Abstract
Antigen preparations in the form of detergent-solubilized cell lysates could, in principle, render membrane proteins (MPs) compatible with in vitro antibody engineering technologies. To this end, detergent-solubilized cell lysates were coupled with the yeast surface display platform to affinity mature an anti-transferrin receptor (TfR) single-chain antibody (scFv). Lysates were generated from TfR-expressing HEK293 cells by solubilization with detergent-containing buffer after undergoing plasma membrane-restricted biotinylation. Lysate-resident TfR was then combined with a mutagenic anti-TfR scFv library in a competitive, dissociation rate screen, and scFvs were identified with up to 4-fold improved dissociation rates on the surface of yeast. Importantly, although the lysates contained a complex mixture of biotinylated proteins, the engineered scFvs retained their TfR binding specificity. When secreted by yeast as soluble proteins, mutant scFvs bound to cell surface TfR with 3–7-fold improvements in equilibrium binding affinity. Although a known MP antigen was targeted for purposes of this study, employing biotin tagging as a means of antigen detection makes the lysate-based approach particularly flexible. We have previously shown that yeast display can be used to identify lead antibodies using cell lysate-resident MP antigens, and combined with this work showing that antibodies can also be quantitatively engineered using cell lysates, these approaches may provide a high-throughput platform for generation and optimization of antibodies against MPs.
Keywords: affinity maturation, antibody, detergent, membrane protein, yeast surface display
Introduction
Membrane proteins (MPs) represent a large and growing class of drug targets as a result of MP accessibility to circulating drugs and MP involvement in the regulation of disease states (Yildirim et al., 2007). One substantial investment in MP-targeted protein therapeutics has been the development of monoclonal antibodies (mAbs) and antibody derivatives (Reichert, 2010). Although in vivo methods of antibody engineering (e.g. cloning of antibodies from antigen-stimulated immune cells) have produced the majority of the mAbs currently approved by the FDA, in vitro display technologies offer significant advantages in throughput, and can be used to rapidly and controllably fine-tune properties such as affinity, stability and specificity (Bradbury et al., 2011; Bhatt et al., 2012). Moreover, techniques like yeast surface display have provided a powerful means of engineering antibodies with otherwise unattainable properties (Bradbury, et al., 2011). However, in vitro engineering of antibodies against MPs can be problematic, due to MP propensity for aggregation and denaturation upon removal from the lipid bilayer (White and Wimley, 1999; Privé, 2007; Lin and Guidotti, 2009). Consequently, in vitro engineering approaches have been largely hampered by the need to generate soluble MP antigens for screening.
Several approaches have been employed to overcome the solubility issues inherent to MP antigens. The use of whole cells is an effective approach for lead antibody identification; however, antibody engineering typically requires a soluble antigen (Poul et al., 2000; Wang and Shusta, 2005; Lipes et al., 2008). The most common approach to generating MP antigens in soluble form is to reduce full-length MPs to extracellular domains or peptide fragments, often a laborious, hit-or-miss process. To facilitate immobilization and/or detection, epitope tags, IgG Fc regions or biotinylation sites are routinely added to the protein termini during cloning (Powers et al., 2001; Young et al., 2012). After heterologous expression and purification, the truncated MP must be appropriately presented for screening (Freigassner et al., 2009; Young, et al., 2012). Often, this entails immobilization of the purified, soluble MP on a solid-phase support. This strategy has found widespread use in the affinity maturation of antibodies using both phage (Schier et al., 1996; Kwong et al., 2008) and ribosome (Finlay et al., 2009) display. Alternatively, antibodies against a recombinant, purified MP can be screened directly using cell-based display technologies in tandem with flow cytometry. For instance, using bacterial display, short peptide ligands against the vascular endothelial growth factor receptor were evolved for high-affinity binding (Kenrick and Daugherty, 2010). Yeast surface display has also been used for single-chain antibody (scFv) affinity maturation against MPs, including scFvs against the tumor marker carcinoembryonic antigen (Graff et al., 2004), and the epidermal growth factor receptor (Lippow et al., 2007). The yeast display platform enables quantitative screening and affinity determination directly on the yeast surface (Boder and Wittrup, 2000; Van Antwerp and Wittrup, 2000). Thus, antibody engineering via yeast surface display would be particularly powerful if it could be combined with a facile means of obtaining soluble MP antigens.
We recently demonstrated an augmented yeast display system for the discovery of MP-targeted antibodies, where MP antigens are presented in detergent-solubilized cell lysates (Cho and Shusta, 2010). Detergents have long been used in the proteomics field for extracting MPs in native or near-native states, and maintaining MP stability in solution (Linke, 2009). Solubilization of MPs directly from mammalian cell cultures eliminates the need for heterologous expression and protein purification while simultaneously presenting the antigen in solution phase. Using this approach, scFvs against MPs were isolated, antibody–antigen pairs were identified and targeted screens were performed to find scFvs against the rat transferrin receptor (TfR) (Cho and Shusta, 2010). Additional methods were detailed by which lysates could also be used to directly compare the equilibrium binding affinity of isolated scFv clones on the yeast surface (Tillotson et al., 2012).
In the current study, we demonstrate that in addition to facilitating antibody discovery, detergent-solubilized cell lysates also enable quantitative antibody engineering against MP antigens. Affinity maturation of an anti-TfR scFv was carried out by kinetic screening of a yeast surface display library against detergent-solubilized HEK293 cell lysates containing fulllength human transferrin receptor. Antigen detection was accomplished by biotinylation of all cell MPs, including TfR, prior to detergent lysis, making the lysate-based approach generalizable to other MP targets. The antibodies engineered using a lysate-based approach retained TfR specificity, possessed improved dissociation rates and exhibited improved equilibrium binding as soluble scFvs.
Materials and methods
Cells, media and plasmids
HEK293 cells (CRL-1573) were obtained from American Type Culture Collection (ATCC) and maintained at 37°C, 5% CO2, in αMEM media (M4523, Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco), 2 mM l-glutamine, 20 mM HEPES buffer pH 7.3 and 1× antibiotic/antimycotic (PSA, Gibco). Saccharomyces cerevisiae strains EBY100 (Boder and Wittrup, 1997) and AWY100 (Wentz and Shusta, 2007) were used for surface display, while strain YVH10 (Shusta et al., 1998) was used for scFv secretion. The vector pCT-ESO (Piatesi et al., 2006) provided the backbone for all scFv surface-display experiments, while plasmid pRS316-GAL-4420 (Hackel et al., 2006) provided the backbone for scFv secretion. EBY100 was grown in SD-CAA (20 g/l dextrose, 6.7 g/l yeast nitrogenous base, 100 mM sodium phosphate buffer pH 6.0 and 5.0 g/l bacto-casamino acids lacking tryptophan and uracil) AWY100 was grown in SD-CAA supplemented with 40 mg/l uracil. YVH10 were grown in SD-2×SCAA + Trp (20 g/l dextrose, 6.7 g/l yeast nitrogenous base, 100 mM sodium phosphate buffer pH 6.0, 190 mg/l Arg, 108 mg/l Met, 52 mg/l Tyr, 290 mg/l Ile, 440 mg/l Val, 220 mg/l Thr, 130 mg/l Gly and 40 mg/l Trp, lacking leucine and uracil).
Anti-TfR scFv cloning and library construction
The scFv H7 against human transferrin receptor was a kind gift from Dr James Marks at the University of California, San Francisco (Poul et al., 2000). The H7 open reading frame (ORF) was amplified by standard polymerase chain reaction (PCR) from the phage display vector pHEN-1 (Sheets et al., 1998). Plasmid pESO-H7 was constructed by homologous recombination in EBY100 yeast (Ma et al., 1987) using the H7 amplicon and (NheI/BamHI) linearized pCT-ESO backbone. Transformed EBY100 clones were screened for full-length surface display of scFv H7 using flow cytometry. Plasmid (pESO-H7) from positive clones was rescued using the Zymoprep-II yeast miniprep kit (Zymo Research) and the H7 sequence verified by bi-directional sequencing using the primers BTSeqF (5′-CTGCTCCGAACAATAAAGATTCTAC-3′) and BTSeqR (5′-GTATGTGTAAAGTTGGTAACGGAAC-3′), which encompass the entire surface display construct from Aga2p start codon to the C-terminal c-myc epitope tag. pESO-H7 was the basis for a recombinant library of scFv H7 variants. Mutagenesis was carried out by error-prone PCR (Zaccolo et al., 1996) using the nucleotide analogs 2′-deoxy-p-nucleoside-5′-triphosphate and 8-oxo-2′-deoxyguanosine-5′-triphosphate (TriLink Biotech), and primers (ESO-Forward-2 5′-GTGGAGGAGGCTCTGGTG-3′ and ESO-Reverse-2 5′-TATCAGATCTCGAGCTATT-3′) specific to pESO-H7 sequences flanking the H7 ORF. The H7 library was subsequently created by homologous recombination of the mutant scFv H7 PCR product and (NheI/BamHI) linearized pESO acceptor vector in yeast (Swers et al., 2004). Library size was determined by plating and counting of colonies. Ten random yeast colonies were selected and the frequency of mutations in the H7 scFv gene was ascertained by plasmid rescue and sequencing as described above. DNA sequencing revealed a mean of eight nucleotide substitutions per variant, a nucleotide mutation rate of ∼1%.
The wild-type and the control scFvs were subcloned into the pESO backbone as NheI/HindIII fragments using standard methods, and transformed into AWY100 yeast by the LiCl/ssDNA/PEG method (Gietz and Schiestl, 2007). AWY100 contains the LEU2 auxotrophic marker rather than the URA3 marker found in EBY100, thus the control yeast should not propagate in SD-CAA media without supplemental uracil helping to avoid wild-type or control scFv contamination of libraries during sorting (e.g. via sample carryover on the cytometer). The negative control for surface display measurements was an anti-hen egg lysozyme antibody, scFv D1.3 (VanAntwerp and Wittrup, 1998).
Creation of detergent-solubilized whole-cell lysates
HEK293 cells were prepared for lysis by washing (U-lysate), or alternatively, by biotinylation and a series of washing/quenching steps (B-lysate). Cells were grown to 80–90% confluence in 75 cm2 tissue culture-treated T-Flasks. A poly-d-lysine (P6407, Sigma-Aldrich) coating was applied, to facilitate cell attachment during washing and biotinylation steps. Ten milliliters of poly-d-lysine solution at 50 μg/ml (diluted in αMEM) were added to a the flask, incubated at 37°C for 1 h, aspirated and the flask was rinsed twice with sterile ddH2O. Cells from a single, subconfluent T-75 flask were used to create 1 ml of HEK293 cell lysate. For biotinylation of plasma MPs, cells were washed twice with 10 ml phosphate-buffered saline (PBS) containing 1 mM CaCl2 and 0.5 mM MgSO4 (PBSCM, pH7.4). Washed cells were incubated with 10 ml of a 1 mg/ml solution of Sulfo-NHS-LC-Biotin (Thermo-Fisher, dissolved in PBSCM) at 37°C for 30 min. The biotinylation solution was removed and the cells were washed twice with PBSCM containing 100 mM glycine to quench any excess biotinylation reagent, then once with PBS (pH7.4). Cells not undergoing biotinylation were prepared for lysis by washing twice in PBSCM and once in PBS.
Cell lysis buffer was prepared by supplementing PBS with 1% (v/v) Triton X-100 (Thermo-Fisher), 2 mM EDTA and 1× protease inhibitor cocktail (P8340, Sigma-Aldrich). Cell lysates were created by the addition of 1 ml of ice-cold lysis buffer to each prepared T-75 flask of HEK293 cells (∼1 × 107 cells). Working at 4°C, cells were scraped from the growth surface using a cell scraper, and collected by micropipette into a 1.5-ml microfuge tube. The lysed cells were briefly vortexed, rotated at 4°C for 15 min and then centrifuged at 4°C, 14 000 rpm for 30 min to remove insoluble material. The supernatant comprising detergent-solubilized whole-cell lysate was collected to a new 1.5-ml tube and used immediately or stored overnight at 4°C for use the following day.
Lysate-based kinetic screening using fluorescence-activated cell sorting
Yeast for surface display were grown in the appropriate medium at 30°C, 260 rpm, overnight. The following day, cultures were diluted to an OD600 nm of 0.3 and grown for ∼4h until the OD600 nm reached 1.0. Induction of surface display was accomplished by replacing SD-CAA with an equivalent volume of SG-CAA (SD-CAA with 20 g/l d-galactose instead of dextrose) and incubating at 20°C, 260 rpm for 16–20 h. Finally, induced yeast were washed three times in ice-cold PBS containing 1 g/l bovine serum albumin (PBS-BSA). Unless specifically noted, PBS-BSA was the default buffer used for all washing, dilution and resuspension steps.
For the first round of sorting, the starting library (R0) contained roughly 5 × 107 members and was sorted without kinetic competition to obtain a library enriched in lysate-binders (Fig. 3a(iii)). A total of 8 × 107 induced yeast cells were incubated in 2 ml B-lysate for 2 h to promote scFv binding of biotinylated transferrin receptor (B-TfR); 2 × 106 control yeast (H7 or D1.3) were incubated with 50 µl B-lysate. This ratio (50 μl B-lysate per 2 × 106 yeast) was held constant through all rounds of sorting. Also, all incubations with lysate were carried out at room temperature, on a rotating mixer. Following incubation with B-lysate, the yeast were washed two times in PBS containing 1% Triton X-100 (PBSTX) to remove non-specifically bound lysate. After washing, yeast were prepared for fluorescence-activated cell sorting (FACS); all subsequent washing and immunolabeling steps were carried out at 4°C to prevent further antigen dissociation. Yeast were washed twice in PBS-BSA to remove detergent and incubated with detection antibodies. Full-length expression was detected using a rabbit polyclonal antibody against the c-myc epitope tag (Thermo-Fisher, diluted 1:1000), followed by a goat anti-rabbit allophycocyanin (APC)-conjugated secondary antibody (Invitrogen, diluted 1:500). Biotinylated antigen binding was detected by a mouse monoclonal anti-biotin antibody (Labvision, clone BTN.4, diluted 1:50) followed by a goat anti-mouse Alexa488-conjugated secondary antibody (Invitrogen, diluted 1:500).
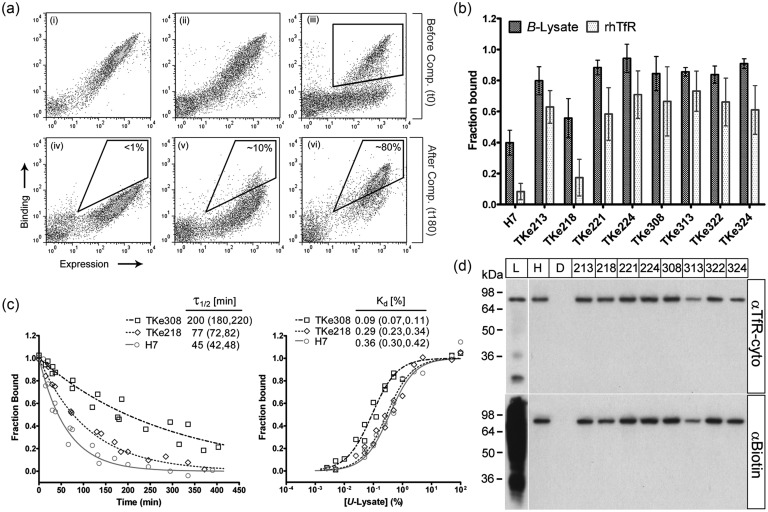
Fig. 3.
Lysate-based screening of a mutagenic H7 library for scFv having improved dissociation rate. (a) Flow cytometric dot plots depict the behavior of the various scFv populations after labeling with B-lysate before (top row) and after 180 min of competition with U-lysate (bottom row). TfR binding was monitored by anti-biotin antibody and expression monitored by anti-c-myc antibody. (i and iv) Wild-type H7; (ii and v) R1 library; (iii) library R0 with gate indicating the population isolated to yield the R1 library; (vi) library KS4. In the bottom row, a sample sort gate with inset percentages illustrates the enrichment of clones that retain binding to B-lysate after competition. (b) Individual clones isolated from the KS4 pool (TKe clones) were assayed for their relative single-time-point dissociation using flow cytometry. The black bars indicate the fraction of B-lysate remaining bound to yeast displayed scFv after 70 min competition with U-lysate. The gray bars indicate the fraction of recombinant human TfR remaining bound after 70 min competition with diferric human transferrin. Error bars represent the 95% confidence interval (CI) from five independent experiments. All clones possessed an increased fraction bound after 70 min in both assays compared with the wild-type (P< 0.05), except for TKe218 as determined by an unpaired Student's t-test. (c) Quantitative dissociation kinetics and equilibrium binding of selected scFv clones using detergent-solubilized lysate as source of TfR as measured on the yeast surface. For the dissociation rate experiments, TfR binding was determined by anti-biotin antibody as a function of competition time with U-lysate. Data from two independent experiments are plotted along with the fitted monoexponential dissociation curves. At minimum, two independent experiments were used to calculate the fit and the inset dissociation half-times and (95% CI). For the equilibrium titrations, the anti-TfR antibody (H68.4) was used to track TfR binding in U-lysate, and the data were normalized to a wild-type Kd of 0.36% as determined in Fig. 2c. Data from two independent experiments are plotted along with each fitted equilibrium binding isotherm. Kd and (95% CI) were computed using data from five independent experiments. (d) Yeast display immunoprecipitation of TfR from HEK293 cell lysate. Clones in the TKe cohort (numbered columns), along with H7 (H) and irrelevant scFv, D1.3 (D), were used to immunoprecipitate TfR from detergent-solubilized B-lysate (L). All TKe clones pull down the same ∼84 kDa band as H7, which corresponds to biotinylated TfR as evidenced by western blotting with either anti-TfR (H68.4) or anti-biotin antibodies.
Following isolation of the enriched library (R1), four rounds of sorting were carried out (KS1-KS4) which included a kinetic competition step to isolate binders with improved dissociation rates. For KS1, the R1 pool was incubated with B-lysate. All yeast were washed two times with PBSTX, then yeast were incubated with excess soluble competitor (100 μl U-lysate per 2 × 106 yeast) for the calculated optimal competition time (180 min, see Results). After washing twice with PBSTX, the yeast were labeled for FACS as described above. Sorting rounds KS2-KS4 were carried out in exactly the same manner as KS1, using yeast recovered during the previous round of sorting.
The screening procedures described above were used with minor modifications to measure single-time point-dissociation, relative off-rates and relative affinities. For single-time-point dissociation experiments, individual clones were saturated with B-lysate then competed with U-lysate for 70 min instead of the full competition time. To measure relative off-rates, a large volume of yeast was saturated with B-lysate. Excess U-lysate was applied, and small aliquots (1 × 106 yeast) were removed at various competition times ranging from 10 to 400 min. At each time point, the yeast were washed with ice-cold PBSTX, pelleted and stored on wet ice until the final time point. Labeling antibodies were identical to those used in the screening process above. Analytical flow cytometry (BD FACSCalibur) was used to measure fluorescently labeled yeast cells and geometric mean fluorescence intensities of the antigen-binding population (MFI, background fluorescence from non-displaying yeast subtracted) were quantified with the FlowJo software package. For the single-time-point experiments, fraction bound was calculated by dividing the MFIt0 by MFIt70. For relative off-rates, the MFI values at each time point were fit to a mono exponential decay model to determine the dissociation rate constant (koff).
Equilibrium binding affinities in lysate were measured on the yeast surface essentially as described previously (Tillotson et al., 2012). For the initial measurement of scFv H7 affinity, B-lysate was diluted serially in PBSTX from 100% down to 0.005%. After 2 h incubation, yeast were washed twice with PBSTX and labeled for FACS as described above. To determine the equilibrium dissociation constant (Kd), MFIs of the antigen-binding population were fit to a simple, monovalent, ligand/receptor-binding model. After library sorting was completed, relative binding affinities were measured by the same procedure except for U-lysate was used and TfR binding was detected by mouse anti-human TfR MAb H68.4 (Invitrogen, diluted 1:250) which binds to an epitope in the cytoplasmic tail of the transferrin receptor, followed by goat anti-mouse Alexa488-conjugated secondary (Invitrogen, diluted 1:500). This anti-TfR antibody was also used as a diagnostic tool to specifically confirm antigen binding at various points in the screening process. For all calculations in this manuscript, data from multiple independent experiments on each of a minimum of 2 days were collectively used to determine quantitative parameters and their associated 95% confidence intervals. The resultant total number of independent experiments for each assay is denoted in the respective figure legend.
Recombinant TfR labeling and holo-Tf competition
Recombinant human transferrin receptor, soluble, extra-cellular portion (rhTfR) was obtained from R&D systems, along with a mouse anti-human TfR antibody recognizing the TfR extra-cellular region (MAb2474, clone 29806). Human holo-transferrin (diferric, iron-saturated) was obtained from Sigma-Aldrich. For the competition assay, yeast cells were prepared as described above. A total of 1 × 106 prepared yeast were incubated with 50 µl of 40 nM rhTfR, rotating, at room temperature, for 2 h. Yeast were washed three times in PBS-BSA and incubated in 50 µl of 10 µM holo-Tf competitor for 70 min at room temperature with rotation. As before, immunolabeling was carried out on ice to prevent further antigen dissociation. rhTfR binding was detected by mouse anti-hTfR MAb2474 (R&D Systems, diluted 1:200) followed by goat anti-mouse Alexa647-conjugated secondary antibody (Invitrogen, diluted 1:500). ScFv expression was detected using a rabbit polyclonal antibody against the c-myc epitope tag (Thermo-Fisher, diluted 1:1000), followed by a goat anti-rabbit Alex488-conjugated secondary antibody (Invitrogen, diluted 1:500). MFI of the rhTfR binding populations were quantified as described above, and the fraction bound was calculated by dividing the MFIt0 by MFIt70.
Secretion and purification of soluble scFv
ScFv-containing plasmids were isolated by yeast miniprep as described above, and the ORFs were subcloned into pRS316-GAL-4420 as NheI/HindIII restriction fragments. The resulting order of elements, from N- to C-terminus is: synthetic pre-pro- leader, scFv heavy chain, G4S linker, scFv light chain, c-myc and 6-histindine tag. Miniprepped plasmid DNA was transformed into the YVH10 yeast strain by the LiCl/ssDNA/PEG method (Gietz and Schiestl, 2007). For scFv secretion, single clones were inoculated into SD-2×SCAA + Trp medium for 72 h. Secretion was induced by replacing SD-2×SCAA + Trp with an equivalent volume of SG-2×SCAA + Trp (SD-2×SCAA + Trp with 20 g/l d-galactose instead of dextrose) and incubating at 20°C, 260 rpm, for 72 h. At the end of the induction period, cultures were centrifuged and the scFv-containing supernatant was removed. The supernatant was dialyzed against TRIS-buffered saline (TBS, 25 mM Tris, 150 mM NaCl, 2 mM KCl, pH 7.9), and purified by nickel-ion affinity chromatography (IMAC) essentially as described (Hackel et al., 2006), with the exception that all IMAC solutions were buffered with 50 mM TRIS, pH 7.9, rather than phosphate. ScFv production and purity were analyzed by SDS-PAGE and Western blotting (Supplementary Fig. S1a), while total protein concentration was measured using the Bradford reagent (Sigma-Aldrich). Purified scFv concentrations were 1–2 mg/l. The absence of aggregated or oligomerized species was confirmed by non-reducing SDS-PAGE and western blotting, and by size-exclusion chromatography of purified scFv (Supplementary Fig. S1b). The anti-FITC scFv 4–4-20, already present on the pRS316-GAL-4420 plasmid was secreted, purified and employed as a negative control.
To assay for specific activity of the purified scFv, Streptavidin-coated COMPEL™ paramagnetic particles (∼6 µm diameter, Bang's Labs) at a concentration of 5 × 106 particles/ml, were incubated with biotinylated mouse anti-c-myc MAb (clone 9E10, Millipore, diluted 1:250) in TRIS-buffered saline containing 1 g/l BSA (TBSA, 25 mM TRIS, 150 mM NaCl, pH 7.2) overnight, at room temperature. 9E10-coated particles were washed three times in TBSA, divided into aliquots (5 × 104 particles each) and dispensed into 96-well plates. All subsequent steps were completed using ice-cold reagents with the plate resting on ice. Two hundred microliters of purified scFv at a concentration of 200 nM (an amount known to saturate available 9E10 on the bead surface) were added to each experimental well and allowed to equilibrate for 2 h. The particles were washed once with TBSA, resuspended in 200 µl of rhTfR (40 nM, saturating) and equilibrated for 1 h. Particle-bound scFv was detected using a rabbit anti-his6 polyclonal antibody (Thermo-Fisher, diluted 1:100) followed by goat anti-rabbit APC conjugate (Invitrogen, diluted 1:500). rhTfR binding was detected by mouse anti-hTfR PE conjugate (R&D systems, diluted 1:10). The activity of each scFv was defined as the ratio of PE fluorescence (TfR binding) divided by the APC fluorescence (scFv amount). ScFv clones were compared with H7 and the negative control 4-4-20, assayed in the same independent experiment (Supplementary Fig. S1c).
Equilibrium binding analysis and immunocytochemistry
At 72 h prior to experimentation, HEK293 cells were seeded at 5 × 104 cell/well, in 24-well poly-d-lysine-coated tissue culture plates. On the morning of the experiment, cells were washed three times in warmed PBSCM then starved of iron: growth media was removed, and cells were incubated at 37°C in warmed serum-free media (same formulation as growth media, without FBS) containing the iron-chelator 10 µM deferoxamine mesylate (Sigma-Aldrich). After starvation, cells were washed three times in PBSCM containing 10% goat serum (PBSCMG, serum from Sigma-Aldrich) and placed on wet-ice. All subsequent steps were carried out with ice-cold reagents, and the cells remained on wet ice throughout. Soluble, monomeric scFv was serially diluted in PBSCMG. Two-hundred microliters of the appropriate concentration was added to each well of cells, and allowed to equilibrate for 2 h.
For equilibrium binding analysis, the scFv concentration ranged from 0.5 pM to 200 nM. To detect bound scFv, cells were further labeled with a rabbit polyclonal anti-c-myc antibody (diluted 1:500 in PBSCMG) and a goat anti-rabbit APC conjugate (diluted 1:250 in PBSCMG). After washing, 500 µl of SYTOX green solution (diluted 1:5000 in PBS) was added to each well, and the HEK293 cells were detached by scraping with the base of a micropipette tip. Cells were analyzed by flow cytometry. At least 10 000 live-cell events (as determined by absence of SYTOX green staining) were collected. Fraction of cell-surface TfRs bound by scFv was quantified using fluorescence data (MFI) from the APC (anti-c-myc) channel. After subtraction of non-specific binding (due to negative control scFv 4-4-20), MFI values were fit to a bimolecular equilibrium binding model to determine the monovalent dissociation binding constant (Kd). For immunocytochemistry, the scFv concentration was 200 nM. Labeling was identical to the equilibrium binding analysis (anti-c-myc followed by APC conjugate). After washing, labeled cells were fixed in 4% paraformaldehyde and examined using a fluorescent microscope (Olympus IX70).
Yeast-display immunoprecipitation
Yeast were prepared, and yeast-display immunoprecipitation (YDIP) was carried out essentially as described before (Cho et al., 2009). Briefly, 3 × 107 yeast were incubated with 200 µl B-lysate at room temperature for 2 h. Antigens were eluted with 30 µl of 0.2 M glycine pH 2.0, and neutralized to pH 7.2 with 5 µl of 1 M TRIS-base. Ten microliters of neutralized YDIP eluent were diluted in reducing sample buffer, denatured by boiling for 5 min and separated by SDS-PAGE on 12% Tris-Glycine gels. After transfer to nitrocellulose, western blots were probed with anti-biotin horseradish peroxidase (HRP) conjugate (Cell Signaling, diluted 1:1000), or either of the mouse anti-TfR antibodies (H68.4 or MAb2474, diluted 1:1000) followed by anti-mouse HRP conjugate (Sigma-Aldrich, diluted 1:2000) and developed using electrochemiluminescence reagents and Hyperfilm (GE Healthcare).
Results
Strategy for antibody screening with cell lysate-based TfR
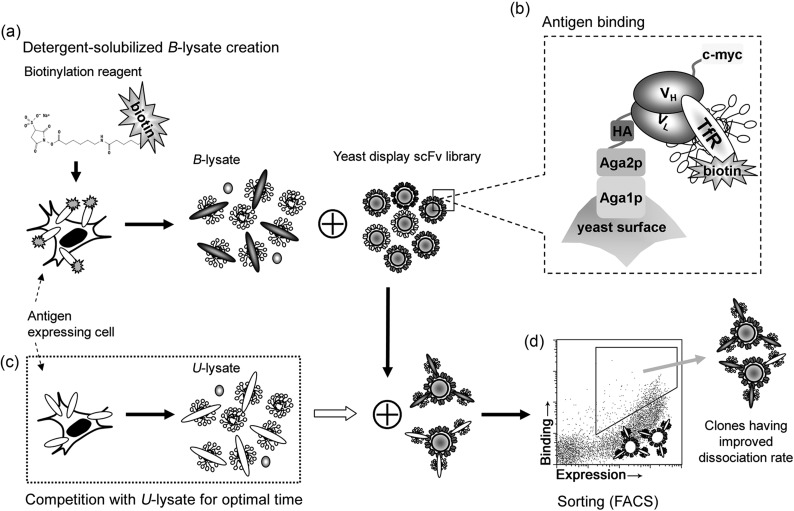
The MP antigen chosen for this study was the human TfR, a transmembrane glycoprotein that mediates iron uptake through binding to its ligand, the iron-carrier protein transferrin (Tf) (Ponka and Lok, 1999). The anti-TfR antibody selected for affinity maturation was scFv H7, previously identified by Poul et al. (2000) by phage panning against human breast cancer cells. After subcloning the H7 scFv into the yeast surface display vector, the wild-type scFv H7 was used to validate the lysate-based screening framework, and to generate quantitative screening parameters. Figure 1 depicts the core steps of lysate-based screening: (a) lysate creation and target MP biotinylation, (b) binding of detergent-solubilized antigen by yeast-displayed scFv, (c) screening by kinetic competition with unlabeled lysate, and (d) FACS to isolate improved clones. Since the H7 scFv is directed against human TfR, the human embryonic kidney 293 cell line (HEK293) was used as a source for lysate creation (Fig. 1a). In addition, HEK293 cells are adherent which is advantageous as they remained immobilized during biotinylation. Biotinylation of MPs was employed as a flexible means of detecting detergent-solubilized MP antigens in the cell lysate. HEK293 cells were either biotinylated using a cell-impermeable reagent to allow MP antigen detection (B-lysate) or left unlabeled to create solubilized MP competitor (U-lysate) for use in the kinetic screen. Detergent lysis and extraction of soluble MPs was performed in 1% (w/v) Triton-X100 buffer, which is known to extract TfR (Schuck et al., 2003).
Fig. 1.
Affinity maturation of an anti-TfR scFv by dissociation rate screening of a mutagenic, yeast display scFv library with antigens presented in a detergent-solubilized cell lysate. (a) Lysate creation consisted of plasma membrane-selective biotinylation and subsequent lysis in a buffered detergent solution. A cell-impermeable biotinylation reagent was used to tag plasma MPs, including the desired antigen (TfR), yielding biotin-tagged lysate (B-lysate). If the antigen-expressing cells were not biotinylated prior to detergent lysis, unlabeled cell lysate (U-lysate) that can act as the soluble competitor in a kinetic screen resulted. (b) An anti-TfR yeast display library was allowed to bind antigen present in the B-lysate. Antigen binding was detected using antibodies or streptavidin conjugates against the biotin tag, or antibodies against TfR. Full-length scFv expression was detected by an antibody directed against the C-terminal c-myc tag. (c) A saturating concentration of B-lysate was applied in step (b), and after washing, an excess of U-lysate was applied in step (c) for a predetermined competition time. (d) ScFvs that retain B-lysate binding, as distinguished by both biotin and c-myc signals, were isolated by flow cytometry to recover mutant scFvs that have a reduced dissociation rate.
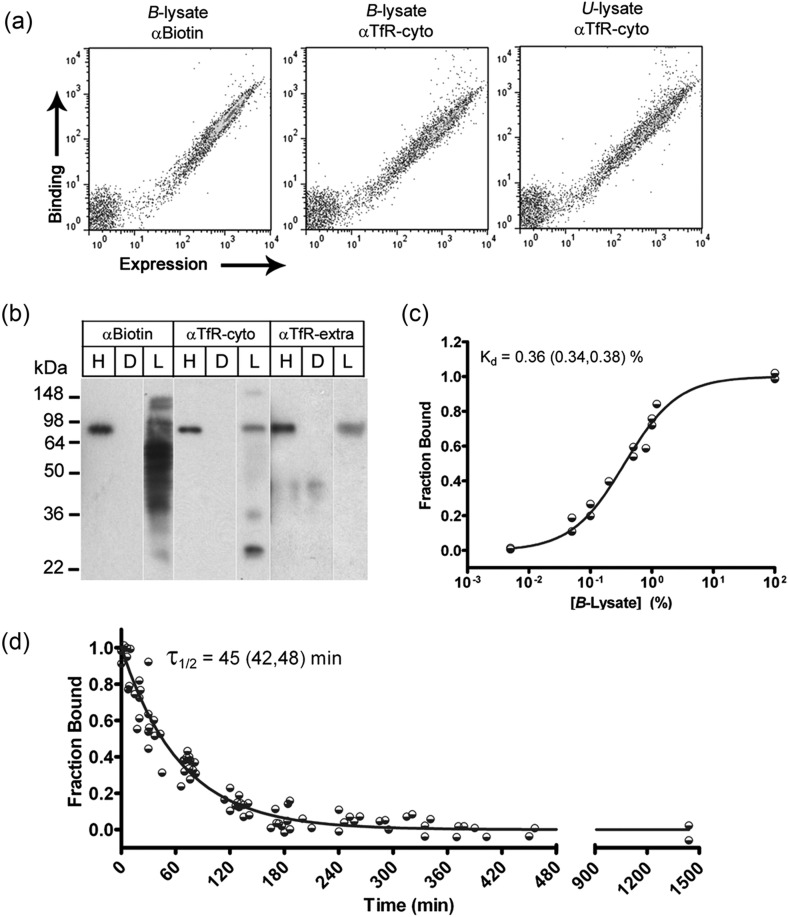
Next, yeast cells displaying H7 on their surface were mixed with either B-lysate or U-lysate to assay for TfR binding (Fig. 1b). Importantly, H7 captured biotinylated TfR from cell lysates and the biotinylation did not negatively affect TfR capture. B- and U-lysate demonstrated similar levels of binding to H7 as measured by an anti-TfR antibody recognizing the cytoplasmic tail of TfR (Fig. 2a). To further elucidate the TfR binding specificity of H7 in the context of a complex cell lysate having many different biotinylated proteins, YDIP was performed (Cho et al., 2009). Surface-display of H7 and scFv D1.3 (recognizing irrelevant antigen, hen-egg lysozyme) were used in a YDIP reaction with B-lysate. Western blotting with anti-biotin and two different anti-TfR antibodies confirmed that H7 specifically binds TfR in the B-lysate (Fig. 2b). Also, based on the expected YDIP product size of 84 kDa, and the fact that the anti-TfR antibodies recognize distal epitopes (H68.4 recognizes an epitope in the TfR cytoplasmic tail, and MAb2474 recognizes an extracellular epitope), the captured TfR was most likely intact. It should be noted that while anti-TfR antibodies could be used as a means for H7 binding detection on the surface of yeast and hence, for screening (Fig. 1a), here they were used solely as a confirmatory diagnostic tool of the more generalizable biotinylation approach.
Fig. 2.
Wild-type H7 scFv was displayed on the surface of yeast and bound its antigen (TfR) in Triton X100-solubilized HEK293 cell lysates. (a) Using flow cytometry, full-length H7 display was detected by c-myc expression, and binding to TfR was assessed in both B-lysate and U-lysate, using either an anti-biotin antibody (αBiotin) or an anti-TfR antibody directed against the cytoplasmic tail of TfR (H68.4, αTfR-cyto). (b) YDIP was performed using yeast displaying H7 (H), or irrelevant scFv, D1.3 (D). Western blot of immunoprecipitation products from B-lysate (L) was performed using the indicated antibodies, including an antibody recognizing the extracellular domain of TfR (MAb2474, αTfR-extra). An ∼84 kDa band corresponding to TfR monomer was detected in the H7 YDIP product using each of the detection antibodies, but TfR was not detected in the YDIP product of irrelevant scFv D1.3. (c) Equilibrium titration of detergent-solubilized TfR using yeast-displayed H7. B-lysate was used as the antigen source, and antigen binding was detected by anti-biotin antibody. Data from two independent experiments are shown along with a line representing the fitted equilibrium binding isotherm. The plotted fit along with inset Kd and (95% confidence interval, CI) were computed using data from eight independent experiments. (d) Dissociation kinetics of the H7–TfR binding interaction. Data from 10 independent experiments are shown along with line representing the fitted solution of a mono-exponential dissociation curve that yields the inset half-time of dissociation and (95% CI).
As with yeast display affinity maturation procedures employing soluble antigens, both equilibrium and kinetic screening are possible using the lysate-based approach. Equilibrium binding affinity in terms of percent lysate was estimated by titrating B-lysate on yeast-displayed H7 and directly analyzing the yeast by flow cytometry (Fig. 2c). The equilibrium binding affinity (Kd) was ∼0.36% lysate (v/v) on the surface of yeast. Secreted and purified H7 scFv bound the TfR residing on HEK-293 cells with an affinity of 1.25 nM (Table I). Knowledge of Kd was an important decision point for the development of an affinity maturation screen, and the Kd measurements above pointed toward a kinetic screen since optimal equilibrium screening would require a nearly 3000-fold dilution of the lysate, leading to excessively large reaction volumes to remain at conditions of molar TfR excess (Boder and Wittrup, 1998). Moreover, it was previously proposed that antibodies with an equilibrium binding affinity (Kd) <10 nM are optimally screened by dissociation kinetics (Boder and Wittrup, 1998). Given these rationale, a kinetic screen was chosen for this study. Screening based on ligand dissociation kinetics depends on a second soluble antigen that can act as a competitor (Boder et al., 2000). As described earlier, lysate-based screening provides a straightforward solution, as the U-lysate (prepared without biotinylation as depicted in Fig. 1a) can act as the soluble competitor. Fig. 1c depicts the competitive binding portion of the screen. As biotinylated TfR dissociates from H7, excess U-lysate containing TfR, but lacking a biotin tag, competes for the H7 binding sites, and the anti-biotin signal decreases as a function of dissociation time. Using flow cytometry, the H7-associated biotin-TfR signal was quantitatively monitored, yielding a dissociation half-time of 45min (Fig. 2d, koff = 0.022/min). Taking into account signal-to-background fluorescence values, and targeting a modest 4-fold improvement in mutant H7 dissociation rate to help ensure the recovery of improved clones, the optimal incubation time with soluble competitor (tcomp) was calculated to be 180min (Boder and Wittrup, 1998). Having validated the lysate-based screening strategy and associated screening parameters, we proceeded to create a mutagenic library of scFv H7 variants.
Table I.
Mutation and binding properties of the TKe cohort
| Amino acid | CDR | H7 | TKe213 | TKe218 | TKe221 | TKe224 | TKe308 | TKe313 | TKe322 | TKe324 |
|---|---|---|---|---|---|---|---|---|---|---|
| H45 | L | P | P | P | ||||||
| H56 | H2 | N | K | D | ||||||
| H57 | H2 | K | R | |||||||
| H102 | H3 | Y | H | |||||||
| H108 | L | P | ||||||||
| L8 | P | |||||||||
| L66 | K | R | Q | E | R | Q | Q | Q | ||
| L77 | S | G | ||||||||
| L95a | L3 | T | A | A | A | |||||
| Kd (nM) (95% CI) | 1.25 (1.17, 1.33) | 0.23 (0.19, 0.27) | 1.03a (0.86, 1.20) | 0.21 (0.19, 0.23) | 0.37 (0.29, 0.45) | 0.18 (0.15, 0.21) | 0.25 (0.22, 0.28) | 0.27 (0.23, 0.30) | 0.38 (0.32, 0.44) |
Residues in VH and VL were numbered according to the Kabat convention in the first column, with CDRs noted in the second column. Not depicted in the table is a Met to Val mutation at the N-terminus of the TKe213, TKe218 and TKe224 scFvs that is located in the multicloning site and is not part of the germline antibody sequence. Equilibrium dissociation constants (Kd) and associated 95% confidence intervals (95% CI) are given for each clone. All Kd values were derived by fitting a bimolecular equilibrium binding model with data derived from, at minimum, six independent experiments.
aKd for TKe218 was not statistically significant versus H7, while all other mutants had statistically improved Kd as determined by an unpaired Student's t-test (P < 0.05).
Library generation and screening
The entire wild-type H7 ORF was randomly mutagenized using nucleoside analogs (see Materials and methods), yielding a yeast display library of ∼5 × 107 H7 mutants. The yeast surface display library was incubated with B-lysate and analyzed for TfR binding by flow cytometry. The initial H7 library (R0) exhibited both binding to B-lysate (anti-biotin antibody, Fig. 3a(iii)) and specific binding to TfR (anti-TfR antibody H68.4, data not shown). However, a sizeable portion of the R0 library, while expressing full-length scFv (detected by anti-c-myc primary antibody), exhibited loss of binding to B-lysate upon mutagenesis (Fig. 3a(iii)). Thus, a single round of FACS was used to isolate the roughly 35% of double-positive yeast (B-lysate binding and full-length expression, gated region in Fig. 3a(iii)). The resulting population (library pool R1, 3a-ii), containing approximately 1.5 × 107 members, was the starting pool employed for kinetic screening. Four rounds of competitive kinetic selection (termed as KS1 through KS4) using the optimal competition time with B-lysate were used to isolate a pool of H7 mutants with a putative slower dissociation rate than wild-type. The flow cytometric dot plots in Fig. 3 illustrate pre- and post-competition B-lysate binding signal for H7 (Fig. 3a(i) and (iv)) and library pool R1 (Fig. 3a(ii) and (v)), with a sample sort gate indicating the improved population isolated by flow cytometry. After four rounds of kinetic selection were completed, the resultant KS4 pool was substantially enriched in mutants capable of retaining much of their TfR binding capacity after 180min of competitive dissociation (Fig. 3a(vi)).
Characterization of recovered KS4 clones
A total of 48 clones were randomly selected from the KS4 pool for additional characterization. Initial characterization involved single-time-point competitive dissociation experiments using the lysate-based framework. We measured the B-lysate binding signal at a U-lysate competition time of 70min, offering a snapshot of the dissociation rate at a shorter time where we would expect to see even modestly improved clones above the wild-type background (Fig. 3b). Of the 48 clones, 39 exhibited a decrease in 70min dissociation (Fig. 3b), echoing the enrichment observed in the KS4 binding pool (Fig. 3a(vi)). These improved mutants are referred to as TKe mutants (TfR binding, kinetically enhanced). DNA sequence data were also considered in the initial characterization of the KS4 pool. Of the 48 clones subjected to the lysate-based competition assay, 15 were found to have unique amino acid sequences. In addition to several mutations occurring in the complementarity-determining regions (CDRs), there were three positions where mutations occurred with high frequency (Table I). Lysine at position L66 which lies between CDRL2 (L50-L56) and CDRL3 (L89-L97) was mutated in 20/48 clones. Another frequently encountered mutation, occurring in 17/48 clones was L(H45)P, which is located adjacent to CDRH2 (H50-H65). The third high-abundance mutation (14/48 clones) T(L95a)A occurred in CDRL3. The TKe cohort represented in Fig. 3b and Table I is comprised of the eight clones from the KS4 pool which best represented the observed mutations, especially the high-frequency and CDR mutations. Of the eight clones, seven exhibited significantly reduced dissociation rates in the single-time-point assay (80–95% lysate binding maintained after 70min competitive dissociation compared with the wild-type H7 at 40%). TKe218 was the one clone that did not exhibit a statistically significant change in dissociation for the single-time-point assay (P > 0.05).
A second characterization assay, decoupled from the cell lysate-based approach, was also used to evaluate the dissociation characteristics of the TKe cohort. Holo-transferrin (Tf) has been shown to compete with H7 for binding to TfR (Poul et al., 2000). Thus, we reasoned that holo-Tf, along with recombinant, soluble human TfR extracellular domain (rhTfR), could be used in a competitive dissociation assay analogous to that performed with lysate-derived TfR. Indeed, holo-Tf was able to competitively dissociate soluble rhTfR from H7 displaying yeast as well as from the TKe mutants (Fig. 3b). While the dissociation proceeded more quickly than that driven by lysate-derived TfR competition, the TKe cohort exhibited the same general trend of reduced dissociation rate in both assays. Similar to the lysate-based dissociation experiment, seven of the eight TKe scFv showed substantially decreased dissociation from rhTfR during this timeframe, with TKe218 again being statistically indistinguishable from wild-type H7 (Fig. 3b, P > 0.05). We further verified that the single-time-point measurements were an estimate of relative dissociation rates by using the lysate-based framework to monitor the quantitative dissociation kinetics and equilibrium binding characteristics of clones TKe218 and TKe308 relative to H7 (Fig. 3c). TKe308 had a 4.4-fold improved off-rate while TKe218 had a reduced 1.7-fold improved off-rate, correlating well with the single-time-point data. Again using lysate-based TfR in the yeast display format, the relative equilibrium binding affinity was measured to determine if improvements in off-rate would correlate to affinity. Indeed, the Kd of TKe308 decreased by 4.2-fold while TKe218 experienced a modest, but statistically indistinguishable, 1.2-fold decrease (P > 0.05).
Finally, antigen specificity was assayed. Not only is the lysate a crude mixture which contains many biotinylated proteins, but the H7 library contains scFvs with random mutations. Thus, the possibility existed that isolated scFvs could have lost some of their antigen specificity or could have bound a biotinylated MP other than TfR. Although the soluble rhTfR could be competed off of the mutant scFvs with Tf, suggesting maintenance of the binding of the mutant H7s for TfR, the YDIP assay was used to confirm that the TKe cohort had retained the same TfR specificity observed with wild-type H7. Immunoprecipitation experiments using the various TKe mutants clearly demonstrated that each clone specifically bound biotinylated TfR from B-lysate, and the YDIP elution products were free of non-specifically bound, biotinylated proteins (Fig. 3d).
Production and characterization of soluble scFv
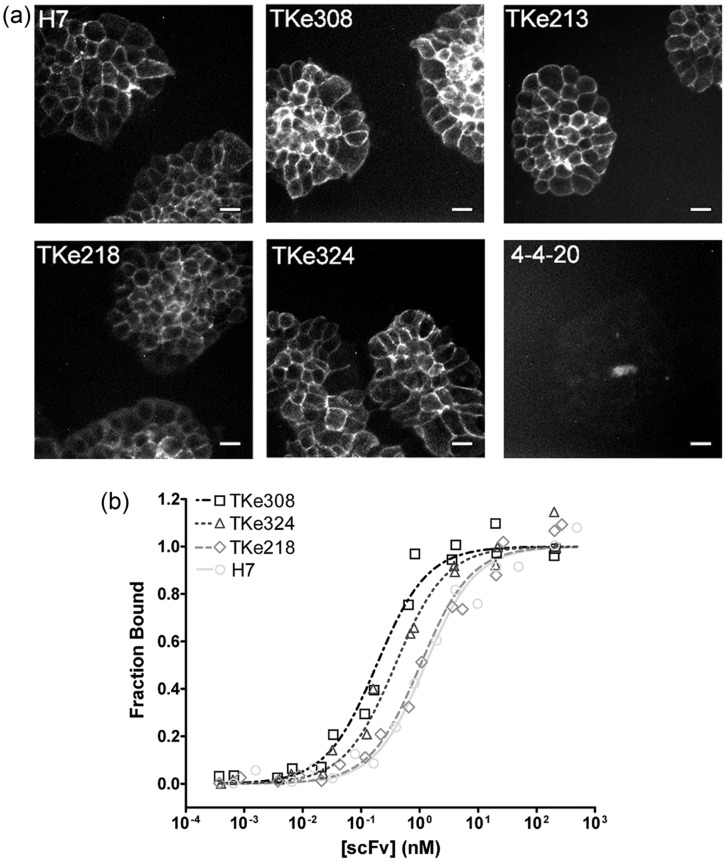
Although the TKe cohort had improved dissociation kinetics and equilibrium binding affinity on the yeast surface, it was important to confirm the effects using soluble scFv in a physiological context, and in the absence of detergent. Each member of the TKe cohort was produced as secreted, soluble scFv protein using yeast, and purified by IMAC for further testing (see Materials and methods and Supplementary Fig. S1a). The purified scFv was monomeric as verified by size exclusion chromatography combined with Western blotting, and the specific activity of the mutant scFv was indistinguishable from wild-type H7 (see Materials and methods and Supplementary Fig S1b and c). Purified TKe and H7 scFv were then used to immunolabel TfR on the surface of live HEK293 cells. Fig. 4a shows representative immunocytochemistry images with the TKe mutants yielding analogous immunolabeling patterns to that of H7. Affinity titrations of purified scFvs on HEK293 cells revealed that the dissociation rate engineering using cell lysate translated to a 3-7-fold improvement in scFv binding affinity under physiological conditions and in the absence of detergent (Fig. 4b and Table I). As suggested by the lysate-based equilibrium binding data, TKe218 has a binding affinity that is not statistically different from H7. The other members of the TKe cohort that possessed substantially improved dissociation rates on the yeast surface had monomeric binding affinities ranging from TKe324 at 0.38 nM (3.4-fold improvement) to TKe308 at 0.18 nM (a 7-fold improvement) as soluble proteins.
Fig. 4.
Measurement of soluble scFv properties in detergent-free conditions. (a) Immunofluorescent detection of scFv binding to live HEK293 cells. Monomeric scFvs at saturating 200 nM concentrations were incubated with the HEK293 cells on ice to prevent internalization of the scFv–TfR complex. Surface immunolabeling was detected by an anti-c-myc antibody and fluorescent secondary antibody, and the anti-fluorescein scFv, 4-4-20, was used as a negative control. Scale bar represents 20 μm. (b) Equilibrium binding of soluble scFv to the HEK293 cell surface. Purified, monomeric scFv was titrated on live HEK293 cells. Data from two independent experiments are plotted along with each fitted equilibrium binding isotherm derived from, at minimum, six independent experiments. Quantitative Kd results for the entire TKe cohort can be found in Table I.
Discussion
Detergent-solubilized cell lysates represent a versatile source of MP antigens that can be readily incorporated into the yeast surface display platform for quantitative antibody engineering. Here we performed dissociation rate-based affinity maturation of the anti-TfR scFv, H7, using detergent-solubilized TfR derived directly from cultured cells. The evolved scFvs exhibited slower dissociation rates and improved affinities for lysate-resident TfR. During the engineering process, the scFvs retained their binding specificity for TfR despite the presence of numerous, non-antigenic, biotinylated proteins in the lysate. When expressed and purified, the engineered TKe cohort exhibited similar improvements under physiological conditions, binding to TfR-expressing cells with 3-7-fold improved affinity. These improved binding properties were conferred by a small number of amino acid mutations (≤3) that resided in both the CDR and framework regions. Taken together, the lysate-based antibody engineering strategy can be implemented in much the same way as previously demonstrated with soluble antigens.
Cell lysates augment antibody engineering by yeast display, allowing key advantages of the platform to be leveraged against MP targets. Two particular benefits offered by yeast display are: (i) quantitative discrimination of improved clones inline with the screening process and (ii) rapid clone characterization directly on the yeast surface. Beginning with yeast surface-displayed scFv H7, the dissociation rate and equilibrium binding were quantitatively determined. Not only did this inform the decision to perform a kinetic screen, but it also generated the necessary screening parameters, specifically the optimal competition time. Kinetic selection required both a soluble TfR antigen, and a soluble competitor to screen for scFvs with a decreased rate of dissociation. This requirement was met by using B- and U-lysate preparations. Provided the affinity of the lead antibody was sufficiently poor (Kd > 10 nM), one could also perform equilibrium screens using the lysate-based approaches given the capability to perform relatively fine discrimination of equilibrium binding affinity using cell lysate-derived antigens (Fig. 3c; Cho et al., 2009). Finally, the rapid characterization of the relative dissociation rate and affinity improvements among TKe clones was conducted directly with yeast-displayed scFv (Figs 1–3). These data indicated that the expected quantitative improvements in dissociation rate were realized and that they translated into a 2-4-fold improved binding affinity on the surface of yeast.
Another key advantage of the lysate-based approach reported here is that the MP target of interest does not require truncation, fusion, heterologous expression or purification. Rather, mammalian cells are simply lysed with non-denaturing detergent providing a soluble source of MP antigen. Since TfR is a well-studied MP, literature was available to indicate that TfR was effectively solubilized in 1% Triton X-100 (Schuck et al., 2003). Importantly, the concentration of Triton X-100 was above the critical micelle concentration (CMC) where MPs are known to be more completely solubilized (le Maire et al., 2000). Without this information, lysate-based engineering could still be accomplished by simply screening detergent solubility of less well-characterized MPs directly using yeast surface-displayed scFvs, coupled with flow cytometry (e.g. biotinylated lysate titrations as in Fig. 2c but using different detergents). In our experience, yeast-displayed scFvs tend to retain their capacity to bind antigen in the presence of non-denaturing detergents such as Triton X-100, CHAPS and n-octyl-β-d-glucopyranoside, enabling lysate-based screens (Cho et al., 2009). Also, because of the non-denaturing character of the employed detergents, the scFv properties resulting from lysate-based screens can translate to non-detergent environments. This was demonstrated here with 3-7-fold improvements in TKe binding affinity to live cells and previously, with scFvs identified in lysate-based screens retaining binding under physiological conditions (Cho and Shusta, 2010). As a more subtle, but potentially important point, TfR was produced by HEK293 cells, and thus possessed human post-translational modifications. It has been shown that proper glycosylation of TfR is required for production of active TfR (Hayes et al., 1995), and most anti-hTfR antibodies have been discovered using antigen derived from mammalian cells (Haynes et al., 1981; Panaccio et al., 1987; Crépin et al., 2010; Yu et al., 2011). Given the potential for non-mammalian modifications from microbial production systems, it is conceivable that use of an unmodified MP directly from a mammalian host could be more successful in generating antibodies that recognize biologically relevant isoforms.
The magnitude of the improvement in Kd generated in this single round of affinity maturation (as yeast-displayed scFv, 2-4-fold or soluble scFv, 3-7-fold) is consistent with a small number of single mutations acting individually or in limited context with each other (Midelfort and Wittrup, 2006). The amino acid mutations indicated in Table I are largely residues predicted to be directly involved in antigen binding. Residues H56, H57, H102 and L95a all reside in the CDR loops. Moreover, residues H56, H57 and L66 are all predicted to have antigen contact, based on statistics gleaned from structural database mining (Honegger and Plückthun, 2001). The L66 residue was mutated in 7 of 8 TKe clones suggesting its importance in reducing TfR dissociation rate and improving affinity. Indeed, clone TKe322 possesses a single charge-neutralizing mutation at position L66 (K(L66)Q) that offered a significant 4.8-fold affinity improvement. In addition to antigen-binding regions, one mutation with a clear contribution to the affinity improvements, L(H45)P, is located between CDRH1 and CDRH2 and is found in three of eight TKe clones. Clone TKe213 possessed a 5.6-fold improvement in Kd and has L(H45)P as its sole mutation in the scFv coding sequence (Table I). The leucine at position H45 is highly conserved (>99%) in naturally occurring VH chains of human, mouse and rabbit (Kabat et al., 1991). Independent analyses of IgG crystal structures when aligned with amino acid sequence data indicate residue L(H45) plays a prominent role in packing of the VH–VL interface through interactions with at least three other residues: F(L98), P(L44) and W(H103) (Steiner et al., 1979; Chothia et al., 1985; Honegger and Plückthun, 2001; Abhinandan and Martin, 2010). While detailed mechanistic investigation would be required to elucidate the various contributions of the L(H45) mutation, literature has shown that it may be possible for improved VH–VL stability to lead to improved equilibrium binding affinity. For example, a single mutation to another core packing residue at the VH–VL interface (W(H103)L) was identified during affinity maturation of the 4-4-20 scFv and led to significant improvements in both affinity and stability (Midelfort and Wittrup, 2006). Similarly, mutations designed to stabilize scFvs can also lead to concomitant gains in affinity (Jung et al., 1999). In addition, the introduction of proline between CDR loops could lead to affinity improvement via reduction in the plasticity of the antigen-binding site (Wedemayer et al., 1997).
In terms of generalizability of the methodologies, we purposely employed a generic biotinylation strategy when engineering the H7 scFv. These results demonstrated that no target-specific subcloning or tagging of an MP (e.g. epitopes, Avitag and green fluorescent protein) is required for the antibody engineering process. This approach contrasts with a recent report that used detergent-solubilized baculovirus particles harboring enzymatically biotinylated (via the Avitag system) MPs as an antigen source for phage display-based antibody engineering (Hötzel et al., 2011). Moreover, both identification and optimization of lead antibodies carry a risk of antibody cross-reactivity, and YDIP can provide rapid assessment of antibody specificity (Cho et al., 2009). Since the lysate-based MP antigen contains a complex mixture of biotinylated proteins, there is a built-in advantage in terms of assaying for cross-reactivity in tandem with the screening process, simply by performing the YDIP procedure along with western blotting (Fig. 3d). In this study, a commercially available antibody against the cytoplasmic tail of TfR, was used to verify that scFv H7 bound detergent-solubilized, biotinylated TfR in the lysate, both on the surface of yeast and by YDIP/western blotting. Thus, if one has an antibody against an epitope distinct from that recognized by the antibody being engineered, it can be used as a replacement for biotinylation if desired. Of course, this would complicate a kinetic screen as the competitor would also be recognized by the detection antibody, and in this case, the use of B- and U-lysate as described here is recommended. It is important to note that the TfR is a well-expressed single-pass transmembrane protein, leading to efficient detergent solubilization and reasonably high TfR concentrations in lysate. Thus, while biotinylation provides a measure of methodological generalizability, the approach still requires that the target MP is detergent soluble, in reasonable abundance and that the scFv being engineered is stable in such detergents (Cho et al., 2009). Finally, TfR is a homodimer, consisting of two disulfide-bonded 84 kDa subunits, the majority of which (∼74 kDa) comprises the extracellular region where Tf binds (Cheng et al., 2004). Since all selections were done under non-reducing conditions, the detergent-solubilized TfR antigen was likely in a largely dimeric form, meaning that two yeast displayed scFvs could bind a single TfR dimer. This adds some level of avidity into the yeast display measurements, meaning that the yeast surface measurements scFv-TfR binding probably represent apparent dissociation rates and affinities. This factor could play a role in the more substantial Kd improvement of the TKe cohort when measured in soluble form as monomeric scFv versus that observed on the yeast surface (4-fold surface vs. 7-fold soluble). Importantly, the results indicate that lysate-based dissociation rate engineering is possible when targeting multimeric antigens, a common occurrence for membrane-resident receptors.
Maturation of lead antibodies is a vital step in the production of therapeutics and affinity reagents. Although new antibodies are regularly discovered using immunization and the hybridoma platform, the capability for engineering of affinity, stability and specificity has been enhanced by advances in in vitro antibody engineering methods. Most attempts at in vitro maturation of antibodies targeting MPs have side-stepped antigen solubility issues through the use of truncated proteins or linear peptides. A few recent studies, however, have shown that detergent solubilization of MPs can be a more direct strategy for antibody engineering against MP antigens (Milovnik et al., 2009; Silverman et al., 2009; Cho and Shusta, 2010; Hötzel, et al., 2011). The collective use of detergent-solubilized cell lysates as an antigen source for lead antibody identification (Cho and Shusta, 2010) antigen characterization (Cho, et al., 2009), and antibody affinity engineering as presented in this study, renders yeast display with lysates a promising platform for antibody discovery and optimization.
Supplementary data
Funding
I.F.L acknowledges Ministerio de Ciencia e Innovación and Gobierno Vasco (Spain) for the research fellowship (PR2008-0168) and travel grants. This work was supported by National Institutes of Health grants NS056249, NS071513 and AI072435.
Supplementary Material
Acknowledgements
The authors acknowledge the staff at the UW-Madison Carbone Cancer Center flow cytometry facility for their expertise and assistance in sorting yeast display libraries. B.J.T. acknowledges Will Deipholz for assistance in sequencing scFvs in the H7 library.
References
- Abhinandan K.R., Martin A.C.R. Protein Eng. Des. Sel. 2010;23:689–697. doi: 10.1093/protein/gzq043. doi:10.1093/protein/gzq043. [DOI] [PubMed] [Google Scholar]
- Bhatt R.R., Haurum J.S., Davis C.G. Technologies for the generation of human antibodies development of antibody-based therapeutics. In: Tabrizi M.A., Bornstein G.G., Klakamp S.L., editors. Development of Antibody-based Therapeutics. New York: Springer; 2012. pp. 33–63. [Google Scholar]
- Boder E.T., Wittrup K.D. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. doi:10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Biotechnol. Prog. 1998;14:55–62. doi: 10.1021/bp970144q. doi:10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Yeast surface display for directed evolution of protein expression, affinity, and stability. In: Jeremy Thorner S.D.E., John N.A., editors. Methods in Enzymology. Academic Press; 2000. pp. 430–444. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Midelfort K.S., Wittrup K.D. Proc. Natl Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. doi:10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A.R.M., Sidhu S., Dubel S., McCafferty J. Nat. Biotechnol. 2011;29:245–254. doi: 10.1038/nbt.1791. doi:10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Zak O., Aisen P., Harrison S.C., Walz T. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. doi:10.1016/S0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Cho Y.K., Shusta E.V. Protein Eng. Des. Sel. 2010;23:567–577. doi: 10.1093/protein/gzq029. doi:10.1093/protein/gzq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.K., Chen I., Wei X., Li L., Shusta E.V. J. Immunol. Methods. 2009;341:117–126. doi: 10.1016/j.jim.2008.11.005. doi:10.1016/j.jim.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Novotný J., Bruccoleri R., Karplus M. J. Mol. Biol. 1985;186:651–663. doi: 10.1016/0022-2836(85)90137-8. doi:10.1016/0022-2836(85)90137-8. [DOI] [PubMed] [Google Scholar]
- Crépin R., Goenaga A.-L., Jullienne B., Bougherara H., Legay C., Benihoud K., Marks J.D., Poul M.-A. Cancer Res. 2010;70:5497–5506. doi: 10.1158/0008-5472.CAN-10-0938. doi:10.1158/0008-5472.CAN-10-0938. [DOI] [PubMed] [Google Scholar]
- Finlay W.J., Cunningham O., Lambert M.A., et al. J. Mol. Biol. 2009;388:541–558. doi: 10.1016/j.jmb.2009.03.019. doi:10.1016/j.jmb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Freigassner M., Pichler H., Glieder A. Microb. Cell Factories. 2009;8:69. doi: 10.1186/1475-2859-8-69. doi:10.1186/1475-2859-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. doi:10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Graff C.P., Chester K., Begent R., Wittrup K.D. Protein Eng. Des. Sel. 2004;17:293–304. doi: 10.1093/protein/gzh038. doi:10.1093/protein/gzh038. [DOI] [PubMed] [Google Scholar]
- Hackel B., Huang D., Bubolz J., Wang X., Shusta E. Pharm. Res. 2006;23:790–797. doi: 10.1007/s11095-006-9778-7. doi:10.1007/s11095-006-9778-7. [DOI] [PubMed] [Google Scholar]
- Hayes G.R., Williams A., Costello C.E., Enns C.A., Lucas J.J. Glycobiology. 1995;5:227–232. doi: 10.1093/glycob/5.2.227. doi:10.1093/glycob/5.2.227. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Hemler M., Cotner T., Mann D.L., Eisenbarth G.S., Strominger J.L., Fauci A.S. J. Immunol. 1981;127:347–351. [PubMed] [Google Scholar]
- Honegger A., Plückthun A. J. Mol. Biol. 2001;309:657–670. doi: 10.1006/jmbi.2001.4662. doi:10.1006/jmbi.2001.4662. [DOI] [PubMed] [Google Scholar]
- Hötzel I., Chiang V., Diao J., Pantua H., Maun H., Kapadia S. Protein Eng. Des. Sel. 2011;24:679–689. doi: 10.1093/protein/gzr039. doi:10.1093/protein/gzr039. [DOI] [PubMed] [Google Scholar]
- Jung S., Honegger A., Plückthun A. J. Mol. Biol. 1999;294:163–180. doi: 10.1006/jmbi.1999.3196. doi:10.1006/jmbi.1999.3196. [DOI] [PubMed] [Google Scholar]
- Kabat E.A., Wu T.T., Perry H.M., Gottesman K.S., Foeller C. Sequences of Proteins of Immunological Interest. 5th edn. Bethesda, MD: U.S. Department of Health and Human Services, Public Health Service, National Institutes of Health; 1991. [Google Scholar]
- Kenrick S.A., Daugherty P.S. Protein Eng. Des. Sel. 2010;23:9–17. doi: 10.1093/protein/gzp065. doi:10.1093/protein/gzp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong K.Y., Baskar S., Zhang H., Mackall C.L., Rader C. J. Mol. Biol. 2008;384:1143–1156. doi: 10.1016/j.jmb.2008.09.008. doi:10.1016/j.jmb.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M., Champeil P., Møller J.V. Biochim. Biophys. Acta (BBA) Biomembranes. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. doi:10.1016/S0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Lin S.-H., Guidotti G. Purification of membrane proteins. In: Richard R.B., Murray P.D., editors. Methods in Enzymology. Academic Press, Amsterdam; 2009. pp. 619–629. [DOI] [PubMed] [Google Scholar]
- Linke D. Detergents: an overview. In: Richard R.B., Murray P.D., editors. Methods in Enzymology. Academic Press, Amsterdam; 2009. pp. 603–617. [DOI] [PubMed] [Google Scholar]
- Lipes B.D., Chen Y.H., Ma H., Staats H.F., Kenan D.J., Gunn M.D. J. Mol. Biol. 2008;379:261–272. doi: 10.1016/j.jmb.2008.03.072. doi:10.1016/j.jmb.2008.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippow S.M., Wittrup K.D., Tidor B. Nat. Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. doi:10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P.J., Botstein D. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. doi:10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Midelfort K.S., Wittrup K.D. Protein Sci. 2006;15:324–334. doi: 10.1110/ps.051842406. doi:10.1110/ps.051842406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovnik P., Ferrari D., Sarkar C.A., Plückthun A. Protein Eng. Des. Sel. 2009;22:357–366. doi: 10.1093/protein/gzp011. doi:10.1093/protein/gzp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaccio M., Zalcberg J.R., Thompson C.H., Leyden M.J., Sullivan J.R., Lichtenstein M., McKenzie I.F.C. Immunol. Cell Biol. 1987;65:461–472. doi: 10.1038/icb.1987.55. doi:10.1038/icb.1987.55. [DOI] [PubMed] [Google Scholar]
- Piatesi A., Howland S.W., Rakestraw J.A., et al. Protein Expr. Purif. 2006;48:232–242. doi: 10.1016/j.pep.2006.01.026. doi:10.1016/j.pep.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Ponka P., Lok C.N. Int. J. Biochem. Cell Biol. 1999;31:1111–1137. doi: 10.1016/s1357-2725(99)00070-9. doi:10.1016/S1357-2725(99)00070-9. [DOI] [PubMed] [Google Scholar]
- Poul M.A., Becerril B., Nielsen U.B., Morisson P., Marks J.D. J. Mol. Biol. 2000;301:1149–1161. doi: 10.1006/jmbi.2000.4026. doi:10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- Powers D.B., Amersdorfer P., Poul M.-A., Nielsen U.B., Shalaby M.R., Adams G.P., Weiner L.M., Marks J.D. J. Immunol. Methods. 2001;251:123–135. doi: 10.1016/s0022-1759(00)00290-8. doi:10.1016/S0022-1759(00)00290-8. [DOI] [PubMed] [Google Scholar]
- Privé G.G. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. doi:10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Reichert J.M. MAbs. 2010;2:695–700. doi: 10.4161/mabs.2.6.13603. doi:10.4161/mabs.2.6.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier R., Bye J., Apell G., McCall A., Adams G.P., Malmqvist M., Weiner L.M., Marks J.D. J. Mol. Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. doi:10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. Proc. Natl Acad. Sci. USA. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. doi:10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets M.D., Amersdorfer P., Finnern R., et al. Proc. Natl Acad. Sci. USA. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. doi:10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shusta E.V., Raines R.T., Pluckthun A., Wittrup K.D. Nat. Biotechnol. 1998;16:773–777. doi: 10.1038/nbt0898-773. doi:10.1038/nbt0898-773. [DOI] [PubMed] [Google Scholar]
- Silverman A.P., Levin A.M., Lahti J.L., Cochran J.R. J. Mol. Biol. 2009;385:1064–1075. doi: 10.1016/j.jmb.2008.11.004. doi:10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner L.A., Pardo A.G., Margolies M.N. Biochemistry. 1979;18:4068–4080. doi: 10.1021/bi00586a003. doi:10.1021/bi00586a003. [DOI] [PubMed] [Google Scholar]
- Swers J.S., Kellogg B.A., Wittrup K.D. Nucleic Acids Res. 2004;32:e36. doi: 10.1093/nar/gnh030. doi:10.1093/nar/gnh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson B.J., Cho Y.K., Shusta E.V. Methods. 2012 doi: 10.1016/j.ymeth.2012.03.010. http://dx.doi.org/10.1016/j.ymeth.2012.03.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanAntwerp J.J., Wittrup K.D. J. Mol. Recognit. 1998;11:10–13. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<10::AID-JMR381>3.0.CO;2-H. doi:10.1002/(SICI)1099-1352(199812)11:1/6<10::AID-JMR381>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Van Antwerp J.J., Wittrup K.D. Biotechnol. Prog. 2000;16:31–37. doi: 10.1021/bp990133s. doi:10.1021/bp990133s. [DOI] [PubMed] [Google Scholar]
- Wang X.X., Shusta E.V. J. Immunol. Methods. 2005;304:30–42. doi: 10.1016/j.jim.2005.05.006. doi:10.1016/j.jim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Wedemayer G.J., Patten P.A., Wang L.H., Schultz P.G., Stevens R.C. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. doi:10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- Wentz A.E., Shusta E.V. Appl. Environ. Microbiol. 2007;73:1189–1198. doi: 10.1128/AEM.02427-06. doi:10.1128/AEM.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.H., Wimley W.C. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. doi:10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Yildirim M.A., Goh K.I., Cusick M.E., Barabasi A.L., Vidal M. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. doi:10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Young C.L., Britton Z.T., Robinson A.S. Biotechnol. J. 2012;7:620–634. doi: 10.1002/biot.201100155. doi:10.1002/biot.201100155. [DOI] [PubMed] [Google Scholar]
- Yu Y.J., Zhang Y., Kenrick M., et al. Sci. Transl. Med. 2011;3:84ra44. doi: 10.1126/scitranslmed.3002230. doi:10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
- Zaccolo M., Williams D.M., Brown D.M., Gherardi E. J. Mol. Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. doi:10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.