Abstract
The fifth component (CSN5) of the mammalian COP9 signalosome complex plays an essential role in cell proliferation and senescence, but its molecular mediator remains to be determined. Here, we searched for interactors among various cell cycle regulators, and found that CSN5, but not the CSN holo-complex, bound to CDK2 in vivo and in vitro. Depletion of CSN5 enhanced phosphorylation of CDK2 by Akt, resulting in cytoplasmic accumulation of CDK2 together with cyclin E in a leptomycin B-resistant manner, and impaired phosphorylation of the retinoblastoma protein. Additional knockdown of CDK2, which reduced the expression of cyclin E to the normal level, did not restore cell proliferation, but significantly suppressed senescence in CSN5-depleted cells. Enforced expression of cytoplasmic cyclin E induced premature senescence in immortalized cell lines. These results show that CSN5 functions through CDK2 to control premature senescence in a novel way, depending on cyclin E in the cytoplasm.
The COP9 signalosome (CSN) complex is composed of 8 subunits (CSN1-8) and well conserved in all eukaryotes from yeast to humans1,2. Although originally discovered as a suppressor of photomorphogenesis in higher plants3, the CSN is now known to be involved in various biological responses4. The most well studied biochemical function of the CSN is the deneddylation of the cullin subunit of cullin-RING ubiquitin ligases (CRLs), the largest family among the ubiquitin ligases, thereby regulating the protein expression by controlling proteolysis through the 26S proteasome5,6,7,8,9. The fifth subunit of the CSN (CSN5, also known as Jab1. See below.) plays an important role in the deneddylation activity, and the integrity of the JAMM domain located in the N-terminus is essential to the function as a deneddylase1,4,8, presumably acting as a catalytic center of the isopeptidase (deneddylase) enzyme. However, the fact that the CSN5 polypeptide alone is deficient in this enzymatic activity indicates that the deneddylation reaction requires the holo-CSN complex.
Among the 8 components of the CSN, CSN5 is unique in many ways. Mammalian CSN5 was originally identified as a protein binding to the transcription factors c-Jun and JunD, and so termed Jun-activation-domain-binding protein (Jab) 110. Thereafter, it was repeatedly isolated as an interactor of factors regulating signal transduction and cell proliferation/survival1,4. Besides being a key component of the deneddylase, CSN5 is also suggested to be involved in other biochemical functions, (i) determination of the specificity of transcription factors such as c-Jun, JunD, and E2F-110,11, (ii) mediation of the phosphorylation of c-Jun, NFκB, and p53 by CSN-associated kinases12,13, and (iii) control of the intracellular distribution of signaling molecules such as p27, COP1, p53, etc.14,15 by ill-defined mechanisms. The multi-functionality of CSN5 may stem from the fact that it exists as a monomer or a smaller complex outside of the holo-CSN complex and is sometimes suggested to act as an individual factor as well as the core of the CSN complex16,17,18,19,20,21. However, the molecular identity and the precise function of the smaller form remain to be investigated.
During the last decade, substantial evidence has accumulated demonstrating that the function of the CSN and its components, especially CSN5, is critical to the proliferation and survival of mammalian cells. First, CSN5 is an oncogene. A high level of CSN5 has been found in many human cancers, and is often correlated with a poor prognosis4 (and see references within). Knockdown of CSN5 inhibits the proliferation of human tumor cells22,23, suggesting that overexpression of CSN5 not only serves as a marker of malignant transformation, but also actually contributes to tumor cell proliferation. In fact, it was shown that an intact CSN was required for the growth of Ras-transformed cells24. Second, ectopic expression of a stable form of CSN5 in mice induces the development of myeloproliferative disorders with expansion of the stem cell population25. Thus, CSN5 functions in favor of cell proliferation/survival and eventual tumorigenesis. Third, mice deficient in a CSN component (CSN2, CSN3, CSN5, or CSN8) die at a very early stage of embryonic development26,27,28,29,30. Knocked-out cells are incapable of proliferation, and undergo accelerated apoptotic cell death, which is frequently accompanied by elevated levels of cyclin E, the CDK inhibitor p27, and the tumor suppressor p53. Conditional knockout of CSN5 in mouse embryonic fibroblasts (MEFs) revealed that (i) the JAMM domain-dependent function of CSN5 is required at multiple points during the cell cycle (e.g., G1, S, and G2/M), (ii) CSN5-depletion increases the population of cells with higher ploidy (4n and more), and (iii) CSN5-depletion induces cellular senescence even in the p53-null background and eventual cell death31. Cellular senescence is the process, by which cells cease to proliferate and eternally withdraw from the cell cycle32. Senescence has multiple causes, known collectively as Hayflick factors, including telomere shortening, accumulation of DNA damage, and derepression of the INK4a/ARF locus, which are induced in overpassaged cultures of primary cells, by activation of oncogenic pathways, and by the generation of reactive oxygen species (ROS)33, and recently accepted as a part of the defense mechanism against cancer34. In a normal setting, the tumor suppressor p53 plays an essential role, but recent findings suggest that a p53-independent mechanism is also involved in the control of senescence35,36,37,38,39. The precise molecular mechanism that initiates the senescence program remains unclear. CSN5-null MEFs exhibited more neddylated Cullins 1 and 4, and contained an increased amount of various proteins including cyclins, CDKs, and CDK inhibitors31. However, the actual molecular targets by which CSN5-depletion affects progression of the cell cycle, ploidy control, and p53-independent senescence are not clear yet. In this study, we sought for a more direct connection between CSN5 and cell cycle regulators, and found that CDK2 but not cyclins formed a stable complex with CSN5 outside of the holo-CSN complex. CSN5-depletion increased the levels of CDK2 and cyclin E in the cytoplasm, and concomitant knockdown of CDK2 reduced the level of cytoplasmic cyclin E and rescued induction of senescence but not the defect of cell proliferation and increase of ploidy. Cytoplasmic cyclin E was detected in mouse embryonic fibroblasts (MEFs) that entered proliferative senescence. Enforced expression of cytoplasmic cyclin E in immortalized mouse and human cells induced senescence. This is the first identification of a factor (CDK2) interacting with CSN5 outside of the CSN holo-complex, which regulates cellular senescence in a novel fashion. CSN5 may function other than as a catalytic core of the CSN deneddylase, playing an essential role in mammalian cell proliferation.
Results
CSN5 interacts with CDK2
Mammalian CSN5 is closely involved with regulation of the cell cycle31. It is now accepted that a strictly-regulated proteolysis involving the ubiquitin-proteasome system plays an essential role in progression of the cell cycle40,41,42, and the CSN is the upstream regulator of the largest family of ubiquitin ligases, CRLs4,5,6,7,8. Therefore, one might assume that the CSN and its catalytic-core subunit, CSN5, participate in regulation of the cell cycle through proteasome-mediated proteolysis. In addition to this, however, we decided to explore the possibility of a more direct connection between CSN5 and the cell cycle machinery.
Besides being a catalytic center of the CSN deneddylase8, CSN5 exists as a monomer (in plants) or smaller complexes (in mammals) and is speculated to act as an individual factor outside the CSN complex16,17,18,19,20,21. As a tool to analyze CSN5-associated complexes, we previously developed two antibodies recognizing CSN5 and CSN1, respectively19. The αCSN5 antibody does not crossreact with other CSN subunits and immunoprecipitates a monomeric or a smaller form of CSN5 free of other CSN subunits, while the αCSN1 antibody, which does not crossreact with other CSN subunits either, recognizes a holo-CSN complex (containing all other CSN subunits). Fig. 1A shows a part of the characterization of the antibodies. αCSN1 immunoprecipitates contained all CSN subunits, CSN1-8 (mammalian CSN consists of two CSN7 subtypes, CSN7a and CSN7b, and both were included in αCSN1 immunoprecipitates.), whereas no other CSN subunits were detected in αCSN5 immunoprecipitates. In addition, αCSN1-immunoprecipitates, but not αCSN5-immunoprecipitates, contained the components of the Cullin RING ubiquitin ligase, Skp2, Cul1, and Cul4 (Fig. 1A). Using this differential immunoprecipitation method, we searched for a novel protein interacting with CSN5 among the various cell cycle regulators. As shown in Fig. 1B, we eventually found that Cyclin-Dependent Kinase-2 (CDK2) stably formed a complex with CSN5, but not with the holo-CSN complex (at least not in αCSN1 immunoprecipitates) in mouse fibroblasts. Mouse cells harbor two types of CDK2 proteins with markedly different molecular weights (p39 and p33) produced by alternative splicing43, the faster migrating form being the major form, both of which were found in αCSN5-immunoprecipitates. However, we detected neither of the activating partners of CDK2, cyclins E and A, in αCSN5 immunoprecipitates (Fig. 1B), although we detected both cyclins in αCDK2-immunoprecipitates (Fig. 1B), indicating that CSN5 interacted exclusively with a monomeric form of CDK2.
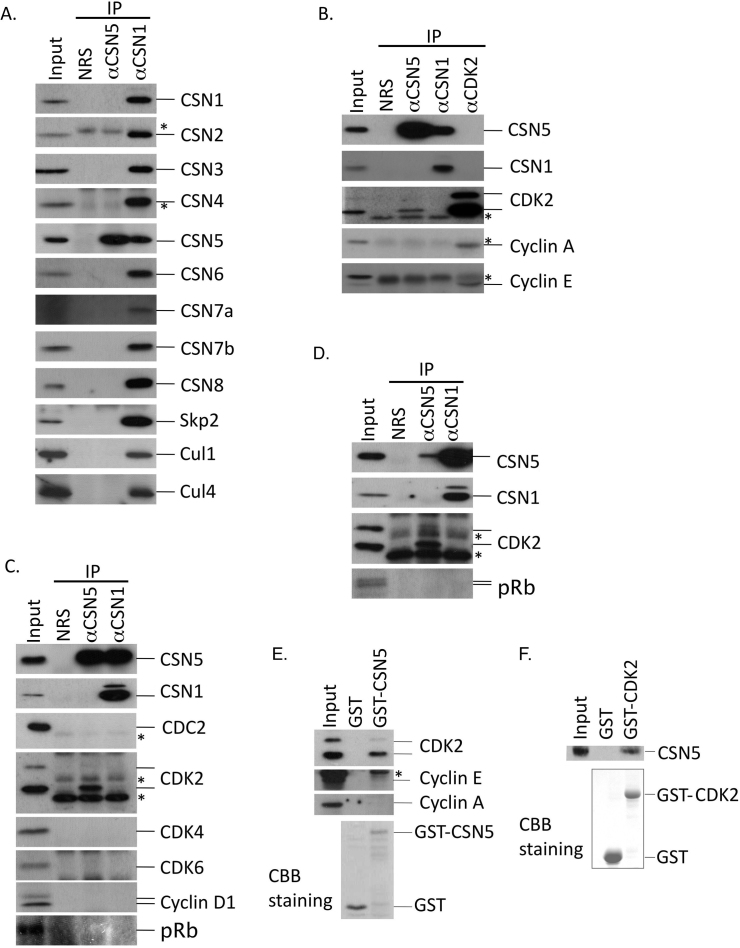
Figure 1. CSN5 specifically interacts with CDK2.
(A) Lysate from proliferating NIH3T3 cells was immunoprecipitated with normal rabbit serum (NRS) and rabbit antibodies to CSN5 and CSN1 (αCSN5 and αCSN1, respectively). The immune complex was analyzed by Western blotting using antibodies against CSN1, CSN2, CSN3, CSN4, CSN5, CSN6, CSN7a, CSN7b, CSN8, Skp2, Cul1, and Cul4. (B) Lysate from NIH3T3 cells was immunoprecipitated with NRS and rabbit antibodies raised against CSN5, CSN1 and CDK2. The immune complex was analyzed by Western blotting using antibodies against CSN5, CSN1, CDK2, cyclin A, and cyclin E. (C) Lysate from NIH3T3 cells was immunoprecipitated as in A. The immune complex was analyzed by Western blotting using antibodies against CSN5, CSN1, CDC2, CDK2, CDK4, CDK6, cyclin D1, and pRb. (D) Lysate from proliferating C2C12 cells was immunoprecipitated as in A. The immune complex was analyzed by Western blotting using antibodies against CSN5, CSN1, CDK2, and pRb. (E) Immobilized recombinant GST proteins (GST and GST-CSN5) were incubated with lysate from proliferating NIH3T3 cells. Bound proteins were detected by immunoblotting using antibodies to CDK2, and cyclins E and A. GST-proteins used in the assay were visualized by CBB staining (lower panel). (F) Immobilized recombinant GST proteins (GST and GST-CDK2) were incubated with lysate from proliferating NIH3T3 cells. Bound proteins were detected by immunoblotting using antibodies to CSN5. GST-proteins used in the assay were visualized by CBB staining (lower panel). (A–F) Input was 10% of the cell lysate used for immunoprecipitation. *; nonspecific background.
The binding of CSN5 to CDK2 is very specific because we found little or no interaction of CSN5 with other CDKs, including CDC2 ( = CDK1), CDK4 and CDK6, by immunoprecipitation in cell lysates isolated from proliferating mouse fibroblasts (Fig. 1C). Although it was reported that, in Drosophila, the CSN physically interacts with the retinoblastoma (Rb) protein44, we detected little association of mammalian CSN5 and the holo-CSN complex (αCSN1 immunoprecipitates) with the Rb protein or cyclin D1 (Fig. 1C). The interaction of CSN5 with CDK2 was not restricted to fibroblasts. We detected a similar complex of CSN5 and CDK2 in C2C12 mouse myoblasts (Fig. 1D). Finally, we successfully recapitulated the interaction between CSN5 and CDK2 in vitro using the GST-pull down assay (Fig. 1E, F). Again, the major partners of CDK2, cyclins A and E, were not detectable in GST-CSN5-pull downs (Fig. 1E). Thus, while the holo-CSN complex binds to the CRL complex, CSN5 specifically forms a complex with CDK2, free from cyclins and outside of the holo-CSN.
Interaction between CSN5 and CDK2 during the cell cycle
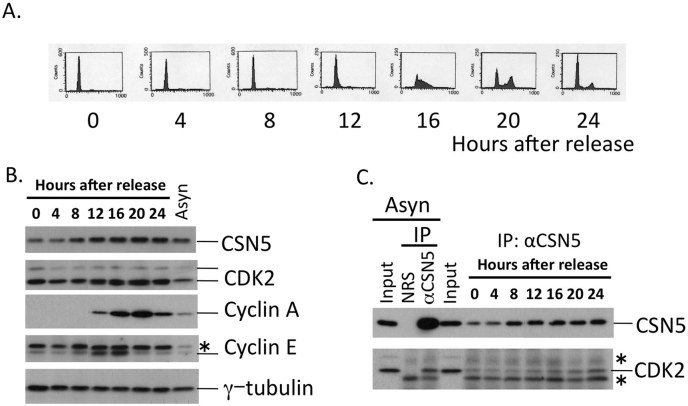
Since each CDK functions at a specific point in the mammalian cell cycle, we analyzed the CSN5-CDK2 interaction during the cell cycle. NIH3T3 mouse fibroblasts were arrested at G0/G1 by depriving them of serum, and allowed to reenter the cell cycle by adding serum. Cells entered the S phase around 12–16 hours after re-addition of the serum, shown by an increase in DNA content (Fig. 2A) and the induced expression of cyclins E and A (Fig. 2B). The expression of both CSN5 and CDK2 was relatively constant throughout the cell cycle with a slight increase after entering the S phase (Fig. 2B). Immunoprecipitation/immunoblotting assays revealed that the complex of CSN5 and CDK2 existed in G0/G1 and throughout the cell cycle with a slight increase later on in accordance with the slightly-up-regulated expression of both proteins (Fig. 2C).
Figure 2. CSN5 interacts with CDK2 throughout the cell cycle.
NIH3T3 cells were synchronized at G0/G1 by contact inhibition for 48 hours, and released to reenter the cell cycle by plating at low density. (A) Cells at indicated times were harvested and analyzed for DNA content by flow cytometry. (B) Lysates isolated from cells at indicated times were analyzed by Western blotting using antibodies against CSN5, CDK2, cyclin A, cyclin E, and γ-tubulin. (C) Lysates isolated from cells at indicated times and asynchronously proliferating cells were immunoprecipitated with NRS and a rabbit antibody to CSN5. The immune complex was analyzed by Western blotting using antibodies against CSN5 and CDK2. Input was 10% of the cell lysate used for immunoprecipitation. * in (B) and (C); nonspecific background.
Conditional knockout of CSN5 in MEF cells
To better understand the physiological relevance of CSN5's functions including binding to CDK2 in proliferating cells, we conditionally knocked out the CSN5 gene in an inducible way in cultured cells because cells are not well maintained for a long time after knocking out the CSN5 allele26,27,31. Mouse embryonic fibroblasts (MEFs) containing the floxed allele of the CSN5 gene27,31 and immortalized by the null allele of p53 were infected with a retroviral vector containing inducible CRE recombinase, which was fused with one or two copies of the estrogen receptor (ER) (CRE-ER and ER-CRE-ER, respectively)45. Addition of 4-Hydroxytamoxifen (4OHT, (Z)-4-(1-[4 -(Dimethylaminoethoxy) phenyl] -2-phenyl-1 -butenyl)phenol) into the medium allowed the ER-fused CRE protein to translocate into the nucleus, excise the genomic DNA fragment flanked with the floxed sequence, and inactivate the CSN5 locus. Fig. 3A shows that both CRE-ER and ER-CRE-ER converted the floxed allele of CSN5 into the deleted allele, CRE-ER being more efficient than ER-CRE-ER. We, therefore, chose to use CRE-ER in the subsequent experiments.
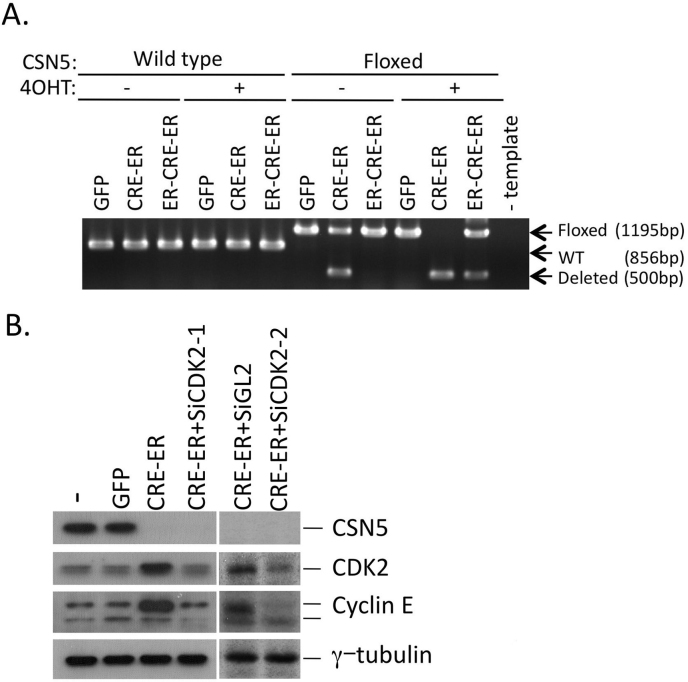
Figure 3. Inducible conditional knockout of the CSN5 locus.
(A) Wild-type and CSN5-floxed MEFs immortalized by the null allele of p53 were infected with viruses containing GFP alone and together with ER-fused CRE (CRE-ER and ER-CRE-ER), and selected for the GFP signal. High molecular weight DNA was extracted from cells cultured with and without tamoxifen (4OHT), and the genotypes were determined by genomic PCR as described31. (B) CSN5-floxed p53−/− MEFs were infected with viruses containing GFP and GFP plus CRE-ER, or co-infected with viruses containing CRE-ER and siRNA (siGL2 for negative control, and two types of siCDK2, siCDK2-1 and 2-2). GFP-positive cells were selected, treated with 4OHT, and harvested. Cell lysates were analyzed by Western blotting using antibodies against CSN5, CDK2, cyclin E, and γ-tubulin. -; mock infection.
After treatment with 4OHT of CSN5-floxed MEF cells infected with CRE-ER (MEF CSN5-Floxed CRE-ER), the level of CSN5 protein declined. Consistent with the previous results26,31, the levels of CDK2 and cyclin E proteins were higher in CSN5-depleted cells (Fig. 3B). To elucidate the physiological meaning of CDK2's up-regulation in CSN5-depleted cells, we knocked down CDK2 on top of the CSN5-gene-knockout. MEFs harboring the floxed CSN5 allele were infected with the CRE-ER virus together with the virus containing siRNA specific to mouse CDK2 (siCDK2-1 and siCDK2-2), and treated with 4OHT (Fig. 3B). Treatment with CDK2-specific siRNA neutralized CDK2's upregulation due to depletion of CSN5, thereby allowing the CDK2 protein to return to a level equivalent to that in normally proliferating cells. Interestingly, the level of cyclin E, which increased in accordance with CDK2's upregulation, also returned to that in normally proliferating cells, indicating that CSN5 regulates the level of cyclin E through CDK2.
Because the main action point of CDK2 in association with cyclins E and A is in the G1 and S phases46, we examined the specific effect of CSN5-depletion on progression through G1. To do this, we arrested MEFs at G0/G1 by contact inhibition and synchronously moved the cell cycle by replating. Because it takes 7 days after gene knockout to eliminate the CSN5 protein31 and completely arrest the cell cycle, we cultured cells for 4 days in a normal medium after gene depletion by the treatment with 4OHT, arrested them by contact inhibition for 48 hours, and allowed them to synchronously reenter the cell cycle. In the absence of 4OHT, cells entered the S phase around 16–20 hours after release (Fig. 4A). However, the majority of cells treated with 4OHT failed to progress through the cell cycle and only a small portion entered S phase, at a significantly delayed time point, approximately 28 hours and later (Fig. 4A).
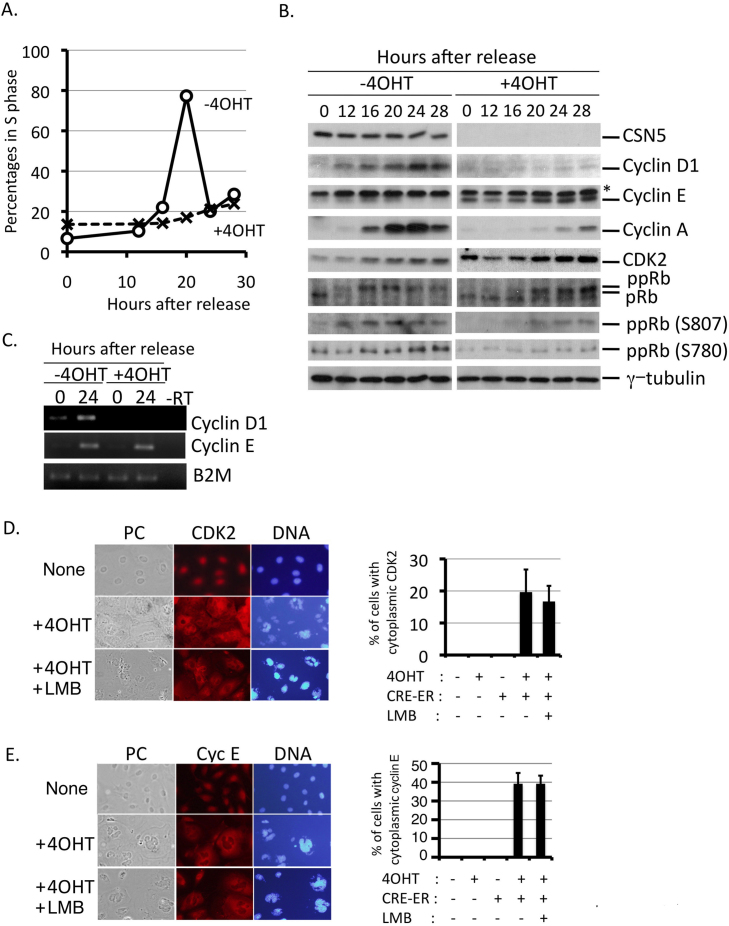
Figure 4. G1 progression of the cell cycle is markedly hampered in CSN5-depleted cells.
CSN5-floxed p53-null MEFs containing CRE-ER (as in Fig. 3) were cultured with and without 4OHT, and passaged in a normal medium for 4 days. Cells were cultured at high density to arrest them in G0/G1 by contact inhibition for 48 hours, and replated at low density to reenter the cell cycle. (A) Cells at indicated times were harvested and analyzed for DNA content by flow cytometry. Percentages of cells in S phase are shown. (B) Lysates isolated from cells at indicated times were analyzed by Western blotting using antibodies against CSN5, cyclin D1, cyclin E, cyclin A, CDK2, pRb, Rb phosphorylated at Ser 807 and Ser 780, and γ-tubulin. (C) Total RNA was extracted from cells at indicated times and analyzed by quantitative RT-PCR using a pair of primers specific to cyclin D1, cyclin E, and β2 microglobulin (B2M). (D), (E) Cells were cultured with and without 4OHT in the absence and presence of LMB, fixed and stained with an antibody to CDK2 (D) and cyclin E (E). Cells with cytoplasmic signals were enumerated (right panels). DNA was visualized by staining with Hoechst 33342. Original magnification; x200.
Mammalian cells contain two types of G1 cyclins, cyclins D and E, which collaboratively play a critical role in progression through the G1 phase46. The major D-type cyclin in fibroblasts is cyclin D1, and was not expressed in G0-arrested fibroblasts. Reentry into the cell cycle of wild-type cells rapidly induced sequential expression of cyclins D1, E, and A during G1, namely cyclin D1 up until 12 hours post-release, and cyclins E and A at around 12–20 hours and around 16–28 hours post-release, respectively (Fig. 4B, left panels, 2nd, 3rd, and 4th lanes). In CSN5-depleted cells, however, only a trace amount of cyclin D1 was induced to express after the release (Fig. 4B, right panels, 2nd lane). Quantitative RT-PCR analysis revealed that the lack of cyclin D1 expression in CSN5-depelted cells at G0/G1 was due to the loss of transcription from the cyclin D1 locus (Fig. 4C, 1st lane). Thus, depletion of CSN5 affects multiple mechanisms for cyclin D regulation47; it stabilizes the cyclin D1 protein in cycling cells31, and inhibits initiation of new transcription. Although we currently do not know the mechanism which suppressed cyclin D1 expression in CSN5-depleted cells, the levels of some known activators of cyclin D1 transcription (NF-κB, β-catenin, and c-Jun) were indistinguishable between wild-type and CSN5-depleted cells (negative data not shown), and, therefore, it is feasible that some transcriptional suppressors for the cyclin D1 promoter may have stabilized.
In contrast to cyclin D1, the levels of cyclin E and CDK2 proteins were higher in CSN5-depleted cells, which is consistent with previous results26,31. Even G0-arrested cells (at time point 0) contained a significantly higher level of cyclin E and CDK2, and the levels were maintained or even higher after the release (Fig. 4B, 3rd and 5th lanes). However, the phosphorylation of retinoblastoma protein was markedly hampered, which was shown by the mobility shift of the Rb protein by immunoblotting (Fig. 4B, 6th lane from the top) and by immunoblotting using antibodies recognizing site-specific phosphorylation of the Rb protein (Fig. 4B, 7th and 8th lanes from the top), indicating that cyclin E-CDK2 kinase was not active or not functional in CSN5-null cells. Immunofluorescence with a specific antibody revealed that CDK2 was located in the cytoplasm, together with cyclin E (Fig. 4D–E). Leptomycin B (LMB) exhibited little effect on the cytoplasmic location of CDK2 (Fig. 4D, E), suggesting that this is not due to the accelerated nuclear export but rather to the retention in the cytoplasm, most likely by the block of the nuclear import. In addition to the impaired cyclin E-CDK2 kinase complex, failure of Rb phosphorylation and progression through G1 in CSN5-depleted cells may also be attributed to the defect in cyclin D1 expression because phosphorylation of Rb by the cyclin D-CDK complex is necessary for recognition by the cyclin E-CDK2 complex as a substrate48. Thus, knockout of CSN5 resulted in different effects on the two G1 cyclin complexes, enforced localization of CDK2 and cyclin E to the cytoplasm and prevention of cyclin D1 transcription. So far, we found the followings; (i) G0-arrested CSN5-null cells expressed cyclin E but not cyclins D1 and A, (ii) CSN5 directly binds to CDK2, and (iii) CDK2-knockdown cancelled upregulation of cyclin E. Taking these into consideration, the cytoplasmic relocation of CDK2 and cyclin E seems to be the more direct and specific effect of CSN5 rather than an indirect effect through the loss of the CSN complex.
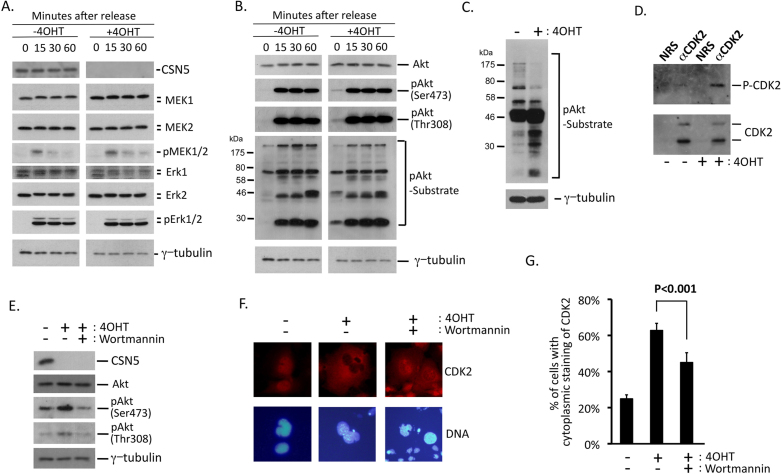
Phosphorylation of CDK2 at the site of Akt kinase was enhanced in CSN5-depleted cells
Because progression through G1 phase of the mammalian cell cycle is strictly regulated by extracellular signals46, we examined signaling pathways in CSN5-null cells. We especially focused on MAK kinase- and PI3 kinase-pathways because we previously found that CSN5 functions downstream of these two cascades in Bcr-Abl-transformed cells49. For this purpose, we cultured MEFs in low serum for 48 hours, treated them in serum for 15–60 minutes, and analyzed by immunoblotting using pan and phospho-specific antibodies to MEK, Erk and Akt kinases. Because it takes 7 days to eliminate the CSN5 protein after gene knockout, MEFs were treated with 4OHT 4 days prior to starvation. We saw little difference in the kinetics of activation of the MAP kinase cascade (MEK1/2 and Erk1/2) between wild-type and CSN5-null cells (Fig. 5A). In the PI3 kinase-pathway, activation of Akt kinase was similarly induced by serum in both wild-type and CSN5-nulls cells. However, a certain phosphorylated Akt substrate was sustained in starved cells (Fig. 5B). In an asynchronous population of cells, the patterns of substrates phosphorylated by Akt kinase were different between wild-type and CSN5-depleted cells (Fig. 5C). Encouraged by this observation, and considering that CDK2 is also a substrate of Akt kinase50, we analyzed the Akt-mediated phosphorylation of CDK2 in CSN5-null cells by immunoprecipitation and immunoblotting, and found it to be markedly increased (Fig. 5D). Akt-mediated phosphorylation of CDK2 at threonine 39 is known to relocate CDK2 to the cytoplasm50. In wild-type cells, CSN5 bound to CDK2 (Figs. 1 and 2). In the absence of CSN5, phosphorylation of CDK2 at the site of Akt kinase was enhanced (Fig. 5D), and CDK2 was relocated to the cytoplasm (Fig. 4D). Other CDKs did not bind to CSN5, were not phosphorylated by Akt in CSN5-null cells, and were not relocated to the cytoplasm (negative data not shown). From these observations, we concluded that CSN5 bound to CDK2 and prevented Akt-mediated phosphorylation and cytoplasmic relocation.
Figure 5. Akt-mediated phosphorylation of CDK2 was enhanced in CSN5-depleted cells.
(A), (B) CSN5-floxed p53-null MEFs containing CRE-ER (as in Fig. 3) were cultured with and without 4OHT, and passaged in a normal medium for 4 days. Cells were incubated in low serum for 48 hours, and stimulated with serum. Lysates isolated from cells at indicated times after serum stimulation were analyzed by Western blotting using antibodies against CSN5, MEK1, MEK2, phosphorylated MEK1/2, Erk1, Erk2, phosphorylated Erk1/2, Akt, phosphorylated Akt (Ser473 and Ser 308), phosphorylated Akt substrate and γ-tubulin. (C) CSN5-floxed p53-null MEFs containing CRE-ER were cultured with and without 4OHT, and then in a normal medium for 6 days. Cell lysate was isolated and analyzed by Western blotting using antibodies against phosphorylated Akt substrate and γ-tubulin. (D) Cell lysate as in C was immunoprecipitated with NRS and rabbit antibodies raised against CDK2. The immune complex was analyzed by Western blotting using antibodies against CDK2 and phosphorylated Akt substrate. (E–G) CSN5-floxed p53-null MEFs containing CRE-ER were cultured with and without 4OHT, and then in a normal medium for 6 days in the presence and absence of wortmannin. Cell lysate was isolated and analyzed by Western blotting using antibodies against Akt, phosphorylated Akt (Ser473 and Ser 308), and γ-tubulin (E). Cells were fixed and stained with an antibody to CDK2. DNA was visualized by staining with Hoechst 33342 (Original magnification; x200)(F). Cells with cytoplasmic staining of CDK2 were enumerated (G).
To further confirm that the PI3 kinase-Akt pathway participates in the regulation of CDK2's subcellular localization in CSN5-depleted cells, we treated cells with a chemical inhibitor to PI3 kinase, wortmannin. MEFs containing the floxed allele of CSN5 and inducible CRE-ER were treated with 4OHT and incubated in the presence and absence of wortmannin. Fig. 5E shows that, in the absence of CSN5, Akt was phosphorylated at the site of activation and was inhibited in the presence of wortmannin. In cells treated with wortmannin, retention of CDK2 in the cytoplasm was significantly suppressed (Fig. 5F, G). Thus, activation of the PI3 kinase-Akt pathway is required for retention of CDK2 in the cytoplasm in the absence of CSN5.
CDK2-knockdown in CSN5-knockout cells
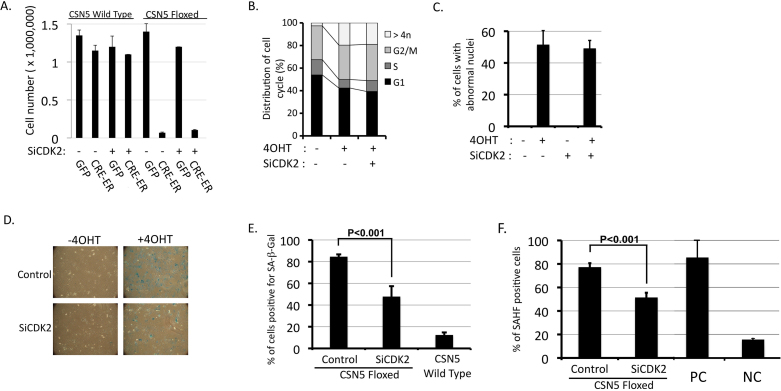
To elucidate the physiological meaning of CDK2's accumulation in the cytoplasm on CSN5's depletion, we examined the effect of CDK2-knockdown in CSN5-null cells, which, as we showed in Fig. 3B, allowed the level of cyclin E to return to that in normally proliferating cells. Previously, we showed that CSN5-depletion resulted in three phenotypes, (1) cell cycle arrest at multiple points, (2) an increase in ploidy, and (3) induction of senescence in a p53-independent manner31. We, therefore, examined whether CDK2-upregulation by CSN5-depletion is connected with any of these three phenotypes. First, we examined the effect on proliferation. Wild-type and CSN5-floxed p53-null MEFs infected and prepared as in Fig. 3B were treated with 4OHT and, after 14 day's incubation for recovery, were enumerated. Fig. 6A shows that CDK2 knockdown did not rescue the proliferative defect of CSN5-null cells. The flow cytometric measurement of DNA content revealed that CSN5-depletion increased the ploidy (cells with >4n DNA content) of a significant population but did not alter the ratio of cell populations in G1, S, and G2/M, and that additional CDK2-knockdown exhibited little effect on the cell cycle profile (Fig. 6B). Enumeration of cells with abnormal nuclei (multinuclei), examined by staining cells on glass slides with the hoechst dye, gave similar results (Fig. 6C). These results indicate that cytoplasmic accumulation of CDK2 and cyclin E was not essential for blockage of the cell cycle, nor the increase in ploidy.
Figure 6. Upregulation of cytoplasmic CDK2 is required for induction of senescence in CSN5-depleted cells.
CSN5-wild type and -floxed MEFs infected with CRE-ER and siCDK2 (siCDK2-1) viruses prepared in Fig. 3B were used in the following experiments. (A) Cells (10,000) were treated with 4OHT and, after 7 days, live cells were enumerated. (B), (C) Cells were cultured with and without 4OHT. After 7 days, DNA content was analyzed by flow cytometry (B), and cells were stained with Hoechst 33342 and observed under a fluorescence microscope. Cells with abnormal nuclei were enumerated (C). (D), (E) Cells were cultured with and without 4OHT in the presence and absence of siCDK2. After 7 days, cells were assayed for SA-β-Gal activity (Original magnification; x50)(D). Cells with SA-β-Gal activity were enumerated and percentages are shown (E). (F) Cells were treated as in D, stained with Hoechst 33342 and observed under a fluorescence microscope. SAHF-like-structure-positive cells were enumerated and percentages are shown (C). For controls, we used wild-type MEFs in the late passage for the positive control (PC), and p53-null fibroblasts for the negative control (NC).
When we looked at the senescence marker, senescence-associated β-galactosidase (SA-β-gal) activity, in these cells, however, we detected a significant difference. CSN5-floxed p53-null MEFs infected with CRE-ER and CDK2-siRNA (siCDK2) viruses were treated with 4OHT and assayed for SA-β-gal activity 7 days later. Fig. 6D shows that nearly all CSN5-depeleted cells exhibited SA-β-gal activity, whereas markedly fewer cells were positive for SA-β-gal after siCDK2 treatment. Quantification of the data shows that knockdown of CDK2 reduced the number of SA-β-gal-positive cells by approximately 50% (Fig. 6E). Considering the level of CDK2 knockdown, the 50% decrease in SA-β-gal-positive cells is significant. When we examined another senescence marker, senescence-associated heterochromatin foci (SAHF), a similar result was obtained (Fig. 6F). Thus, accumulation of cytoplasmic CDK2 and concomitant cyclin E was required for induction of p53-independent senescence in CSN5-null cells.
Cytoplasmic cyclin E, but not CDK2, is essential for induction of p53-independent cellular senescence
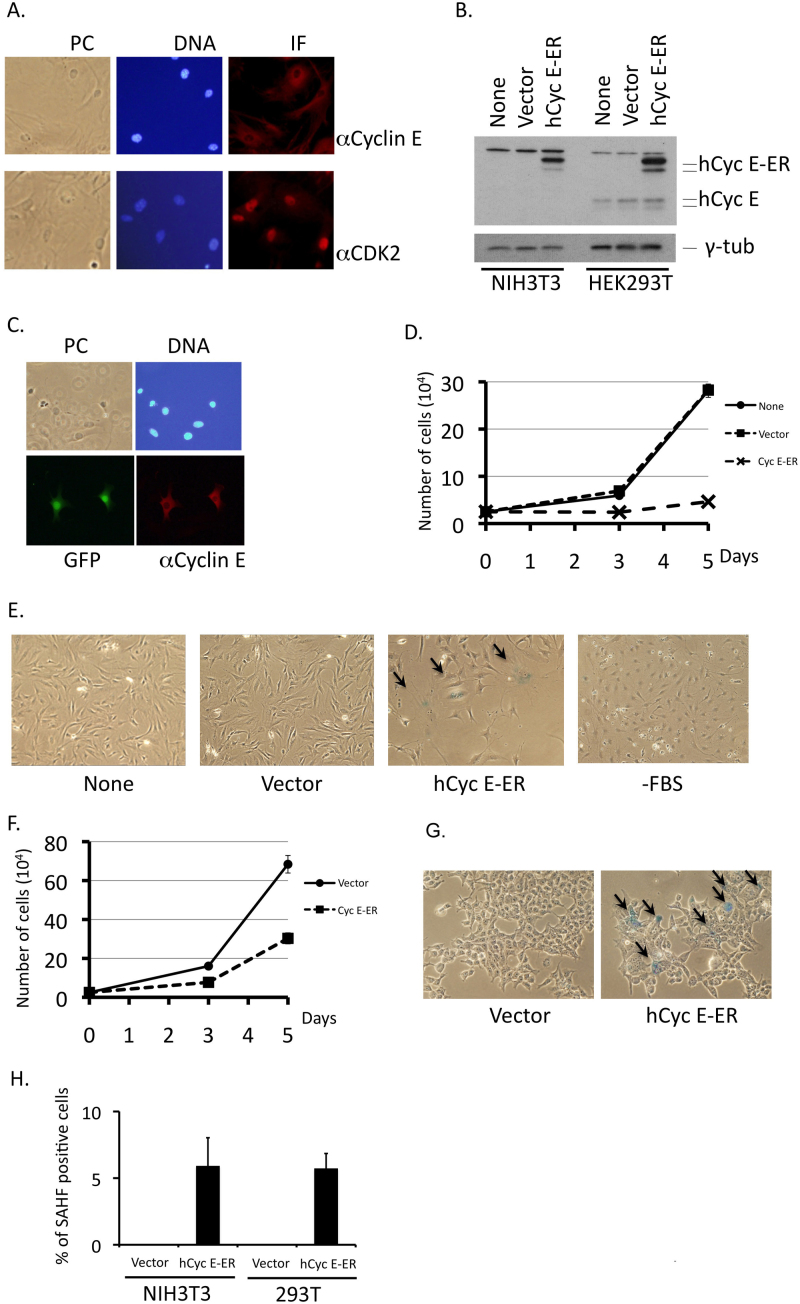
To investigate the potential role of cytoplasmic CDK2 and cyclin E in senescence, we first examined the subcellular localization of CDK2 and cyclin E in late-passaged MEFs, which had already entered proliferative senescence. In proliferating cells, CDK2 and cyclin E occur largely in the nucleus and to some extent in the cytoplasm (for example, see Fig. 4D and E). Subcellular localization of CDK2 changed little after the cells entered proliferative senescence, though a portion of cells contained cyclin E in the cytoplasm (Fig. 7A), suggesting that cytoplasmic cyclin E is a key element for the induction of senescence.
Figure 7. Cytoplasmic cyclin E is responsible for induction of senescence.
(A) Wild-type MEFs were cultured in vitro for more than 10 passages. Cells were fixed and stained with antibodies to mouse cyclin E and CDK2. (B) NIH3T3 and HEK293T cells were mock-transfected (None) or transfected with an empty vector (Vector) and a mammalian expression vector containing human cyclin E fused with ER (hCycE-ER). Transfectants were sorted for the GFP-positive signal 4 days after transfection. Lysates isolated from transfected cells were analyzed by Western blotting using antibodies specifically recognizing human cyclin E and γ-tubulin. (C) NIH3T3 cells transfected with hCycE-ER together with GFP were fixed and stained with antibody specific to human cyclin E. Chromosomal DNA was visualized by staining with Hoechst 33342. (D) NIH3T3 cells transfected and prepared as in B were plated (2.5 × 104 cells per plate) and counted after 3 and 5 days. (E) NIH3T3 cells transfected and prepared as in B were plated and stained for SA-β-Gal activity. As a control, NIH3T3 cells arrested in G0/G1 by serum starvation for 48 hours were assayed (-FBS) likewise. (F) HEK293T cells transfected and prepared as in B were plated (2.5 × 104 cells per plate) and counted after 3 and 5 days. (G) HEK293T cells transfected and prepared as in B were plated and stained for SA-β-Gal activity. (H) Transfectants were stained with Hoechst 33342 and observed under a fluorescence microscope. SAHF-like-structure-positive cells were enumerated and percentages are shown. Original magnification; x200 for A and C, and x50 for (E) and (G).
To further confirm that the cytoplasmic relocation of cyclin E is sufficient for induction of senescence, we forced cyclin E expression in the cytoplasm in immortalized cultured cells. Because our data indicate that cyclin E was retained in the cytoplasm, but not exported from the nucleus, in senescent cells, we took advantage of the estrogen receptor (ER)-tag, which retains the fused protein in the cytoplasm in the absence of its ligand such as tamoxifen, rather than adding extra-NES sequences, which translocates the protein already present in the nucleus into the cytoplasm. We fused human cyclin E with the ER-tag to distinguish the endogenous protein from the ectopic one and transfected it into mouse NIH3T3 fibroblasts and human HEK293T cells. Fig. 7B shows that the ER-fused cyclin E was stably expressed in transfected cells without significant degradation. Use of an antibody specific to human cyclin E enabled us to distinguish human and mouse proteins (Fig. 7B, left 3 lanes) and immunofluorescent staining using this antibody showed that ER-fused cyclin E (Cyc E-ER) was exclusively located in the cytoplasm (Fig. 7C). NIH3T3 cells expressing Cyc E-ER proliferated very poorly (Fig. 7D), and were positive for SA-β-gal activity (Fig. 7E). Similar results were obtained in HEK293T cells (Fig. 7F and G). Introduction of ER-fused CDK2 did not induce senescence in these cells (data not shown), suggesting specificity for cyclin E. Thus, the cytoplasmic distribution of cyclin E is sufficient to induce senescence. Interestingly, simple growth arrest such as deprivation of serum from the medium failed to induce senescence (Fig. 7, right panel), indicating that an unknown function of cyclin E in the cytoplasm is connected to the induction of senescence.
Discussion
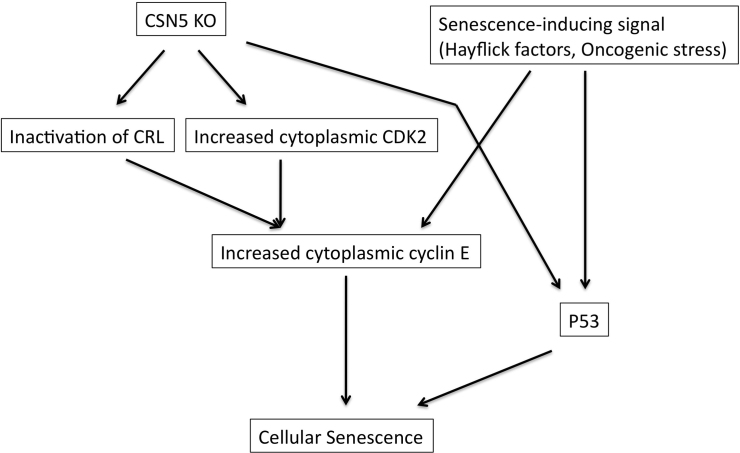
The function of the CSN is critical to mammalian cell proliferation and tumorigenesis, and the expression of CSN5 is frequently elevated in human cancers4, indicating that CSN5 possesses an important function that is closely connected with tumor cell proliferation. In this paper, we, for the first time, show that CSN5, but not the whole CSN complex, interacts with a key regulatory element of the cell cycle, CDK2. The interaction between CSN5 and CDK2 may participate in control of premature senescence, which is recently proposed as a powerful anti-cancer mechanism33,34, rather than the cell cycle itself. Depletion of CSN5 induces senescence in p53-null fibroblasts31, but knockout of the CSN5 gene upregulates p53 expression26, indicating that CSN5 occurs upstream of both p53-dependent and -independent senescence pathways (Fig. 8) and that interaction between CSN5 and CDK2 may take part in the p53-independent pathway.
Figure 8. A model of the control of senescence mediated by CSN5 and cytoplasmic cyclin E.
Interestingly, we placed a sufficient amount of CDK2 in the cytoplasm by overexpressing ER-fused CDK2 in cultured cells, but it failed to induce senescence (data not shown), indicating that additional signal is needed for the efficient induction of senescence. Furthermore, overpassaged MEFs, which entered proliferative senescence, did not express increased levels of cytoplasmic CDK2. Therefore, the relocation of cyclin E to the cytoplasm is a key event, and the physiological relevance of cytoplasmic CDK2 to the authentic senescence pathway remains to be understood. However, cytopasmic redistribution of CDK2 was observed on activation of the Akt pathway50, and in breast epithelial cells in response to TGF-β51, and therefore could be a more general mechanism to control cell proliferation. In CSN5-depleted cells, upregulation of cytoplasmic CDK2 is required for senescence, suggesting that blocking the function of CSN5 to regulate CDK2 by a small molecule may be a novel approach to developing a new type of anti-cancer drug, which will induce senescence of tumor cells regardless of p53 status.
Cyclin E is known to be targeted by two types of ubiquitin ligases; the Fbw7/hCdc4-SCF complex52,53 and an ubiquitin ligase containing Cul354. The former recognizes cyclin E phosphorylated by or in a complex with CDK2, while the latter targets free cyclin E for degradation. In CSN5-null cells, cyclin E was stabilized for two reasons, (1) the Fbw7/hCdc4-SCF complex was not active due to the lack of CSN deneddylase activity, and (2) considering that cyclin E was located in the cytoplasm resistant to LMB, the cyclin E/CDK2 complex was not activated by CAK in the nucleus, and so cyclin E was unphosphorylated. Additional knockdown of CDK2 enabled cyclin E to be ubiquitinated by a ligase that can target free cyclin E as a substrate such as Cul3-containing complex54 or an unidentified ligase. Upregulation of cyclin E was also seen in CSN2-null cells28, but, in CSN8-null T-cells, cyclin E protein was only marginally stabilized30. Interestingly enough, in CSN8-null cells, CSN5 protein level did not diminish30, consistent with the notion that full upregulation of cyclin E requires loss of CSN5, in addition to the inactivation of the CSN complex.
Stabilization of cyclin E and CDK2 in the cytoplasm seems to involve the cytoplasmic retention mechanism rather than enhanced nuclear export. Phosphorylation mediated by Akt kinase is known to affect the substrate protein through cytoplasmic retention50,55,56,57,58. CSN5 specifically interacted with CDK2 in proliferating cells and, in CSN5-null cells, CDK2, free from CSN5, was specifically phosphorylated by Akt kinase. We, therefore, propose that CSN5 binds to CDK2 to prevent phosphorylation by Akt kinase, which results in accumulation of CDK2 and cyclin E in the cytoplasm. This mechanism is unique, and as far as we know, reported here for the first time.
Several pathways and molecules are known to induce senescence in the absence of p53 (e.g., ARF38, simultaneous loss of Skp2 and Pten39, loss of CDK437, p16 in case of telomere shortening35, etc.). Although none of them precisely fit the senescence induced by CSN5-depletion and cytoplasmic cyclin E (because the level of Skp2 expression was maintained, our NIH3T3 line lacks the p16/ARF locus, and expression of CDK4 was maintained), it was reported that inhibition of CDK activity by upregulation of the CDK inhibitors p21 and p27 induces senescence in some cases and mislocation of cyclin E in the cytoplasm may exert a similar effect. In this scenario, enforced arrest of the cell cycle in G0 (e.g., caused by an increase in p21/p27 due to a loss of Skp2, loss of CDK4, and increase in p16) induces dephosphorylation and activation of pRb, which is key to senescence. However, the arrest of the cell cycle in G0 is not sufficient to induce senescence, and therefore, there must be an additional event that pushes cells from the reversible G0 arrest toward eternal withdrawal from the cell cycle. In our experiments, serum deprivation arrested cells in G0/G1, but was not sufficient to induce senescence. Because overexpression of wild-type cyclin E in the nucleus does not induce senescence59, we favor the idea that cyclin E has an unidentified function in the cytoplasm to induce senescence. Importantly, we observed cyclin E, but not CDK2, in the cytoplasm during the proliferative senescence in wild-type MEFs in the late passages, and so we propose that this might be an alternative and novel way of inducing senescence in the absence of p53. Recently, it was shown that senescence is not required for p53 to suppress tumorigenesis60. Thus, the pathway for p53-independent senescence seems to be important as a target for anti-tumor drugs because more than 50% of human cancers have deletions and mutations at the p53 locus. We can now separate the distinct functions of CSN5; cell cycle control and immature senescence, the latter being through CDK2. It would be nice to screen for small molecular weight chemicals that inhibit interaction between CSN5 and CDK2 but do not inhibit the deneddylase activity, which may induce senescence without affecting global proliferation and the cell cycle, leading to the development of anti-cancer drugs with minimum side effects.
Methods
Cell culture, transfection, and retroviral infection
Mouse embryonic fibroblasts (MEFs) harboring null (-) and conditional (floxed, f) alleles of the CSN5/Jab1 gene and immortalized by the null allele of p53 were described previously26,31. NIH3T3 (Arf-null, p53-wild-type) mouse fibroblasts (provided by Drs C. J. Sherr and M. F. Roussel), MEFs, C2C12 mouse myoblasts, and 293T human embryonic kidney (HEK) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin (GIBCO/BRL). Transfection with expression vectors was performed by the calcium phosphate-DNA precipitation method61. For viral production, the plasmid was co-transfected into 293T cells together with a plasmid encoding an ecotropic helper virus containing a defective virion-packaging (ϕ2) sequence. Culture supernatants containing retroviruses harvested 48~72 h after transfection were used to infect proliferating NIH3T3 cells and MEFs in the presence of polybrene (4 μg/ml)62. Infected cells were selected for resistance to puromycin (5 μg/ml) or G418 (1 mg/ml) depending on the type of vector used for infection and sorted for GFP-positive signals with a FACS Aria flow cytometer (Becton Dickinson). For the activation of inducible CRE proteins, MEFs containing the conditional allele of CSN5 and null allele of p53 and infected with the CRE-ER virus were treated with 4OHT (200 nM) for 1–4 hours. (Note that we confirmed that incubation in a higher concentration of 4OHT and for much longer time did not cause any effects on the cell cycle, proliferation, and survival of cells containing no floxed alleles.) High molecular DNA was extracted from the cells and the genotypes were determined as described31. In some experiments, cells were incubated in the presence of wortmannin (100 nM) before harvest.
Plasmid construction
The GFP-fused protein expression vectors (pMSCV-puro-GFP and pMSCV-IRES-GFP, the latter a gift from Dr. Owen Witte), into which CRE-ER and ER-CRE-ER45 cDNAs (purchased from Addgene Inc.) were subcloned, were described previously62. The vectors for RNA interference (RNAi) were constructed with the pSUPER RNAi system (pSUPER.retro, pSUPER.retro.neo+gfp, and pSUPER.retro.neo, purchased from OligoEngine) according to the manufacturer's instructions. The sequence used for the knockdown of CDK2 was obtained from the published results63.
Cell cycle
Mouse fibroblasts were arrested at the G0/G1 phase by serum depletion (0.5%) or contact inhibition for 48 hours. Cells were simultaneously reentered into the cell cycle by re-addition of the serum or by re-plating at low cell density. The cell cycle was monitored by flow cytometric analysis as described previously19,62.
Quantitative RT-PCR
Total RNA was isolated using the ISOGEN reagent (Nippon Gene) and reverse transcribed using RNase-free Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative RT–PCR was performed within a linear range as described previously62,64 and the data were normalized by the expression level of β2 microglobulin (B2M). We confirmed that the reactions were quantitatively performed within a linear range by several control experiments. The following oligonucleotide primers specific to mouse B2M, cyclin D1 and cyclin E1 were used: B2M, 5′-CTG TAT GCT ATC CAG AAA ACC CCT CAA-3′ (sense) and 5′-CAT GTC TCG ATC CCA GTA GAC GGT-3′ (antisense); cyclin D1, 5′-GCA GAG AGC TAC AGA CTC CG-3′ (sense) and 5′-GTC CAG GTA GTT CAT GGC CAG-3′ (antisense); cyclin E1, 5′-CCC TCT GAC CAT TGT GTC CT-3′ (sense) and 5′-TCG CAC CAC TGA TAA CCT GA-3′ (antisense).
Protein analyses
Cell lysis, immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were performed as described62,65. For immunoprecipiotation of CDK2, cell lysate in an SDS-sample buffer (40 mM Tris-HCl, pH 6.8, 0.1 M DTT, 1% SDS, 10% glycerol, and 0.05% Bromophenol Blue) was diluted (1:20 or more) in a hypotonic buffer (50 mM Tris-HCl pH8.0 and 10 mM MgCl2) and incubated with an excess amount of antibody against CDK2 (M2) for 2–3 nights at 4°C. Immune complexes were collected and analyzed by immunoblotting with antibodies to phosphorylated Akt-substrates (Cell Signaling Technology, Inc.). Rabbit polyclonal antibodies to the CSN subunits, Cul1 and 4, p53, p21, and Skp2 were generated using bacterially produced polypeptides in our laboratory. Mouse monoclonal antibodies to cyclin D1 (72-13G) and CDC2 (17) and rabbit polyclonal antibodies to cyclin E (M20), cyclin A (C19), CDK2 (M2), CDK4 (C22), CDK6 (C21), ppRb (S780) and c-Jun/AP1(D) were obtained from Santa Cruz. Mouse monoclonal antibodies to γ-tubulin (GTU88) and Rb (G3-245) were purchased from Sigma and Pharmingen, respectively. Rabbit polyclonal antibodies to ppRb (S807), Akt (C67E7), phosphorylated Akt (C31E5E and D9E), and phosphorylated Akt-substrates (100B7E and #9611) were acquired from Cell Signaling Technology.
Immunofluorescence staining
Cells grown on coverslips were fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton X-100, stained with rabbit polyclonal and mouse monoclonal antibodies, and incubated with Alexa Fluor 594-conjugated anti-mouse/rabbit IgG (Invitrogen). Chromosomal DNA was stained by incubation in 1 μg/ml of Hoechst 33342 for 2 min. The cell samples were viewed using phase-contrast or fluorescence microscopy to enumerate fluorescence-positive cells.
In vitro binding assay
A cDNA fragment containing the entire coding sequence of mouse CSN5 was inserted into the pGEX vector (Pharmacia) in-frame with Glutathione S-transferase (GST). Expression and purification of GST-fused proteins and the binding conditions were as described62.
β-galactosidase assay
Cells were fixed in 0.25% glutaraldehyde, incubated for 16–24 hours in X-Gal solution containing 0.2% X-Gal, 2 mM MgCl2, 5 mM K4Fe(CN)6, and 5 mM K3Fe(CN)6, and viewed with a phase-contrast microscope.
Author Contributions
JyK and NY-K wrote the main manuscript text and AY and JyK prepared figures. All authors reviewed the manuscript.
Acknowledgments
We thank Drs. CJ Sherr and MF Roussel for the NIH3T3 cell line, Dr. O Witte for the MSCV vector, Drs. Ruggero Pardi and Martina Panattoni for CSN5-floxed mice, and Ms I Nakamae for excellent technical assistance. This work was supported by Grants-in-aid for Scientific Research and for Cancer Research from the Ministry of Education, Science, and Culture of Japan.
References
- Wei N. & Deng X. W. The COP9 signalosome. Annu Rev Cell Dev Biol 19, 261–286 (2003). [DOI] [PubMed] [Google Scholar]
- Wei N. & Deng X. W. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet 15, 98–103 (1999). [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. & Isono E. The COP9 signalosome and its role in plant development. Eur J Cell Biol 89, 157–162 (2010). [DOI] [PubMed] [Google Scholar]
- Kato J. Y. & Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells 14, 1209–1225 (2009). [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. & Deng X. W. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol 11, 420–426 (2001). [DOI] [PubMed] [Google Scholar]
- Bech-Otschir D., Seeger M. & Dubiel W. The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci 115, 467–473 (2002). [DOI] [PubMed] [Google Scholar]
- von Arnim A. G. On again-off again: COP9 signalosome turns the key on protein degradation. Curr Opin Plant Biol 6, 520–529 (2003). [DOI] [PubMed] [Google Scholar]
- Cope G. A. & Deshaies R. J. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671 (2003). [DOI] [PubMed] [Google Scholar]
- Wolf D. A., Zhou C. & Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol 5, 1029–1033 (2003). [DOI] [PubMed] [Google Scholar]
- Claret F. X., Hibi M., Dhut S., Toda T. & Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 383, 453–457 (1996). [DOI] [PubMed] [Google Scholar]
- Hallstrom T. C. & Nevins J. R. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes Dev 20, 613–623 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D. et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. Embo J 20, 1630–1639 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M. et al. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. Faseb J 12, 469–478 (1998). [PubMed] [Google Scholar]
- Tomoda K., Kubota Y. & Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398, 160–165 (1999). [DOI] [PubMed] [Google Scholar]
- von Arnim A. G. & Deng X. W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045 (1994). [DOI] [PubMed] [Google Scholar]
- Kwok S. F. et al. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10, 1779–1790 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S. et al. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol 9, 1187–1190 (1999). [DOI] [PubMed] [Google Scholar]
- Oron E. et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129, 4399–4409 (2002). [DOI] [PubMed] [Google Scholar]
- Tomoda K. et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem 277, 2302–2310 (2002). [DOI] [PubMed] [Google Scholar]
- Fukumoto A., Tomoda K., Kubota M., Kato J. Y. & Yoneda-Kato N. Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett 579, 1047–1054 (2005). [DOI] [PubMed] [Google Scholar]
- Sharon M. et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17, 31–40 (2009). [DOI] [PubMed] [Google Scholar]
- Adler A. S. et al. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet 38, 421–430 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto A., Tomoda K., Yoneda-Kato N., Nakajima Y. & Kato J. Y. Depletion of Jab1 inhibits proliferation of pancreatic cancer cell lines. FEBS Lett 580, 5836–5844 (2006). [DOI] [PubMed] [Google Scholar]
- Luo J. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Yoneda-Kato N., Yoshida A. & Kato J. Y. Stable form of JAB1 enhances proliferation and maintenance of hematopoietic progenitors. J Biol Chem 283, 29011–29021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K., Yoneda-Kato N., Fukumoto A., Yamanaka S. & Kato J. Y. Multiple functions of jab1 are required for early embryonic development and growth potential in mice. J Biol Chem 279, 43013–43018 (2004). [DOI] [PubMed] [Google Scholar]
- Panattoni M. et al. Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J Exp Med 205, 465–477 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K. et al. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23, 6790–6797 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. et al. COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol 23, 6798–6808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S. et al. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol 8, 1236–1245 (2007). [DOI] [PubMed] [Google Scholar]
- Yoshida A., Yoneda-Kato N., Panattoni M., Pardi R. & Kato J. Y. CSN5/Jab1 controls multiple events in the mammalian cell cycle. FEBS Lett 584, 4545–4552 (2010). [DOI] [PubMed] [Google Scholar]
- Hayflick L. & Moorhead P. S. The serial cultivation of human diploid cell strains. Exp Cell Res 25, 585–621 (1961). [DOI] [PubMed] [Google Scholar]
- Collado M., Blasco M. A. & Serrano M. Cellular senescence in cancer and aging. Cell 130, 223–233 (2007). [DOI] [PubMed] [Google Scholar]
- Acosta J. C. & Gil J. Senescence: a new weapon for cancer therapy. Trends Cell Biol 22, 211–219 (2012). [DOI] [PubMed] [Google Scholar]
- Jacobs J. J. & de Lange T. Significant role for p16INK4a in p53-independent telomere-directed senescence. Curr Biol 14, 2302–2308 (2004). [DOI] [PubMed] [Google Scholar]
- Xu M. et al. Beta-catenin expression results in p53-independent DNA damage and oncogene-induced senescence in prelymphomagenic thymocytes in vivo. Mol Cell Biol 28, 1713–1723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X. et al. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev 16, 2923–2934 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha L. et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci U S A 104, 10968–10973 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. K. et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature 464, 374–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D. M., Harper J. W. & Elledge S. J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97, 431–434 (1999). [DOI] [PubMed] [Google Scholar]
- King R. W., Deshaies R. J., Peters J. M. & Kirschner M. W. How proteolysis drives the cell cycle. Science 274, 1652–1659 (1996). [DOI] [PubMed] [Google Scholar]
- Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol 9, 788–799 (1997). [DOI] [PubMed] [Google Scholar]
- Kwon T. K., Buchholz M. A., Jun D. Y., Kim Y. H. & Nordin A. A. The differential catalytic activity of alternatively spliced cdk2 alpha and cdk2 beta in the G1/S transition and early S phase. Exp Cell Res 238, 128–135 (1998). [DOI] [PubMed] [Google Scholar]
- Ullah Z., Buckley M. S., Arnosti D. N. & Henry R. W. Retinoblastoma protein regulation by the COP9 signalosome. Mol Biol Cell 18, 1179–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T. & Cepko C. L. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A 104, 1027–1032 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J. Induction of S phase by G1 regulatory factors. Front Biosci 4, D787–792 (1999). [DOI] [PubMed] [Google Scholar]
- Kato J. Y. & Yoneda-Kato N. New twist in the regulation of cyclin D1. BioMol Concepts 1, 403–409 (2010). [DOI] [PubMed] [Google Scholar]
- Lundberg A. S. & Weinberg R. A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol 18, 753–761 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K. et al. The Jab1/COP9 signalosome subcomplex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell-cycle progression. Blood 105, 775–783 (2005). [DOI] [PubMed] [Google Scholar]
- Maddika S. et al. Akt-mediated phosphorylation of CDK2 regulates its dual role in cell cycle progression and apoptosis. J Cell Sci 121, 979–988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A., Roberts R. L., Arteaga C. L. & Law B. K. Transforming growth factor-beta induces Cdk2 relocalization to the cytoplasm coincident with dephosphorylation of retinoblastoma tumor suppressor protein. Breast Cancer Res 6, R130–139 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D. M. et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 (2001). [DOI] [PubMed] [Google Scholar]
- Strohmaier H. et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413, 316–322 (2001). [DOI] [PubMed] [Google Scholar]
- Singer J. D., Gurian-West M., Clurman B. & Roberts J. M. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 13, 2375–2387 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglietto G. et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med 8, 1136–1144 (2002). [DOI] [PubMed] [Google Scholar]
- Liang J. et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med 8, 1153–1160 (2002). [DOI] [PubMed] [Google Scholar]
- Shin I. et al. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med 8, 1145–1152 (2002). [DOI] [PubMed] [Google Scholar]
- Zhou B. P. et al. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol 3, 245–252 (2001). [DOI] [PubMed] [Google Scholar]
- Arata Y., Fujita M., Ohtani K., Kijima S. & Kato J. Y. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J Biol Chem 275, 6337–6345 (2000). [DOI] [PubMed] [Google Scholar]
- Li T. et al. Tumor Suppression in the Absence of p53-Mediated Cell-Cycle Arrest, Apoptosis, and Senescence. Cell 149, 1269–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. & Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7, 2745–2752 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda-Kato N., Tomoda K., Umehara M., Arata Y. & Kato J. Y. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. Embo J 24, 1739–1749 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar K. et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118, 477–491 (2004). [DOI] [PubMed] [Google Scholar]
- Matsumoto N. et al. Elevated MLF1 expression correlates with malignant progression from myelodysplastic syndrome. Leukemia 14, 1757–1765 (2000). [DOI] [PubMed] [Google Scholar]
- Kato J. Y., Nakamae I., Tomoda K., Fukumoto A. & Yoneda-Kato N. Preparation and characterization of monoclonal antibodies against mouse Jab1/CSN5 protein. Hybridoma (Larchmt) 25, 342–348 (2006). [DOI] [PubMed] [Google Scholar]