Abstract
Construction and intracellular targeting of eukaryotic pre-ribosomal particles involve a multitude of diverse transiently associating trans-acting assembly factors, energy-consuming enzymes, and transport factors. The ability to rapidly and reliably measure co-enrichment of multiple factors with maturing pre-ribosomal particles presents a major biochemical bottleneck towards revealing their function and the precise contribution of >50 energy-consuming steps that drive ribosome assembly. Here, we devised a workflow that combines genetic trapping, affinity-capture, and selected reaction monitoring mass spectrometry (SRM-MS), to overcome this deficiency. We exploited this approach to interrogate the dynamic proteome of pre-60S particles after nuclear export. We uncovered assembly factors that travel with pre-60S particles to the cytoplasm, where they are released before initiating translation. Notably, we identified a novel shuttling factor that facilitates nuclear export of pre-60S particles. Capturing and quantitating protein interaction networks of trapped intermediates of macromolecular complexes by our workflow is a reliable discovery tool to unveil dynamic processes that contribute to their in vivo assembly and transport.
Keywords: nuclear export, ribosome assembly, selected reaction monitoring mass spectrometry, targeted proteomics
Introduction
Large macromolecular complexes contribute to diverse anabolic and catabolic processes. Proper assembly, quality control, intracellular transport, and regulation of these complexes are a prerequisite to elicit their cellular function. The construction and targeting of the eukaryotic ribosome is a spectacular example of a highly dynamic and regulated process. The small (40S) and large (60S) ribosomal subunits are assembled from >70 ribosomal proteins (r-proteins) and four different ribosomal RNA (rRNA) species. While the structure of the mature cytoplasmic ribosome is better characterized, our knowledge of the assembly and transport of this universal translating machine is only emerging.
More than 200 non-ribosomal trans-acting factors aid the assembly, maturation and intracellular transport of pre-40S and pre-60S particles as they travel from the nucleolus to the cytoplasm (Tschochner and Hurt, 2003; Strunk and Karbstein, 2009; Kressler et al, 2010). The 35S pre-rRNA produced by RNA polymerase I undergoes co-transcriptional modifications in the nucleolus, and associates with mainly small-subunit r-proteins and trans-acting factors to form the 90S particle (Grandi et al, 2002). Cleavage of the 35S pre-rRNA releases the pre-40S particle and permits the remaining pre-rRNA to associate with large-subunit r-proteins and assembly factors to form pre-60S particles (Grandi et al, 2002). Pre-60S subunits encounter ∼100 trans-acting factors as they travel through the nucleoplasm towards the nuclear pore complex (NPC) and therefore undergo dynamic compositional changes (Nissan et al, 2002). In contrast, pre-40S particles undergo fewer changes as they travel through the nucleoplasm (Grandi et al, 2002; Schäfer et al, 2003). These changes are most likely induced by transiently associating energy-consuming enzymes, which result in sequential reduction of complexity and acquisition of nuclear export competence (Strunk and Karbstein, 2009; Kressler et al, 2010).
Export competent pre-40S and pre-60S subunits are transported separately through NPCs by shuttling export receptors (Johnson et al, 2002). The essential exportin Xpo1 directly interacts with Phe-Gly (FG) rich nucleoporins lining the NPC transport channel, and recognizes nuclear export sequences (NESs) to mediate nuclear export of pre-60S and pre-40S subunits (Moy and Silver, 1999; Ho et al, 2000; Gadal et al, 2001). Nmd3 is the only known essential NES-containing adaptor that bridges interactions between Xpo1 and pre-60S particles in a RanGTP-dependent manner (Ho et al, 2000; Gadal et al, 2001). Genetic studies in budding yeast have uncovered FG-interacting trans-acting factors (Arx1, Ecm1, and Rrp12) and the mRNA export factor Mex67-Mtr2 in the nuclear export of pre-60S and pre-40S particles (Oeffinger et al, 2004; Bradatsch et al, 2007; Yao et al, 2007, 2010; Hung et al, 2008; Faza et al, 2012). Considering the size of the pre-ribosomal subunits, multiple factors are expected to aid transport of pre-ribosomes through the NPC (Ribbeck and Gorlich, 2001).
Pre-40S and pre-60S particles undergo final maturation steps in the cytoplasm, prior to initiating translation. These steps involve the release of shuttling trans-acting factors and transport factors, incorporation of the remaining r-proteins, and final pre-rRNA processing steps (Panse and Johnson, 2010; Panse, 2011). In the 60S assembly pathway, late maturation is triggered by release factors that transiently associate with pre-60S particles in the cytoplasm. These steps are crucial for the 60S assembly because a failure to release and recycle shuttling trans-acting factors leads to the depletion from their nucleolar/nuclear sites of action, resulting in impaired pre-rRNA processing, assembly defects, and impaired nuclear export of pre-60S subunits. Late maturation steps along the 60S maturation pathway prevent immature translation incompetent pre-60S particles from initiating translation (Panse and Johnson, 2010; Panse, 2011).
While proteomic approaches have greatly expanded the inventory of trans-acting factors that aid the assembly and transport of pre-ribosomal particles, their precise function(s) are only beginning to be unraveled. Precise knowledge of the maturation stage at which the majority of these factors join pre-ribosomal particles still remains scarce. Moreover, the timing of release after fulfilling their role, and the mechanisms/factors required for these steps remain unknown. Typically, analyses of pre-ribosomal particles involve their isolation at different maturation steps followed by western analysis using either epitope tagged factors or antibodies directed against proteins of interest. While this approach is extremely reliable, it is time-consuming and requires generating either strains containing epitope tagged factors that might hinder their function, or protein-specific antibodies. The ability to reliably and rapidly quantitate co-enrichment of multiple trans-acting factors with pre-ribosomal particles presents a major bottleneck towards revealing the function of assembly factors and, in particular, the precise role(s) of >50 energy-consuming enzymes in the complex ribosome assembly pathway.
While shotgun proteomics has proved to be an extremely powerful de novo discovery tool, these approaches are tedious and semi-stochastic in nature, which complicates the reliable quantitation of the co-enrichment of trans-acting factors, especially over multiple pre-ribosomal particles at different maturation stages. Recently, selected reaction monitoring mass spectrometry (SRM-MS) was proposed as a tool capable of overcoming such limitations and enabling the reproducible quantification of predetermined sets of proteins at high sensitivity and precision across a multitude of samples (Anderson and Hunter, 2006; Addona et al, 2009; Bisson et al, 2011; Picotti and Aebersold, 2012). SRM relies on the development of specific mass spectrometric assays for every target protein and their subsequent application to the relative or absolute quantification of the protein in biological samples. The approach starts with the selection of representative peptides for each protein (proteotypic peptides, PTPs). Then, for each PTP, mass spectrometric coordinates (such as precursor/fragment ion pairs, collision energy, and peptide elution time) are established that characterize the PTP and allow its targeted measurement with a chromatography coupled-triple-quadrupole mass spectrometer. Once SRM assays are established for a set of proteins, they become applicable to different samples, akin to sets of antibodies for western analysis.
Here, we have developed a resource of SRM assays that enabled us to reliably and rapidly monitor the co-enrichment of 40 trans-acting factors and 10 r-proteins with maturing pre-60S particles as they are transported from the nucleolus to the cytoplasm, in a single experiment. The developed assays were exploited to interrogate the proteome of genetically trapped cytoplasmic pre-60S particles after nuclear export. Our data revealed trans-acting factors that need to travel with 60S pre-ribosomes to the cytoplasm to be released. Functional analyses identified a shuttling trans-acting factor that is required for efficient nuclear export of pre-60S particles.
Results
Developing SRM assays to investigate the 60S biogenesis pathway
We began our analyses by selecting a target list of trans-acting factors whose association with pre-60S particles was not quantitatively characterized (Supplementary Table 1). In addition, we selected factors that broadly reflected the cellular subterritories and compartments (nucleolus, nucleoplasm, and cytoplasm) through which pre-60S particles pass at different stages of maturation and where their function is likely to be elicited. To assess the reliability of our approach, we included factors whose association with pre-60S particles had been well characterized by western analysis, and 10 r-proteins for normalization purposes.
Next, to develop specific SRM assays for each protein, we selected a set of up to five PTPs from a collection of shotgun proteomic analyses of pre-60S particles at different maturation stages, that exhibit >100 known pre-60S particle-associated factors (Supplementary Table 2). We preferred peptides with high MS signal response and devoid of amino acids prone to modification artifacts. For each peptide, we experimentally tested a set of precursor-to-fragment ion pairs (SRM transitions) and extracted up to three peptides and four transitions per peptide that resulted in the highest signals for each protein, together with peptide elution times. The final set of SRM coordinates are tabulated in Supplementary Table 3. Finally, we generated a label-free multiplexed, time-constrained (scheduled) SRM assay, which concomitantly measures the whole set of 50 target proteins in a single mass spectrometric run of ∼1 h.
Protein interaction dynamics of 60S pre-ribosomes investigated by SRM
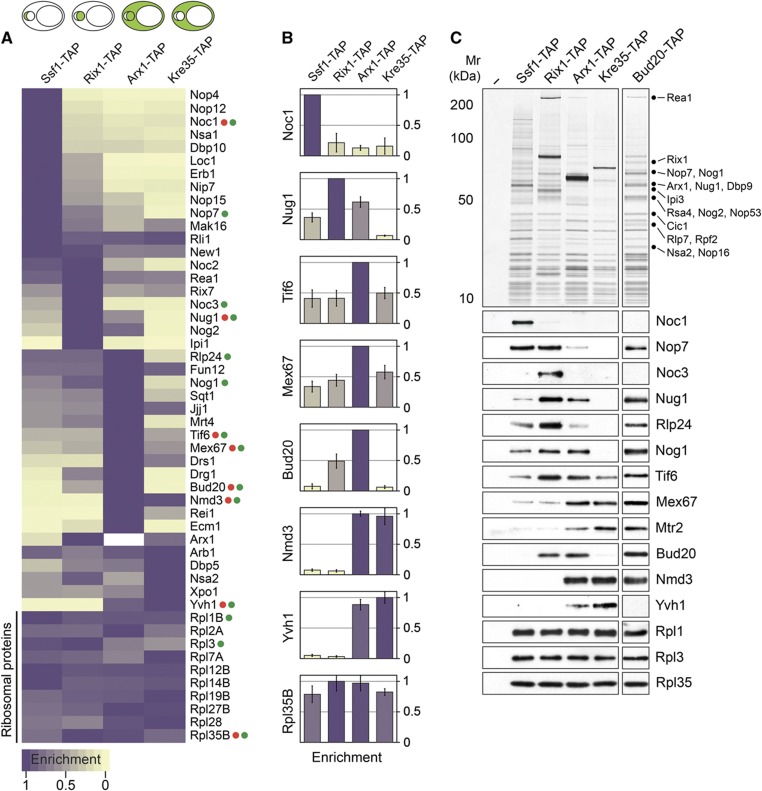
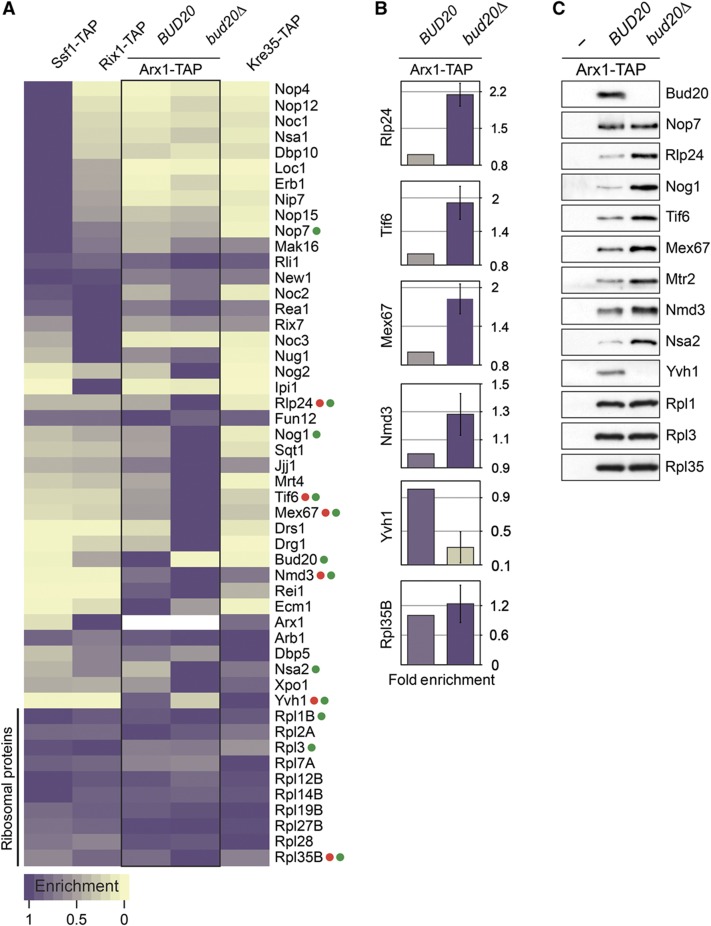
We exploited the multiplexed SRM assay to measure the relative co-enrichment of the 40 trans-acting factors and 10 r-proteins with pre-60S particles at different maturation stages. For these analyses, we affinity-purified Ssf1-TAP, an early nucleolar pre-60S particle, Rix1-TAP, an intermediate nucleoplasmic pre-60S particle, Arx1-TAP, a late export competent pre-60S particle, and finally Kre35-TAP, an exclusive cytoplasmic pre-60S particle (Nissan et al, 2002; Yao et al, 2007). To simplify the visualization of the SRM data, we have employed a color-scale gradient. Maximal to minimal relative co-enrichment of a factor with a pre-60S particle is depicted in purple to gold color-scale, respectively, and is calculated from the average of multiple SRM transition intensities per peptide and multiple peptides per protein (Figure 1A). The relative co-enrichments of trans-acting factors along the 60S maturation pathway monitored by SRM in Figure 1A and B are in good agreement with previous reports (Supplementary Table 1) and western analysis performed in this study (Figure 1C). For example, both SRM and western analyses demonstrate that the factors Noc1 and Noc3 significantly co-enrich with nucleolar pre-60S particles (Ssf1-TAP) and nucleoplasmic particles (Rix1-TAP), respectively (Figure 1A–C) (Nissan et al, 2002; Kressler et al, 2008; Kemmler et al, 2009). The NES-containing Nmd3, and the export receptor Mex67-Mtr2 maximally co-enrich with late pre-60S particles (Arx1-TAP and Kre35-TAP) (Figure 1A–C) (Yao et al, 2007; Kemmler et al, 2009; Faza et al, 2012). Also, Yvh1 that is responsible for the release of shuttling trans-acting factor Mrt4 is found to co-enrich mainly with late pre-60S particles (Figure 1A–C) (Kemmler et al, 2009). Moreover, our analyses placed the different trans-acting factors into four clusters: an early nucleolar, an intermediate nucleoplasmic, a late and finally a cytoplasmic cluster (Figure 1A). Thus, the developed SRM assays reliably monitored the changing abundance of several known trans-acting factors on pre-60S particles, as well as revealed dynamic interactions of several other trans-acting factors as pre-60S particles travel from the nucleolus to the cytoplasm.
Figure 1.
Dynamic association of trans-acting factors with maturing pre-60S subunits revealed by SRM. (A) A representative analysis (three independent biological replicates) of relative enrichment of 40 trans-acting factors with maturing pre-60S particles are depicted using purple to gold color-scale. Maximum enrichment is depicted as purple and minimum enrichment as gold. Each measurement represents the average of different SRM transitions per peptide and different peptides per protein. Akin to western analysis, the acquired intensity of each factor was normalized based on the average intensities of 10 depicted large-subunit r-proteins. (B) The enrichment of selected factors (labeled as red dots in (A)) is represented using histograms with error bars (±s.d.) to assess the precision of our analyses for the relative co-enrichment of individual factors. The color of each column corresponds to the relative enrichment as shown in (A). (C) Western analyses of selected pre-60S trans-acting factors (labeled as green dots in (A)) using TAP purified pre-60S and Bud20 particles. The purifications were separated on 4–12% gradient gels, and subjected to silver staining and western analyses. The large-subunit r-proteins Rpl1, Rpl3, and Rpl35 served as loading controls.
Interrogating the proteome of genetically trapped cytoplasmic 60S pre-ribosomes by SRM
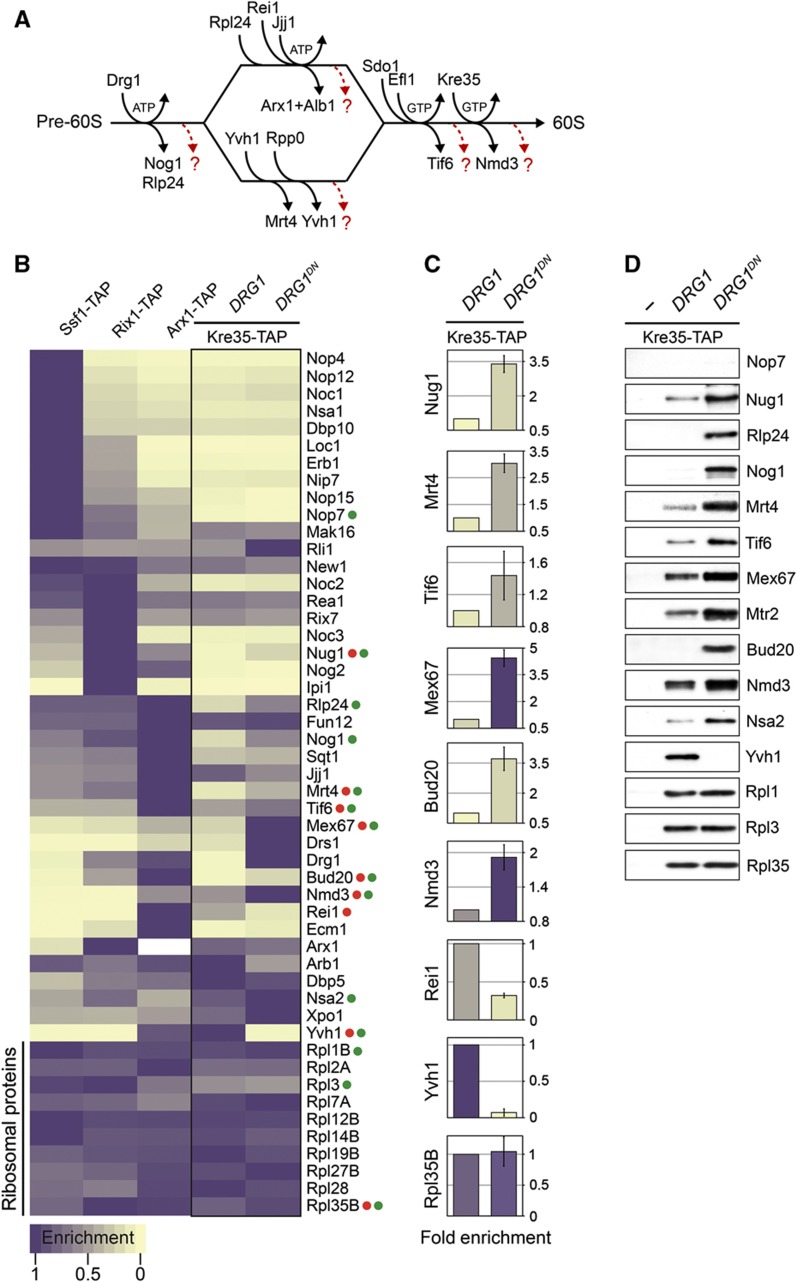
Upon arrival in the cytoplasm, pre-60S particles undergo sequential release of bound trans-acting factors and transport factors, before initiating translation. Till date only genetic approaches in yeast have permitted uncovering few nucleolar/nuclear trans-acting factors that travel with pre-60S particles and are released by cytoplasmic release factors (Figure 2A). A systematic survey of nucleolar/nuclear trans-acting factors that ‘need’ to travel to the cytoplasm with pre-60S particles to be released was never performed. We sought to address this deficit by employing the developed SRM assays to quantitatively investigate the proteome of a genetically trapped pre-60S particle poised to initiate cytoplasmic maturation.
Figure 2.
Proteome of genetically trapped cytoplasmic pre-60S particles revealed by SRM. (A) Proposed pathway of 60S cytoplasmic maturation initiated by Drg1. Maturation events indicated on the pathway only represent the order of action but not necessarily the association with the pre-60S particle. (B) A representative analysis (three independent biological replicates) of the relative enrichment of the depicted factors on the indicated pre-60S particles isolated at different stages of maturation, and genetically trapped cytoplasmic pre-60S particles are shown using purple to gold color-scale. Maximum enrichment is depicted as purple and minimum enrichment as gold. Each measurement represents the average of different SRM transitions per peptide and different peptides per protein. Akin to western analyses, the acquired intensities of each protein were normalized based on a set of 10 large-subunit r-proteins. (C) The relative enrichment of selected proteins (labeled in (B) with red dots) in Kre35-TAP WT (DRG1) and DRG1DN samples is represented using histograms showing the fold change with error bars (±s.d.). The color of each bar corresponds to the enrichment shown in (B). (D) Western analyses of selected pre-60S trans-acting factors (labeled in (B) with green dots) using affinity-purified pre-60S particles. Samples were analyzed on NuPAGE 4–12% gradient gels followed by western analyses. The large-subunit r-protein Rpl1, Rpl3, and Rpl35 served as loading controls.
To trap pre-60S particles after nuclear export, we took advantage of the sequential nature of the 60S cytoplasmic maturation pathway (Figure 2A). Upon arrival in the cytoplasm, the AAA-ATPase Drg1 triggers the earliest maturation step on pre-60S particles. This step, which involves the release of the ribosomal-like protein Rlp24, is a pre-requisite for subsequent cytoplasmic maturation steps of 60S pre-ribosomes (Figure 2A) (Pertschy et al, 2007; Lo et al, 2010). An impaired Drg1 (drg1-ts) or expression of a dominant negative mutant of Drg1 (DRG1DN) results in cytoplasmic localization of a subset of nucleolar/nuclear factors that remain bound to 60S pre-ribosomes (Pertschy et al, 2007; Lo et al, 2010). With the aim of accumulating nucleolar/nuclear trans-acting factors on cytoplasmic pre-60S particles, we transformed an inducible DRG1DN allele into a strain expressing Kre35-TAP that purifies cytoplasmic pre-60S particles. After induction of Drg1DN, Kre35-TAP particles were isolated and subjected to SRM analysis. To better evaluate the compositional changes in Kre35-TAP upon induction of the DRG1DN allele, pre-60S particles at different stages of maturation (Ssf1-TAP, Rix1-TAP, and Arx1-TAP) were included in the data acquisition and analysis. Our SRM analysis revealed known shuttling trans-acting factors belonging to the nucleoplasmic and the late cluster (Tif6, Mrt4, Rlp24, Nog1, Arx1) that accumulated on Kre35-TAP particles when the DRG1DN allele was induced (Figure 2B and C). Additionally, we found several trans-acting factors that accumulated on the Kre35-TAP particle upon induction of Drg1DN (Figure 2B and C). For example, the 60S export receptor Mex67-Mtr2, the trans-acting factors Bud20 and Nsa2, the GTPase Nug1, the ABC-ATPase Rli1, and the RNA helicase Drs1 were significantly enriched on the Kre35-TAP particle upon induction of Drg1DN. Western analyses carried out for some of the trans-acting factors confirmed this enrichment (Figure 2D). In contrast, the nucleolar cluster (Nop4, Nop12, Noc1, Nsa1, etc.) was not found on the Kre35-TAP particle in cells expressing the DRG1DN allele, indicating that these factors might be released during early biogenesis steps. Thus, our analyses of the proteome of a genetically trapped pre-60S particle after nuclear export by SRM uncovered several shuttling factors.
Bud20 shuttles between the nucleus and cytoplasm
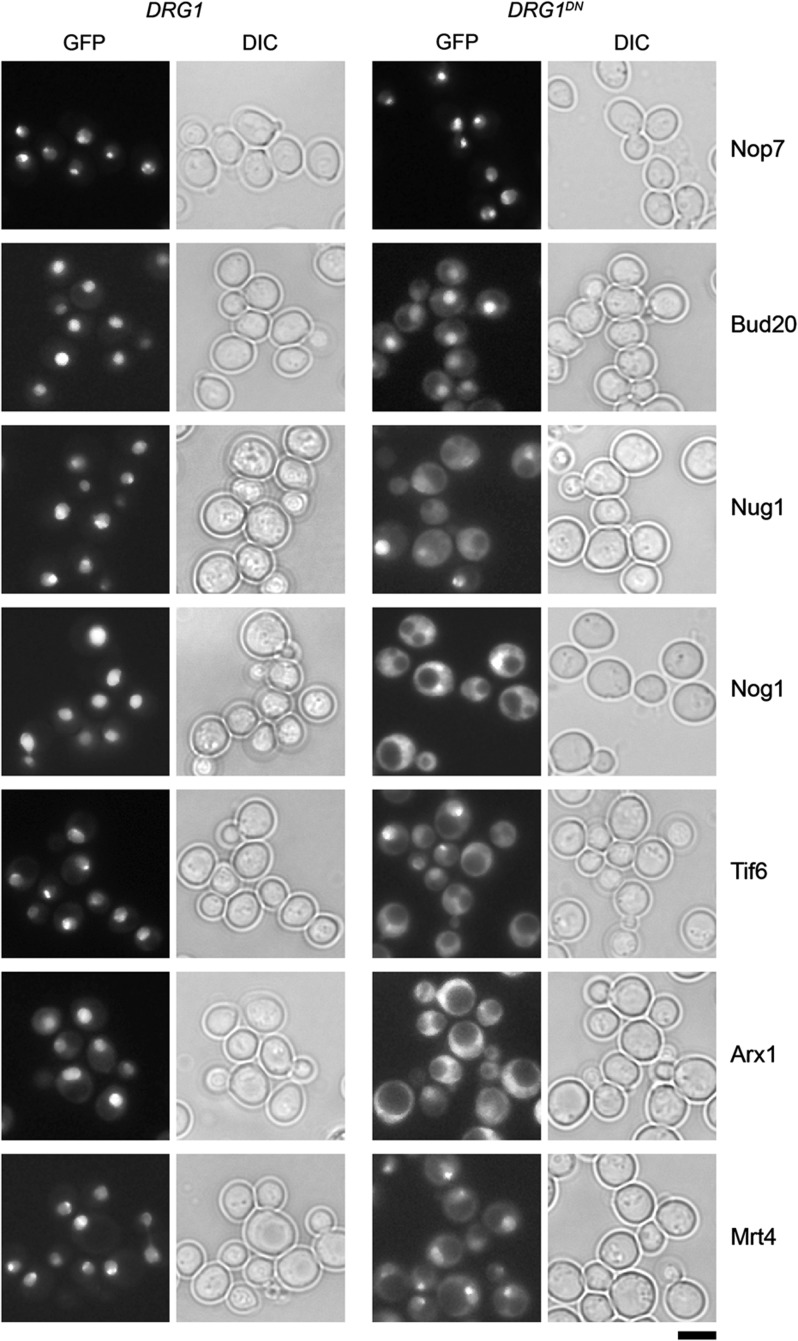
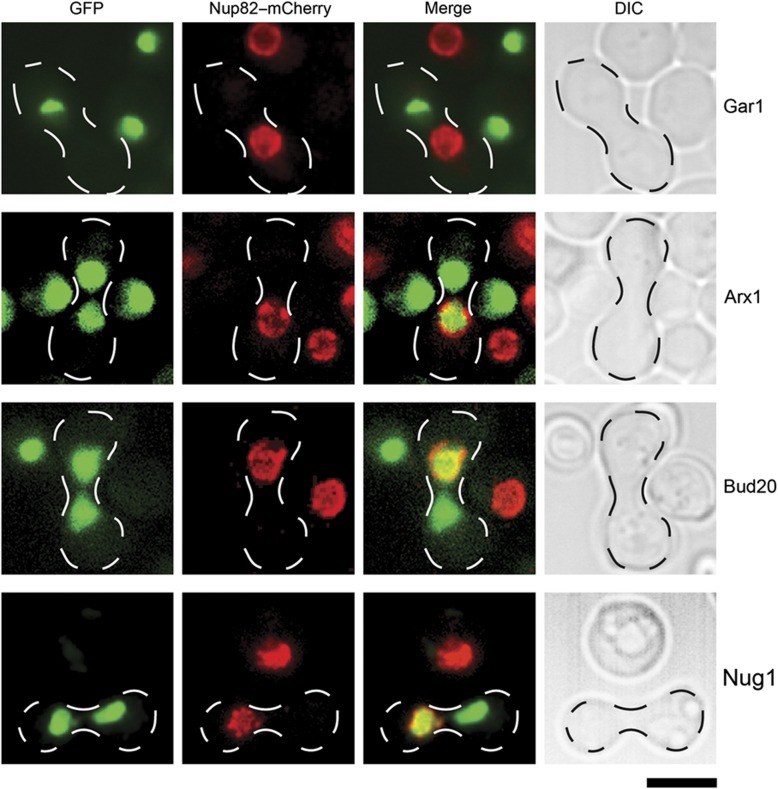
SRM and western analyses of trapped cytoplasmic pre-60S particles indicated that the uncharacterized trans-acting factor Bud20 accompanies pre-60S subunits from the nucleus through the NPC and into the cytoplasm (Figure 2B–D). To further characterize the shuttling behavior of Bud20, we examined the cellular localization of Bud20–GFP upon induction of the DRG1DN allele. In agreement with previous studies, we found that the nucleolar/nuclear-localized factors (Tif6, Mrt4, Nog1, and Arx1), known to travel with pre-60S particles to the cytoplasm (Panse and Johnson, 2010; Panse, 2011), were either strongly or partially mislocalized to the cytoplasm upon induction of the DRG1DN allele (Figure 3). Consistent with SRM and western analyses, we found that Bud20–GFP partially mislocalized to the cytoplasm, upon induction of the DRG1DN allele (Figure 3). In contrast, the nucleolar/nuclear localization of the non-shuttling trans-acting factor Nop7 was unaltered (Figure 3). To further ascertain whether Bud20 can shuttle rapidly between the nucleus and cytoplasm, we resorted to the previously described heterokaryon assay (Belaya et al, 2006). A strain expressing a Bud20–GFP fusion was crossed with a kar1-1 expressing strain, in which mating and cell conjugation is not followed by nuclear fusion, leading to heterokaryon formation. In order to distinguish the two nuclei in the resulting heterokaryon, the nuclear pore protein Nup82 was tagged with mCherry in the kar1-1 expressing strain. As controls we used the shuttling Arx1–GFP and non-shuttling Gar1–GFP strains, respectively. While the non-shuttling Gar1–GFP was never seen in the nucleus of the kar1-1 expressing strain (red signal), Bud20–GFP and the known shuttling factor Arx1–GFP localized to both nuclei (Figure 4). Collectively, these data are consistent with the nucleo-cytoplasmic shuttling of Bud20.
Figure 3.
Overexpression of DRG1DN results in mislocalization of Bud20–GFP and Nug1–GFP. Cells carrying a plasmid containing DRG1DN under the control of CUP1 promoter were grown to early log phase. Expression of DRG1DN was induced by 0.5 mM copper sulfate for 5–7 h. Cells were analyzed by fluorescence microscopy. Empty vector (pYEX4-T1) was used as controls. Scale bar=5 μm.
Figure 4.
Bud20–GFP and Nug1–GFP shuttle between the nucleus and cytoplasm. Cells expressing Arx1–GFP, Gar1–GFP, Bud20–GFP, Nug1–GFP were mated with the kar1-1 overexpressing strain containing Nup82–mCherry. Heterokaryons were analyzed by fluorescence microscopy. Arx1–GFP and Gar1–GFP served as positive and negative controls, respectively. Scale bar=5 μm.
Bud20 co-enriches with late 60S pre-ribosomes
Large-scale proteomic approaches reported the association of the evolutionarily conserved trans-acting factor Bud20 with pre-60S particles (Gavin et al, 2002; Ho et al, 2002). Both SRM and western analyses revealed that Bud20 co-enriches mainly with the intermediate nucleoplasmic (Rix1-TAP) and late pre-60S subunits (Arx1-TAP), but not with early nucleolar pre-60S particles (Ssf1-TAP) or cytoplasmic pre-60S particles (Kre35-TAP) (Figure 1A–C). Next, we directly isolated the Bud20-TAP particle and analyzed its protein composition by MS. We found that Bud20-TAP co-enriched many early-to-late nuclear pre-60S factors (Figure 1C). Notably, western analyses revealed that Bud20-TAP co-enriches export factors Nmd3 and Mex67-Mtr2 (Figure 1C). Together, these analyses show that Bud20 co-enriches with late 60S pre-ribosomes.
Bud20 is required for proper nuclear export of 60S pre-ribosomes
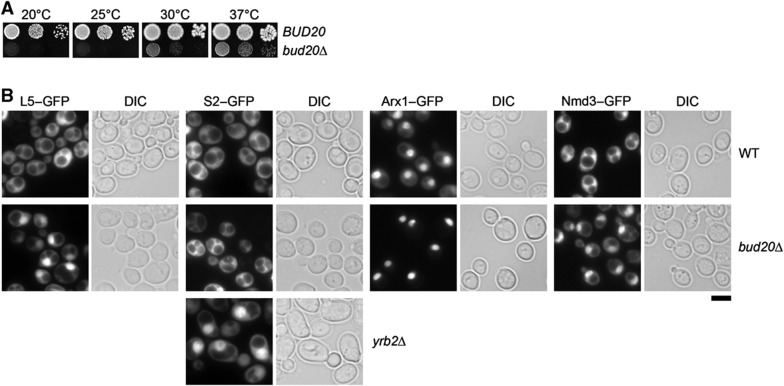
To investigate the uncharacterized role of Bud20 in the 60S maturation pathway, we disrupted BUD20 in wild-type (WT) diploid cells. Tetrad analysis yielded two spores with WT growth rates and two spores with a slow-growth phenotype that carried the BUD20 deletion (bud20Δ). Thus, BUD20 is not an essential gene; however, the bud20Δ mutant is strongly impaired in growth (Figure 5A). Next, we examined the localization of the known 40S and 60S reporters in the bud20Δ mutant. As expected, WT cells showed cytoplasmic localization for both 40S and 60S reporters S2–GFP and L5–GFP, respectively (Figure 5B). In contrast, bud20Δ cells revealed a strong nucleoplasmic accumulation of the 60S reporter L5–GFP whereas the localization of the 40S reporter S2–GFP remained unaffected (Figure 5B). If Bud20 were required for a late maturation and/or export step, we might expect a nuclear accumulation of late associating transport receptors (Arx1 and Nmd3) and shuttling trans-acting factors on pre-60S particles in the bud20Δ mutant. This was indeed the case. Both Arx1–GFP and Nmd3–GFP reporters strongly accumulated in the nucleoplasm in the bud20Δ mutant (Figure 5B). Next, we investigated whether the nuclear accumulation of Nmd3–GFP and Arx1–GFP is a direct consequence of accumulating pre-60S particles in the nucleus. To this end, we purified Arx1-TAP particles from WT and Bud20-deficient cells. To evaluate and compare the dynamic changes in the Arx1-TAP bud20Δ proteome, Ssf1-TAP, Rix1-TAP, and Kre35-TAP pre-60S particles from WT cells were also included in data acquisition and analysis. The relative enrichment of 40 trans-acting factors in the affinity-captured pre-60S particles was quantified by SRM (Figure 6A). These analyses revealed that nucleolar cluster factors (e.g., Nop4 and Nop12) that appear to participate in early biogenesis steps (Sun and Woolford, 1997; Wu et al, 2001) did not accumulate on pre-60S particles purified via Arx1-TAP in bud20Δ cells. In contrast, the transport factors (e.g., Nmd3 and Mex67-Mtr2) and trans-acting factors (e.g., Tif6, Mrt4, and Rlp24) that travel to the cytoplasm to be released were significantly enriched on the Arx1-TAP isolated from bud20Δ cells (Figure 6A and B). Western analyses confirmed this enrichment (Figure 6C). Notably, both SRM and western analyses showed that the cytoplasmic release factor Yvh1 failed to be recruited to Arx1-TAP in the bud20Δ mutant (Figure 6A–C). Collectively, these data show that late pre-60S particles loaded with transport receptors and shuttling trans-acting factors accumulate in the nucleoplasm in bud20Δ cells. Moreover, these data indicate that Bud20 is not required for the recruitment of Nmd3 and Mex67-Mtr2 to pre-60S particles. Thus, Bud20 might participate in either late maturation and/or function in the nuclear export of pre-60S subunits.
Figure 5.
Bud20 is required for proper pre-60S subunit export. (A) The bud20Δ mutant is impaired in growth at different temperatures. BUD20 and bud20Δ cells were spotted in 10-fold dilutions on YPD plates and grown at indicated temperatures for 3–7 days. (B) The bud20Δ mutant is impaired in nuclear export of pre-60S subunits. BUD20 and bud20Δ cells expressing the indicated GFP fusion proteins were grown at 30°C until mid log phase. Cells were analyzed by fluorescence microscopy. The yrb2Δ mutant served as positive control for cytoplasmic S2–GFP localization. Scale bar=5 μm.
Figure 6.
Proteome of late pre-60S particles in the bud20Δ mutant revealed by SRM. (A) A representative analysis (of three independent biological replicates) of relative enrichment of the trans-acting factors in the indicated pre-60S particles and in the bud20Δ mutant is shown using a purple to gold color-scale. Maximum enrichment is depicted as purple and minimum enrichment as gold. Each measurement represents the average of different SRM transitions per peptide and different peptides per protein. The acquired intensities for each protein were normalized based on a set of 10 large-subunit r-proteins. (B) The relative enrichment of selected proteins (labeled in (A) with red dots) in Arx1-TAP WT (BUD20) and bud20Δ samples is represented using histograms showing the fold change with error bars (±s.d.). The color of each bar corresponds to the enrichment shown in (A). (C) Western analyses of selected pre-60S trans-acting factors (labeled in (A) as green dots) using affinity-purified pre-60S particles. Samples were analyzed on NuPAGE 4–12% gradient gels followed by western analyses. The large-subunit r-protein Rpl1, Rpl3, and Rpl35 served as loading controls.
Bud20 functionally overlaps with factors involved in the nuclear export of pre-60S subunits
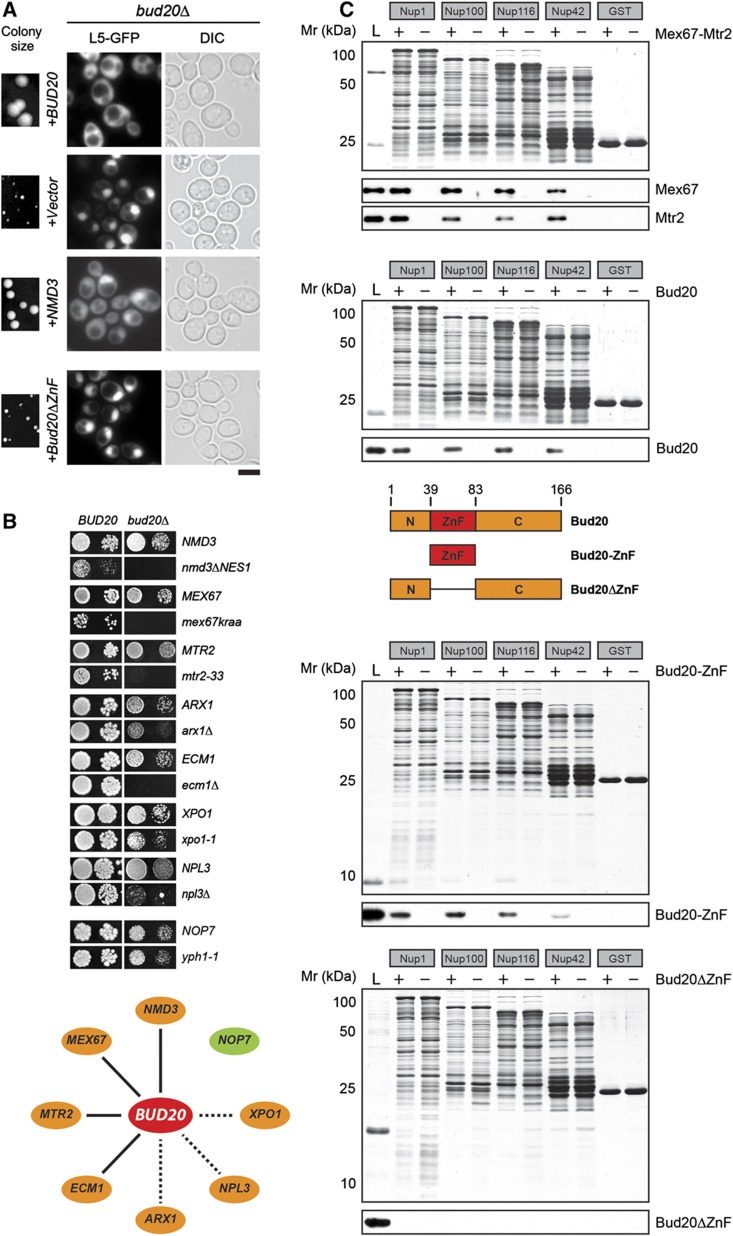
To unravel the role of Bud20 in the late maturation/export of pre-60S subunits, we searched for extra-genic high-copy suppressors of the slow growth of the bud20Δ mutant. In addition to Bud20, we found that overexpression of the NES-containing factor Nmd3 (Ho and Johnson, 1999; Ho et al, 2000) partially rescued the slow growth of the bud20Δ mutant (compare single colony size in Figure 7A). Next, we investigated whether overexpression of Nmd3 could rescue the nuclear accumulation of the 60S reporter L5–GFP exhibited by the bud20Δ mutant. Overexpression of Nmd3 that partially rescued the slow growth of bud20Δ cells also partially rescued the nucleoplasmic accumulation of the L5–GFP seen in the bud20Δ mutant (Figure 7A).
Figure 7.
Bud20 functionally overlaps with pre-60S export factors and interacts with FG repeat containing nucleoporins. (A) Overexpression of NMD3 can partially rescue slow growth and pre-60S export defect of bud20Δ mutants. The bud20Δ strain was transformed with indicated plasmids and grown on SD plates at 30°C. Pictures of single colonies were taken after 4 days. Localization of the L5–GFP reporter in these cells was determined by fluorescence microscopy. Cells were grown in SD medium to mid-log phase before picture acquisition. Scale bar=5 μm. (B) Bud20 genetically interacts with factors required for proper pre-60S subunit export. Synthetic lethality (sl) or synthetic enhancement (se) of the bud20Δ mutant combined with indicated mutants strains. Strains carrying the WT and mutant alleles were spotted in 10-fold dilutions on 5-FOA (SD/SG) plates (when sl) or YPG/YPD plates (when se) and grown at 20–30°C for 3–9 days. Solid line indicates sl, dashed line indicates se. (C) Recombinant Bud20 interacts with FG repeat sequences of different nucleoporins. Different GST-nucleoporin fusion proteins were expressed in E. coli cells and immobilized on glutathione sepharose before incubation with recombinant Bud20, Bud20-ZnF, Bud20ΔZnF, or Mex67-Mtr2 (positive control). Bound proteins were eluted by SDS sample buffer and analyzed by SDS–PAGE followed by Coomassie staining or western analyses. L=Load.
Bud20 co-enriches with late pre-60S particles (Arx1-TAP in Figure 1A–C) that carry factors such as Nmd3 and Mex67-Mtr2 that facilitate nuclear export of pre-60S subunits. Overexpression of Nmd3 partially rescued the slow growth and nuclear accumulation of the 60S reporter observed in bud20Δ cells (Figure 7A). Moreover, Bud20 shuttles between the nucleus and cytoplasm (Figure 4). These findings prompted us to test whether Bud20 genetically interacts with factors directly involved in pre-60S export. Indeed, the bud20Δ strain exhibits synthetic lethal (sl) or synthetic enhanced (se) growth defects when combined with mutants of known pre-60S subunit export factors (nmd3ΔNES1, xpo1-1, arx1Δ, ecm1Δ and npl3Δ) (Figure 7B). Previously, mex67 and mtr2 mutant alleles were isolated that are specifically impaired in 60S subunit, but not in 40S subunit or mRNA export (Baßler et al, 2001; Yao et al, 2007). These alleles (mex67kraa and mtr2-33) were found to be sl when combined with the bud20Δ strain (Figure 7B). On the other hand, a mutant allele of the early pre-60S biogenesis factor Nop7 (yph1-1) (Du and Stillman, 2002) did not genetically interact with Bud20 (Figure 7B). Together, these studies suggest that Bud20 functionally overlaps with factors that directly function in the nuclear export of pre-60S subunits.
Bud20 interacts with FG-rich nucleoporins
The co-enrichment of the shuttling factor Bud20 with late 60S pre-ribosomes (Figure 1A–C) and the different functional interactions (suppression (Figure 7A) and synthetic lethality (Figure 7B)) between Bud20 and pre-60S export factors point to a role of Bud20 in the transport of pre-60S subunits possibly via interactions with the NPC. The slow growth of bud20Δ mutant was rescued by overexpression of the NES-containing Nmd3, but not a mutant of Nmd3 that lacked NES (Nmd3ΔNES1) (Figure 7A and data not shown). This prompted us to test whether Bud20 contains a NES and therefore can bind the general export receptor Xpo1 in the presence of RanGTP in vitro. However, this was not the case suggesting that it does not contain a NES, and therefore is unlikely to function as an adapter for the exportin Xpo1 (Supplementary Figure 1).
Bud20 was reported to co-purify with NPCs (Rout et al, 2000). These data led us to investigate whether Bud20 can directly bind to FG-rich nucleoporins. For this, we incubated purified recombinant Bud20 with immobilized FG-rich domains of different nucleoporins in presence of Escherichia coli cell lysates to saturate non-specific binding sites. As a positive control, we performed the binding assay in parallel with recombinant purified Mex67-Mtr2, an export receptor that is known to interact with several FG nucleoporins (Sträßer et al, 2000; Strawn et al, 2001). As previously reported, we found that recombinant Mex67-Mtr2 efficiently binds FG-rich domains of Nup1, Nup100, Nup116, and Nup42. Like Mex67-Mtr2, we found that Bud20 efficiently binds FG-rich domains of the tested nucleoporins (Figure 7C).
Bud20 contains a central C2H2 zinc finger (ZnF) domain, a common fold among transcription factors and known to directly interact with nucleic acids (Hayes et al, 2008). Additionally, Bud20 exhibits conserved flanking N-terminal and C-terminal charged segments (Figure 7C). We investigated which region of Bud20 interacts with the different FG-nucleoporins. The ZnF domain of Bud20, but not its flanking segments efficiently bound the FG-rich domains of Nup1, Nup100, Nup116, and Nup42 (Figure 7C), and is also required for the function of Bud20 in proper nuclear export of pre-60S subunits (Figure 7A). Thus, the ZnF domain of Bud20 provides the interaction surface for FG-nucleoporin binding.
Discussion
In contrast to a handful of non-essential factors identified in prokaryotes, eukaryotic ribosome assembly and transport is aided by >200 trans-acting factors (Strunk and Karbstein, 2009; Kressler et al, 2010). Here, we developed a multiplexed SRM assay that concomitantly measures in a single MS analysis 40 trans-acting factors and 10 r-proteins, and therefore permits the rapid analysis of the proteome of maturing pre-60S particles. We exploited this assay to analyze reliably the co-enrichment of 40 trans-acting factors with pre-60S particles at different stages of maturation. Our analyses were in good agreement with western analyses (Figure 1A–C).
The main advantage of this approach is that it allowed consistent measurement of the protein set throughout all the chosen biological conditions, with basically no missing data points and a total of 350 protein data measurements acquired (50 proteins in 7 different pre-60S particles). The designed assays and their combination within a single SRM measurement can be reproduced in laboratories equipped with a triple-quadrupole-like instrument mass spectrometer, to enable the quantification of the set of ribosomal trans-acting factors in other samples or conditions of interest. The SRM coordinates are best reproduced in instruments capable of q2-fragmentation and after realignment of peptide elution times to match the chromatographic setup used, in case scheduled SRM acquisition is performed. In this study, label-free SRM-based quantification was used, but in principle SRM assays can also be combined to a variety of stable-isotope labeling techniques (Picotti and Aebersold, 2012). A limitation of targeted SRM analyses is that they do not allow or simultaneous de novo identification of proteins. A hypothesis driven target list needs to be drawn based on experiments performed before assay development or literature knowledge. Efforts aiming at developing collections of SRM assays for complete proteomes are currently underway (Picotti et al, in press), and will allow measuring other pre-ribosome-associated proteins by SRM in the near future.
Diverse energy-consuming enzymes such as GTPases, protein kinases, ATP-dependent RNA helicases, and AAA-type ATPases aid ribosome maturation and confer directionality to the assembly process (Strunk and Karbstein, 2009; Kressler et al, 2010). To unravel the precise role(s) of these energy-consuming enzymes in ribosome assembly/maturation, it is essential to determine their protein substrates and characterize the downstream effects of their activities. The AAA-ATPase Drg1 plays a crucial role in initiating final maturation steps of pre-60S particles upon their arrival in the cytoplasm (Pertschy et al, 2007). These steps involve sequential removal of transport receptors and nucleolar/nuclear trans-acting factors that travel with pre-60S particles to the cytoplasm to be released (Figure 2A). The steady-state nucleolar/nuclear localization of shuttling trans-acting factors (e.g., Tif6 and Mrt4) is very misleading since it does not reveal their need to travel to the cytoplasm for their release from pre-60S particles. Till date, genetic approaches in budding yeast have been instrumental to reveal these cytoplasmic steps. Here, we have coupled our SRM assay with genetic trapping and affinity-capture to uncover the dynamic proteome of pre-60S particles poised to initiate cytoplasmic maturation. The proteome of these genetically trapped pre-60S particles uncovered several nucleolar/nuclear factors that travel with pre-60S particles to the cytoplasm to be released, and therefore might participate in their transport and/or functional proofreading (Figure 2B). Thus, our work adds new steps to the 60S cytoplasmic maturation pathway. Moreover, the multiplexed SRM assay developed for the 40 different trans-acting factors now can be systematically employed to directly uncover the energy-consuming enzymes that trigger their release from maturing 60S pre-ribosomes.
One factor identified by our SRM analysis was the conserved and uncharacterized ZnF domain containing protein Bud20 that co-enriches with late pre-60S particles. Several evidences strongly implicate Bud20 in directly promoting nuclear export of pre-60S subunits. First, we found that the bud20Δ mutant is specifically impaired in the nuclear export of pre-60S subunits as determined by strong nucleoplasmic accumulation of several 60S reporters (Figure 5B). SRM and western analyses revealed several shuttling factors accumulating on late pre-60S particles in the bud20Δ mutant (Figure 6A–C). Second, Bud20 genetically interacts with known components of the 60S subunit export machinery (e.g., Nmd3 and Mex67-Mtr2) (Figure 7B). Third, Bud20 associates with late pre-60S particles that are loaded with factors that are directly involved in nuclear export of pre-60S subunits. Likewise, Bud20-TAP co-enriches several 60S export factors (Figure 1A–C). Fourth, we show that Bud20 shuttles between the nucleus and cytoplasm (Figure 4). Finally, in vitro binding assays showed that the ZnF domain of Bud20 directly binds FG repeats of Nup1, Nup100, Nup116, and Nup42 (Figure 7C). Taking all the data together, we propose that Bud20 could function in concert with the other transport factors (Arx1, Ecm1, Mex67-Mtr2, Npl3, and Xpo1) to efficiently translocate pre-60S particles through the NPC.
When is Bud20 released from cytoplasmic pre-60S particles? Localization of Bud20–GFP remained unaltered in yvh1Δ, jjj1Δ, and rei1Δ strains (Supplementary Figure 2). Bud20–GFP was only partially mislocalized to the cytoplasm after induction of Drg1DN, indicating that Bud20 might not be a direct substrate of the AAA-ATPase Drg1 (Figure 3). Bud20 might be released from pre-60S particles by yet unknown cytoplasmic factors that function downstream of Drg1. Notably, the essential GTPase Nug1 accumulated on Kre35-TAP (Figure 2B–D) and was strongly mislocalized to the cytoplasm upon expression of Drg1DN (Figure 3). Moreover, the heterokaryon assay demonstrated that Nug1 shuttles between the nucleus and cytoplasm (Figure 4). However, Nug1–GFP was not mislocalized in yvh1Δ, jjj1Δ, and rei1Δ strains (Supplementary Figure 2). Curiously, the FG-interacting transport receptor Mex67-Mtr2 accumulated on Kre35-TAP upon the induction of Drg1DN (Figure 2B–D), but not in yvh1Δ, jjj1Δ, and rei1Δ strains (data not shown). Elucidating the mechanisms by which the shuttling trans-acting factors and transport factors identified in this study are released from pre-60S particles remains an important challenge for the future.
Previous work showed that the downstream release of the transport factor Arx1 and the ribosomal-like protein Mrt4 by the cytoplasmic release factors Rei1 and Yvh1, respectively, requires the upstream Drg1-mediated release of the ribosome-like protein Rlp24 (Kemmler et al, 2009; Lo et al, 2009). In agreement with previous analyses, we found Rei1 failed to co-enrich with pre-60S particles upon induction of the DRG1DN allele (Figure 2B and C) (Lo et al, 2010), thereby inducing cytoplasmic mislocalization of Arx1–GFP (Figure 3). However, the reason for mislocalization of Mrt4–GFP upon induction of the DRG1DN allele was not clarified. Here, we found that Yvh1 failed to co-enrich with late pre-60S particles upon induction of the DRG1DN allele (Figure 2B–D), thereby offering a plausible explanation as to why Mrt4 accumulates on Kre35-TAP, and mislocalizes to the cytoplasm upon induction of the DRG1DN allele. Both these analyses indicate that sequential recruitment of release factors might order cytoplasmic maturation of pre-60S subunits.
The translation initiation factor and cytoplasmic GTPase Fun12 promotes binding of tRNAfMet and subunit joining during translation initiation (Pestova et al, 2000; Lee et al, 2002). Recently, the 60S subunit aided by GTPase activity of Fun12 was shown to engage in interactions with pre-40S particles to form 80S-like particles. This interaction was demonstrated to be required for efficient 20S pre-rRNA processing leading to the formation of mature 40S subunits (Lebaron et al, 2012). Our SRM analyses revealed that Fun12 maximally co-enriches with Arx1-TAP (Figure 1A). It appears that late pre-60S particles might be competent to load Fun12, potentially to check their ability to subsequently engage with pre-40S subunits for cytoplasmic maturation and translation initiation.
The ABC-type ATPase Rli1 (Yarunin et al, 2005) is known to be involved in disassembling the 80S particle after translation and 80S-like particles after 20S pre-rRNA cleavage (Pisareva et al, 2011; Shoemaker and Green, 2011; Strunk et al, 2012). Our SRM data show that Rli1 is recruited to early pre-60S particles in the nucleus (Figure 1A). Thus, Rli1 might sense the ability of the 60S subunit to disengage 80S-like particles after 20S pre-rRNA cleavage and terminate translation, already during nuclear pre-60S maturation. Interestingly, Rli1 accumulated on Kre35-TAP upon induction of Drg1DN (Figure 2B). Thus, Rli1 in concert with Tif6 might actively prevent the genetically trapped immature pre-60S particles from interacting with 40S subunits.
In summary, we devised a strategy in which genetic trapping and affinity-capture were combined with the reliability of SRM, to measure dynamic interactions of 50 trans-acting factors and r-proteins with maturing pre-60S particles, in a high-throughput manner. Our strategy represents a significant step towards characterizing dynamic proteomes of pre-ribosomal particles, and thus opens the way to analyze the functional and mechanistic contribution of the >50 diverse energy-consuming factors involved in the complex ribosome assembly process, a goal that is of obvious importance to the ribosome maturation field. Moreover, our targeted approach based on SRM and genetic trapping provides a versatile discovery tool to reveal dynamic interactions of macromolecular assemblies involved in processes such as replication, transcription, cell division, chromatin remodeling, protein and RNA metabolism, and how these interactions might be regulated by diverse energy-consuming enzymes. A reliable quantitative view of dynamic protein interactions should aid modeling efforts aimed at understanding the regulatory aspects of essential anabolic and catabolic processes.
Materials and methods
Sample preparation for mass spectrometry
Pre-60S particles were isolated from various strains by tandem affinity purification (TAP) using established protocols (Kemmler et al, 2009). Tobacco etch virus (TEV) protease treated eluates were precipitated using trichloroacetic acid (TCA) and resuspended in denaturing buffer containing 8 M urea, 50 mM NH4HCO3, and 5 mM EDTA. Next, proteins were reduced with 12 mM dithiothreitol for 30 min at 32°C and alkylated with 40 mM iodoacetamide for 45 min at 25°C. The samples were diluted 1:5 with 0.1 M NH4HCO3 and digested with sequencing-grade porcine trypsin (Promega, Madison, WI, USA) at an enzyme/substrate ratio of 1:100. The digestion was performed overnight and stopped with formic acid to a final concentration of 2%. The peptide mixtures were desalted on Sep-Pak C18 cartridges (Waters, Milford, MA, USA), eluted with 80% acetonitrile, vacuum centrifuged until dryness and resuspended in 0.15% formic acid.
Shotgun proteomic analyses of purified pre-ribosomal particles
The peptide samples were measured in shotgun mode on a 5600 TripleTOF™ (ABSciex, Concord, Canada) with a nano-electrospray ion source. For the chromatographic separation of the peptides, the instrument was coupled with an Eksigent Nano LC system (ABSciex, Foster City, CA, USA) equipped with a 15-cm fused silica column, 75 μm inner diameter (BGB Analytik, Böckten, Switzerland), packed in-house with Magic C18 AQ, 5 μm beads (Michrom Bioresources, Leonberg, Germany). The peptide mixtures (∼3 μg) were loaded from a autosampler cooled to 4°C (ABSciex, Foster City, CA, USA) and separated with a linear gradient of acetonitrile/water containing 0.1% formic acid from 5 to 35% acetonitrile in 120 min, with a flow rate of 300 nl/min. The mass spectrometer was operated in data-dependent acquisition mode. For TOF scans, the accumulation time was set to 0.299995, s, and the mass range to 400–1250 Da. Peptides with 2–5 charges and signals exceeding 150 cps were selected for fragmentation. Per cycle, up to 20 precursor ions were monitored and excluded for 20 s after one occurrence. The product ion scan was performed with an accumulation time of 0.149998, s, and a mass range of 170–1500 Da in a high sensitivity mode. The total cycle time was 3.35 s.
The collected spectra were searched against the Saccharomyces cerevisiae SGD protein database with Sorcerer™-SEQUEST® (Thermo Electron, San Jose, CA, USA). Trypsin was set as the digesting protease with the tolerance of two missed cleavages, one non-tryptic terminus and not allowing for cleavages of KP and RP peptide bonds. The monoisotopic peptide and fragment mass tolerances were set to 50 p.p.m. and 0.8 Da, respectively. Carboxyamidomethylation of cysteines (+57.0214 Da) was defined as fixed modification and oxidation of methionines (+15.99492) as variable modification. Protein identifications were statistically analyzed with ProteinProphet (v3.0) and filtered to a cutoff of 0.9 ProteinProphet probability, which in this case corresponds to a FDR<1%, calculated based on a target-decoy approach (Elias and Gygi, 2007). Raw shotgun MS data are available at http://www.peptideatlas.org/PASS/PASS00115.
SRM assay development
For each protein of interest, up to five peptides were selected from the FDR-filtered Sequest output. Peptides with the highest spectral counts, a length between 8 and 30 amino acids and devoid of methionine residues and missed/mis-cleavages were chosen for SRM assay development. For each peptide, we selected from the product ion spectra acquired in shotgun mode the five most intense SRM transitions in a mass range of 350–1250 Da, using Skyline (v1.3, MacCoss Lab Software, Seattle, WA, USA). For proteins with less than five suitable peptides identified, we chose additional peptides from the PeptideAtlas database (http://www.peptideatlas.org/). Only singly charged product ions were considered. For assay refinement, we experimentally tested the set of transitions in SRM mode using a pool of tryptic digests of the different purified pre-ribosomal particles. Samples were measured on a triple-quadrupole/ion trap mass spectrometer (5500 QTrap, ABSciex, Concord, Canada) equipped with a nano-electrospray ion source. Peptides were loaded and chromatographically separated by the system described above at a flow rate of 350 nl/min. A gradient from 5–35% acetonitrile in 30 min was used, corresponding to a total MS acquisition time of about 1 h. SRM analysis was conducted with Q1 and Q3 operated at unit resolution (0.7 m/z half maximum peak width) with up to 70 transitions per run (dwell time, 30 ms; cycle time <2.5 s). Data were analyzed with Skyline and for the final SRM assays only the top three to four transitions per peptide with no obvious interferences and up to three peptides per protein were retained. Collision energies were calculated according to the formula: CE=0.044 × m/z+5.5 (CE: collision energy, m/z: mass-to-charge ratio of the precursor ion).
SRM-based quantitation and statistical analysis
All transitions were pooled in one scheduled-SRM method with a 30-min gradient, using retention times extracted during the assay refinement. A cycle time of 2.5 s and a retention time window of ±3 min were used. Peak height of the transitions was used for quantitation, after confirming co-elution and shape similarity of the transitions monitored for each peptide. Outlier transitions (e.g., shouldered or noisy transition traces) were not considered in the calculations. Results are presented as average values out of all transitions and peptides per protein, after an intensity normalization step based on a set of 10 large-subunit r-proteins. Relative co-enrichment of each protein in each sample was calculated as a fraction of the maximum intensity acquired for the same protein throughout the sample set. Standard deviations were calculated for the relative co-enrichment or fold enrichment based on the ratios of all transitions for one given protein. Raw SRM-MS data are available at http://www.peptideatlas.org/PASS/PASS00123.
Genetic methods and analysis
Genomic disruptions and C-terminal tagging at the genomic loci were performed as previously described (Faza et al, 2009). Preparation of media, yeast transformations, mating, sporulation of diploids, tetrad analysis, and genetic manipulations were performed according to established procedures. All plasmids and yeast strains used in this study are listed in Supplementary Tables 4 and 5, respectively. Genetics were performed as previously described (Faza et al, 2012).
Biochemical analyses and binding assays
TAPs of pre-ribosomal particles were carried out as previously described (Kemmler et al, 2009). Cells were grown in 2 l cultures OD600=3. For DRG1DN strain, expression of dominant negative DRG1 was induced by 0.5 mM copper sulfate. Two-step purifications were performed using IgG sepharose and calmodulin sepharose in the lysis buffer (50 mM Tris–HCl pH 7.5, 75 mM NaCl, 1.5 mM MgCl2, 0.15% NP-40). Eluates were analyzed by NuPAGE 4–12% Bis-Tris gel (Invitrogen, Zug, Switzerland) followed by silver staining or western analysis.
Recombinant Bud20 and Mex67-Mtr2 were produced in BL21 E. coli strain by IPTG induction and affinity purified using Ni sepharose (GE Healthcare, Uppsala, Sweden). Recombinant GST-tagged proteins were expressed in BL21 E. coli strain upon IPTG induction. Protein pull-down assays were carried out in universal buffer (Künzler and Hurt, 1998). GST-Nucleoporins were immobilized on glutathione sepharose (GE Healthcare, Uppsala, Sweden) in the presence of E. coli lysate and incubated with purified Bud20 or Mex67-Mtr2. Bound proteins were eluted with SDS sample buffer.
Western analyses were performed as previously described (Kemmler et al, 2009). The following primary antibodies were used: α-Bud20 (1:4000; this study), α-Mex67 (1:5000; C Dargemont, Institut Jacques Monod, Paris, France), α-Mtr2 (1:1000; E Hurt, Heidelberg University, Heidelberg, Germany), α-Mex67/Mtr2 (1:3000; this study), α-Nmd3 (1:5000; A Johnson, University of Texas at Austin, Austin, TX, USA), α-Nog1 (1:1000; M Fromont-Racine, Institut Pasteur, Paris, France), α-Nop7 (1:2000; B Stillman, Cold Spring Harbor Laboratory, New York, NY, USA), α-Nug1 (1:1000; this study), α-Rlp24 (1:2000; M Fromont-Racine, Institut Pasteur, Paris, France), α-Tif6 (1:2000; GenWay Biotech, San Diego, CA, USA), α-Yvh1 (1:4000; this study), α-polyHistidine (1:1000; Sigma-Aldrich, Buchs SG, Switzerland), α-Rpl1 (1:10 000; F Lacroute, Centre de Génétique Moléculaire du CNRS, Gif-sur-Yvette, France), α-Rpl3 (1:5000; J Warner, Albert Einstein College of Medicine, Bronx, NY, USA) and α-Rpl35 (1:4000; this study). The secondary HRP-conjugated α-rabbit, α-mouse and α-chicken antibodies (Sigma-Aldrich, St Louis, MO, USA) were used at 1:1000–1:5000 dilutions. Protein signals were visualized using Immun-Star HRP chemiluminescence kit (Bio-Rad Laboratories, Hercules, CA, USA). SRM and western analyses were performed on three independent biological replicates. All TAP purifications and SRM analyses depicted in each figure were performed in parallel.
Fluorescence microscopy and heterokaryon assay
Cells were visualized using DM6000B microscope (Leica, Solms, Germany) equipped with HCX PL Fluotar × 63 1.25 NA oil immersion objective (Leica, Solms, Germany). Images were acquired with a fitted digital camera (ORCA-ER; Hamamatsu Photonics, Hamamatsu, Japan) and Openlab software (Perkin-Elmer, Waltham, MA, USA).
The heterokaryon assay was adapted from previously described (Belaya et al, 2006). In short, strains were grown until OD600 of ∼1. For mating, Arx1–GFP, Gar1–GFP, Bud20–GFP, Nug1–GFP expressing cells were mixed with Nup82–mCherry cells overexpressing kar1-1. The mixture was concentrated onto 0.45 μm nitrocellulose filters and subsequently placed on YPD plates. After 1 h at 30°C, filters were transferred to YPD plates containing 50 μg/ml cycloheximide and incubated for another 1–2 h before cells were analyzed by fluorescence microscopy.
Supplementary Material
Supplementary Figures S1–2, Supplementary Tables S1, S4, and S5
Acknowledgments
We are grateful to E Hurt, M Peter, A Johnson, J Warner, P Milkereit, J Woolford, M Fromont-Racine, F Lacroute, B Stillman, C Dargemont, M Künzler, M Rexach, D Tollervey, H Bergler, G Schlenstedt, and B Zehnder for generously sharing strains, plasmids, and antibodies. We thank J Pfannstiel and Y Feng for mass spectrometry analysis and all members of the Panse laboratory for enthusiastic discussions. P Picotti and VG Panse are supported by grants from the Swiss National Science Foundation and the ETH Zürich. P Picotti was supported by a Promedica Stiftung and a Marie Curie Reintegration Grant (ERG-277147). VG Panse is the recipient of a Starting Grant Award (EURIBIO260676) from the European Research Council.
Author contributions: MA, YC, AM, LO, SS, UR, PP, and VGP designed the experiments; MA, YC, AM, LO, SS, and UR performed the experiments; MA, YC, AM, PP, and VGP analyzed the data; and PP and VGP wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham A-JL, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM et al. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Hunter CL (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 5: 573–588 [DOI] [PubMed] [Google Scholar]
- Baßler J, Grandi P, Gadal O, Leßmann T, Petfalski E, Tollervey D, Lechner J, Hurt E (2001) Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 [DOI] [PubMed] [Google Scholar]
- Belaya K, Tollervey D, Koš M (2006) FLIPing heterokaryons to analyze nucleo-cytoplasmic shuttling of yeast proteins. RNA 12: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson N, James DA, Ivosev G, Tate SA, Bonner R, Taylor L, Pawson T (2011) Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol 29: 653–658 [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgärtel V, Boese G, Bassler J, Wild K, Peters R, Yoneda Y, Sinning I, Hurt E (2007) Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell 27: 767–779 [DOI] [PubMed] [Google Scholar]
- Du Y-CN, Stillman B (2002) Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109: 835–848 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG (2012) Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet 8: e1002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faza MB, Kemmler S, Jimeno S, González-Aguilera C, Aguilera A, Hurt E, Panse VG (2009) Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol 184: 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauß D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A-C, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Baßler J, Petfalski E, Strauß D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, Gavin A-C, Hurt E (2002) 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115 [DOI] [PubMed] [Google Scholar]
- Hayes PL, Lytle BL, Volkman BF, Peterson FC (2008) The solution structure of ZNF593 from Homo sapiens reveals a zinc finger in a predominately unstructured protein. Protein Sci 17: 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH-N, Johnson AW (1999) NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol 19: 2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH-N, Kallstrom G, Johnson AW (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams S-L, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hung N-J, Lo K-Y, Patel SS, Helmke K, Johnson AW (2008) Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol Biol Cell 19: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Lund E, Dahlberg J (2002) Nuclear export of ribosomal subunits. Trends Biochem Sci 27: 580–585 [DOI] [PubMed] [Google Scholar]
- Kemmler S, Occhipinti L, Veisu M, Panse VG (2009) Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J Cell Biol 186: 863–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baßler J (2010) Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Kressler D, Roser D, Pertschy B, Hurt E (2008) The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J Cell Biol 181: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler M, Hurt EC (1998) Cselp functions as the nuclear export receptor for importin α in yeast. FEBS Lett 433: 185–190 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, Granneman S, Watkins NJ, Tollervey D (2012) Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol 19: 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Pestova TV, Shin B-S, Cao C, Choi SK, Dever TE (2002) Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc Natl Acad Sci USA 99: 16689–16694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K-Y, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K-Y, Li Z, Wang F, Marcotte EM, Johnson AW (2009) Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol 186: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy TI, Silver PA (1999) Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev 13: 2118–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Baszler J, Petfalski E, Tollervey D, Hurt E (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Dlakić M, Tollervey D (2004) A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev 18: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG (2011) Getting ready to translate: cytoplasmic maturation of eukaryotic ribosomes. CHIMIA 65: 765–769 [DOI] [PubMed] [Google Scholar]
- Panse VG, Johnson AW (2010) Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 35: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, Högenauer G, Fromont-Racine M, Bergler H (2007) Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol 27: 6581–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 9: 555–566 [DOI] [PubMed] [Google Scholar]
- Picotti P, Clement-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Röst H, Sun Z, Rinner O, Reiter L, Shen Q, Michaelson JJ, Frei A, Alberti S, Kusebauch U, Wollscheid B, Moritz R, Beyer A, Aebersold R (2012) A complete mass spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV, Pisarev AV (2011) Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30: 1804–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex. J Cell Biol 148: 635–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Strausz D, Petfalski E, Tollervey D, Hurt E (2003) The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 22: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R (2011) Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA 108: E1392–E1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K, Baßler J, Hurt E (2000) Binding of the Mex67p/Mtr2p heterodimer to Fxfg, Glfg, and Fg repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol 150: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR (2001) The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem 276: 6445–6452 [DOI] [PubMed] [Google Scholar]
- Strunk BS, Karbstein K (2009) Powering through ribosome assembly. RNA 15: 2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K (2012) A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Woolford JL (1997) The yeast nucleolar protein Nop4p contains four RNA recognition motifs necessary for ribosome biogenesis. J Biol Chem 272: 25345–25352 [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Wu K, Wu P, Aris JP (2001) Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res 29: 2938–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Roser D, Köhler A, Bradatsch B, Baßler J, Hurt E (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
- Yao Y, Demoinet E, Saveanu C, Lenormand P, Jacquier A, Fromont-Racine M (2010) Ecm1 is a new pre-ribosomal factor involved in pre-60S particle export. RNA 16: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC (2005) Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J 24: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–2, Supplementary Tables S1, S4, and S5