone.0020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldmann T, Rebibo-Sabbah A, Overlack N, Nudelman I, Belakhov V, Baasov T, Ben-Yosef T, Wolfrum U, Nagel-Wolfrum K. Beneficial readthrough of a USH1C nonsense mutation by designed aminoglycoside NB30 in the retina. Invest Ophthalmol Vis Sci. 2010;51(12):6671–6680. doi: 10.1167/iovs.10-5741. [DOI] [PubMed] [Google Scholar]
- 36.Goldmann T, Overlack N, Moller F, Belakhov V, van Wyk M, Baasov T, Wolfrum U, Nagel-Wolfrum K. A comparative evaluation of NB30, NB54 and PTC124 in translational read-through efficacy for treatment of an USH1C nonsense mutation. EMBO Mol Med. 2012 doi: 10.1002/emmm.201201438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Belakhov V, Kandasamy J, Baasov T, Li SC, Li YT, Bedwell DM, Keeling KM. The designer aminoglycoside NB84 significantly reduces glycosaminoglycan accumulation associated with MPS I-H in the Idua-W392X mouse. Mol Genet Metab. 2011;105(1):116–125. doi: 10.1016/j.ymgme.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.NCCLS. Fifth information supplement: Approved Standard M100-S5. NCCLS; Villanova, Pa: 1994. National Committee for Clinical Laboratory Standards, Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 39.Leatherbarrow RJ. GraFit. Vol. 5. Erithacus Software Ltd; Horley, U.K: 2001. [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.