Abstract
The evolution of the diverse insect lineages is one of the most fascinating issues in evolutionary biology. Despite extensive research in this area, the resolution of insect phylogeny especially of interordinal relationships has turned out to be still a great challenge. One of the challenges for insect systematics is the radiation of the polyneopteran lineages with several contradictory and/or unresolved relationships. Here, we provide the first transcriptomic data for three enigmatic polyneopteran orders (Dermaptera, Plecoptera, and Zoraptera) to clarify one of the most debated issues among higher insect systematics. We applied different approaches to generate 3 data sets comprising 78 species and 1,579 clusters of orthologous genes. Using these three matrices, we explored several key mechanistic problems of phylogenetic reconstruction including missing data, matrix selection, gene and taxa number/choice, and the biological function of the genes. Based on the first phylogenomic approach including these three ambiguous polyneopteran orders, we provide here conclusive support for monophyletic Polyneoptera, contesting the hypothesis of Zoraptera + Paraneoptera and Plecoptera + remaining Neoptera. In addition, we employ various approaches to evaluate data quality and highlight problematic nodes within the Insect Tree that still exist despite our phylogenomic approach. We further show how the support for these nodes or alternative hypotheses might depend on the taxon- and/or gene-sampling.
Keywords: polyneoptera, zoraptera, dermaptera, plecoptera, data quality
Introduction
The resolution of the Insect Tree of Life has recently improved using phylogenomic data. Here, new data sets resolved the origin of hexapods (Pancrustacea = “Crustacea” + Hexapoda) (Regier et al. 2008; Meusemann et al. 2010; Regier et al. 2010; von Reumont et al. 2012), the sistergroup relationship of Hymenoptera to remaining Holometabola (Savard et al. 2006; Zdobnov and Bork 2007; Simon et al. 2009; Meusemann et al. 2010) and the intra-ordinal relationships within some holometabolan orders; for example, in Hymenoptera (Sharanowski et al. 2010), or in Coleoptera (Hughes et al. 2006). Despite this increase in resolution, several ambiguities within the Insect Tree exist. Recent discussions center around 1) the phylogenetic relationships of the three wingless entognathous orders (Collembola, Protura, and Diplura), 2) the basal pterygote divergence (“Palaeoptera Problem”), 3) the polyneopteran relationships (unresolved polytomy), and 4) the monophyly of Paraneoptera and their position within Neoptera (for review see also Trautwein et al. 2012; Yeates et al. 2012).
One major problem in resolving insect relationships using phylogenomic data is the lack and/or overlap of genomic and/or transcriptomic data. There are more than one million described insect species (Foottit and Adler 2009) but only 172 insect genomes have been sequenced or are in progress (http://www.ncbi.nlm.nih.gov/genome; last accessed April 2012). In addition, 151 of these projects are conducted on the single most derived lineage of Neoptera: Holometabola. For Polyneoptera, comprising 11 orders and representing presumably the earliest splits of the neopteran lineage, no genome project is available.
The polyneopteran lineage still appears in an unresolved polytomy within the Insect Tree and even its monophyly is disputed. Herein, especially the phylogenetic position of Plecoptera (Zwick 2009) and Zoraptera (Yoshizawa 2007) is far from settled (table 1). Both of them belong to the most phylogenetically ambiguous insect orders and even their placement within the polyneopteran lineage is still under discussion.
Table 1.
Existing Phylogenetic Hypotheses within the Polyneopteran Lineages
To further clarify this most controversial problem among the higher systematics of insects, in this study we provide the first transcriptomic data (derived from 454 expressed sequence tag [EST] data) for three representatives of hitherto unsampled polyneopteran orders: Zoraptera (Zorotypus gurneyi?), Plecoptera (Nemurella pitetii), and Dermaptera (Forficula auricularia).
In addition to addressing these phylogenetic questions with new genomic information, we further address several mechanistic problems relevant to phylogenetic reconstruction. These problems include missing data, phylogenetic resolution, and taxon and gene sampling, all of which contribute to the underlying data quality and consequently the resolution of a certain phylogenetic question (Philippe et al. 2005, 2011; Baurain et al. 2007). For example, following a previous study (Simon et al. 2009) that has shown how biological function of the genes might have an impact on data quality, we extended this approach in the current study using dense taxon sampling across the diverse insect lineages. The difficulty inherent in insect systematics and the existence of competing phylogenetic hypotheses offers a great opportunity to explore the source of incongruence in phylogenomic studies more generally. Here, we test several phylogenetic hypotheses within the Insect Tree and explore how support for these hypotheses might be influenced by missing data, matrix selection, gene and taxa number/choice, and the biological function of the genes. Different approaches to reduce missing data and to select an optimal data set to infer the species evolution were compared. We further characterized the strength of support for the concatenated phylogenetic hypotheses using a newly developed approach, RADICAL (Narechania et al. 2012), which allows us to identify the problematic nodes within the Insect Tree and quantify their relative weakness. In sum, this study 1) provides new insights into the evolution of three ambiguous insect orders, 2) highlights the problems in insect systematics despite the use of numerous characters even in the context of this phylogenomic data set, and 3) demonstrates which factors might influence the phylogenetic inference.
Materials and Methods
Sequencing and Assembly
454-pyrosequencing (ROCHE) was used to generate EST sequences from three polyneopteran species (Forficula auricularia, Nemurella pictetii, and Zorotypus gurneyi?). Fresh tissue was preserved in RNAlater and stored at −80°C. For Forficula auricularia (Dermaptera) and Nemurella pictetii (Plecoptera) total RNA extraction (Absolutely RNA kit, Stratagene), cDNA synthesis (Mint kit, Evrogen), and 454 pyrosequencing on a Titanium FLX sequencer were conducted at the Max Planck Institute for Molecular Genetics, Berlin, Germany. Sequence processing and assembly for the two species were conducted as described in von Reumont et al. (2012) at the Center of Integrative Bioinformatics Vienna, Vienna, Austria.
Total RNA of 10 larval specimens (pooled) of Zorotypus gurneyi? (Zoraptera) was extracted (mRNA-Only Eucaryotic mRNA Isolation Kit, Epicentre, Madison, WI) and its corresponding cDNA synthesized (Mint-Universal cDNA Synthesis Kit user manual [Evrogen, Moscow, Russia]) at LGC Genomics GmbH, Berlin, Germany.
Normalization was carried out using the Trimmer Kit (Evrogen, Moscow, Russia). Library generation for the 454 FLX sequencing was carried out according to the manufacturer’s standard protocols (Roche/454 Life Sciences, Branford, CT). The resulting fragment library was sequenced on 5 individual 1/8 picotiterplates on the GS FLX using the Roche/454 Titanium chemistry. Prior to assembly, the zorapteran sequence reads were screened for the Sfi-linker that was used for concatenation, the linker sequences were clipped out of the reads and the clipped reads assembled to individual transcripts using the Roche/454 Newbler software at default settings (454 Life Sciences Corporation, Software Release: 2.5.3 [20101207_1124]).
For all three new EST-projects, the function of genes was analyzed by KOG (Clusters of eukaryotic orthologous groups) (Tatusov et al. 2003) using OrthoSelect (Schreiber et al. 2009).
Ortholog Prediction and Data Set Generation
The Transcriptome Shotgun Assembly projects have been deposited at DDBJ/EMBL/GenBank under the accessions GAAV00000000 (Nemurella pictetii, BioProject PRJNA172454), GAAX00000000 (Forficula auricularia, BioProject PRJNA172453), and GABA00000000 (Zorotypus gurneyi?, BioProject PRJNA172455).
Additional assembled EST contigs were downloaded from http://www.deep-phylogeny.org, last accessed February 25, 2011 (supplementary table S1, Supplementary Material online). We have only chosen taxa for which at least 1,000 EST contigs were available. The data set comprised a total of 78 species consisting of 4 crustacean species (outgroup), 6 primarily wingless hexapods and 68 pterygote species (2 palaeopteran, 9 polyneopteran, 14 paraneopteran, and 43 holometabolan species). For each taxon, identification of orthologous genes was carried out using the HaMStR approach (Ebersberger et al. 2009) (hamstrsearch_local-hmmer3.v7.pl; http://www.deep-phylogeny.org/hamstr/) with the insecta_hmmer3-2 core reference taxa set. For the re-blast of the candidate EST contigs, we used Apis mellifera, Capitella sp., Daphnia pulex, Ixodes scapularis, and Bombyx mori (options -representative -strict). Overall our core ortholog set encompassed 1,579 clusters of orthologous genes, which were used to assign EST contigs to individual genes. A set of PERL scripts was applied to generate a fasta file for each of the orthologous genes and to automatically align group of orthologous amino acid sequences separately with MAFFT L-INS-I (Katoh and Toh 2008). Randomly similar aligned positions were identified with ALISCORE (Misof and Misof 2009) using the default sliding window size, the maximal number of pairwise comparisons and a special scoring for gappy amino acid data (options -e -r). Randomly aligned positions were subsequently removed with ALICUT v2.0 (http://www.utilities.zfmk.de) and the final gene alignments were concatenated using FASconCAT (Kück et al. 2010).
The original matrix consists of 78 taxa, 1,579 genes, 744402 amino acid positions but shows only a density of 34.2%. Therefore, different approaches to reduce the amount of missing data were applied: 1) The first matrix was created using MARE (v0.1.2-rc) (Meyer et al. April 2011) (http://mare.zfmk.de) where genes and taxa are selected based on information content and data availability. Applying this approach the dictyopteran Hodotermopsis sjoestedti, Blattella germanica and Periplaneta americana were defined as taxon-constraints so they were not dropped from the matrix. Following this restriction, we aimed to maintain a number of polyneopteran species to better unravel the phylogenetic position of Dermaptera, Plecoptera, and Zoraptera. In addition, the “palaeopterous” species Ischnura elegans and Baetis sp. were defined as taxon-constraints due to their primitive position within pterygotes. Therefore, we constrained matrix reduction to retain these five species as key taxa. 2) The second matrix was created using a PERL script that calculates different combinations of taxa and genes to reduce the number of missing data (Simon et al. 2009). As selection criterion, we imposed that Baetis sp., Ischnura elegans and the three new EST projects were present in this matrix. Based on this approach, we selected two different matrices, one which maximizes the number of genes (P_matrix_g) and the other which maximizes the number of species (P_matrix_s).
Using these three matrices, we evaluated how different approaches reducing missing data influence our resulting topology and if the selected taxa and genes based on these different approaches have an influence on the inferred phylogeny.
Phylogenetic Analyses and Random Addition Concatenation Analysis
For all matrices, Maximum likelihood (ML) analyses were performed with the Pthreads-parallelized version of RAxML 7.2.8 (Stamatakis 2006; Ott et al. 2007) under a rapid bootstrap analysis (-f a) applying the PROTCATWAGF model. The branching support was assessed by 1,000 bootstrap replicates.
To further assign the relative branch support, we applied RADICAL (Random Addition Concatenation Analysis) (Narechania et al. 2012) to the three data matrices. RADICAL generates a library of trees along a set of random concatenation chains varying from one gene to whole-matrix concatenation. Using this approach, the dynamics of concatenation was monitored by calculating support statistics for candidate test topologies assessed against the library of trees.
We applied 10 randomized chains using a step function of five for all three matrices. This means that for each matrix 10 concatenation paths were conducted sequentially 5 genes added, in which no gene is included more than once, and ending with the total concatenation of all genes. At each concatenation step, ML trees were generated with RAxML. RADICAL attempted in total 680 tree reconstructions for the M_matrix, 580 tree reconstructions for the P_matrix_g, and 200 tree reconstructions for the P_matrix_s, respectively.
Results and Discussion
ESTs and Alignments
An overview of the three new EST projects is given in supplementary table S2, Supplementary Material online. To predict the gene function, KOG analyses were conducted. The gene function of the sequences was predicted through BLAST (blastx, E < e−10) against the KOG database using OrthoSelect (Schreiber et al. 2009). For 7,431 sequences of Forficula auricularia, for 5,627 sequences of Nemurella pictetii and for 2,776 sequences of Zorotypus gurneyi? significant hits were detected and classified into 22 categories according to gene function (supplementary fig. S1, Supplementary Material online).
Our three variants from the original matrix (35% density) successfully reduced the overall amount of missing data. The first matrix which applied MARE (named M_matrix) was comprised of 53 species, 335 genes, 71369 amino acid positions, and increased the density to 70%. The second matrix generated using a PERL script (named P_matrix_g) was comprised of 62 species, 285 genes, 79506 amino acid positions and increased the density to 75%. The third matrix also generated using the PERL script (named P_matrix_s) was comprised of 73 species, 102 genes, 24507 amino acid positions and increased the density to 85%. An overview of represented genes in each matrix is given in supplementary table S3, Supplementary Material online. The overlap of genes in these three matrices is shown in supplementary figure S2, Supplementary Material online.
Compared with previous published studies, the three current data sets have a 90 gene overlap with the data sets of Simon et al. (2009) and a 78 gene overlap with the SOS alignment of Meusemann et al. (2010) (supplementary table S3, Supplementary Material online).
Higher Level Insect Relationships
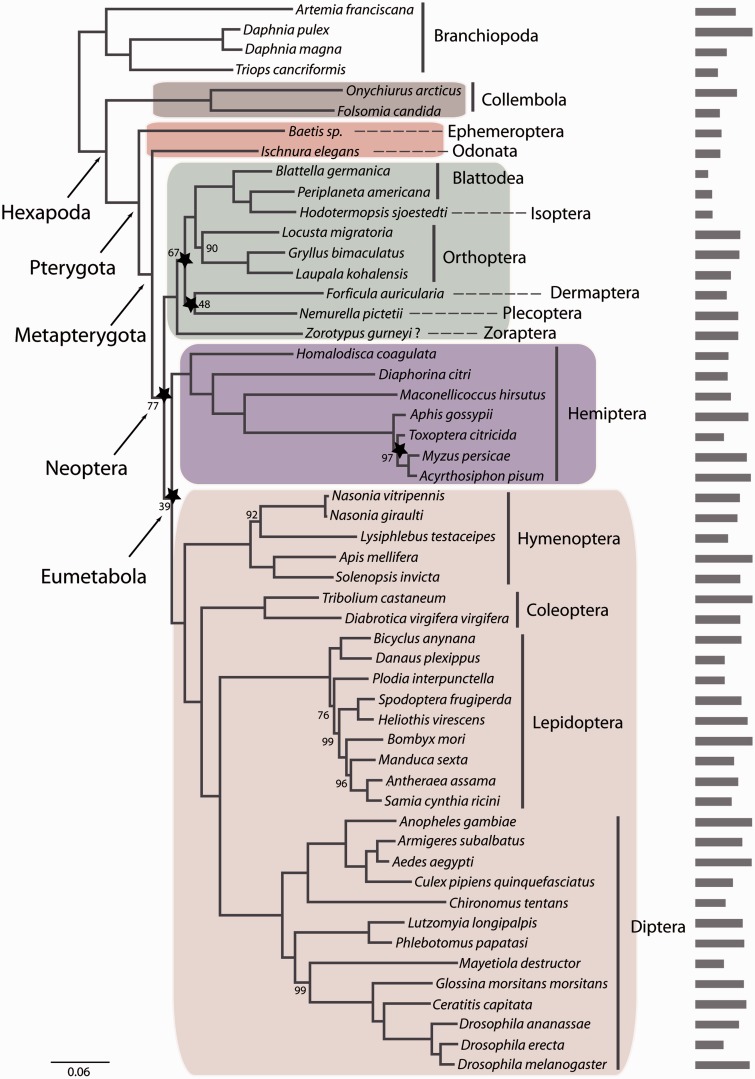
The tree topology shown in figures 1–3 was inferred from the M_matrix, P_matrix_g and P_matrix_s analyses, respectively. The tree topologies are essentially the same except for relationships within Hymenoptera and Lepidoptera. All analyses strongly support the monophyly of the major higher groups, namely Hexapoda, Ectognatha, Pterygota, Polyneoptera, and Holometabola (100–97% bootstrap support). The sistergroup relationship of Odonata to Neoptera, a clade named Metapterygota, was strongly supported in the topology obtained from the M_matrix and the P_matrix_g analyses, whereas the P_matrix_s analyses resulted only in 61% bootstrap support for this clade. The monophyly of Neoptera received strong support in the P_matrix_g and P_matrix_s (both 99%), whereas it was decreased in the M_matrix analyses (77%). Also the monophyly of Paraneoptera was only supported in the M_matrix and the P_matrix_g analyses while the in P_matrix_s analyses the support was inconclusive (33%). This could be a result of the inclusion of the louse Pediculus humanus. A previous study including this species could not recover the monophyly of Paraneoptera and indeed supported a sistergroup relationship of Pediculus humanus to Polyneoptera (Meusemann et al. 2010).
Fig. 1.—
RAxML topology derived from data matrix M_matrix (53 species, 335 genes, 71369 amino acid positions), PROTCATWAGF model. Support values are derived from 1,000 bootstrap replicates. Bootstrap values are only given for nodes that lack maximum support. Stars indicate nodes for which at least 90% of data (=300 genes) have to be concatenated to recover this specific node based on the RADICAL analyses. Also shown is a barplot indicating the number of putative orthologous genes for each of the taxa in this data set. Color code: primarily wingless hexapods, gray; “palaeopteran” insects, red; polyneopteran insects, green, paraneopteran insects, purple; and holometabolan insects, pink.
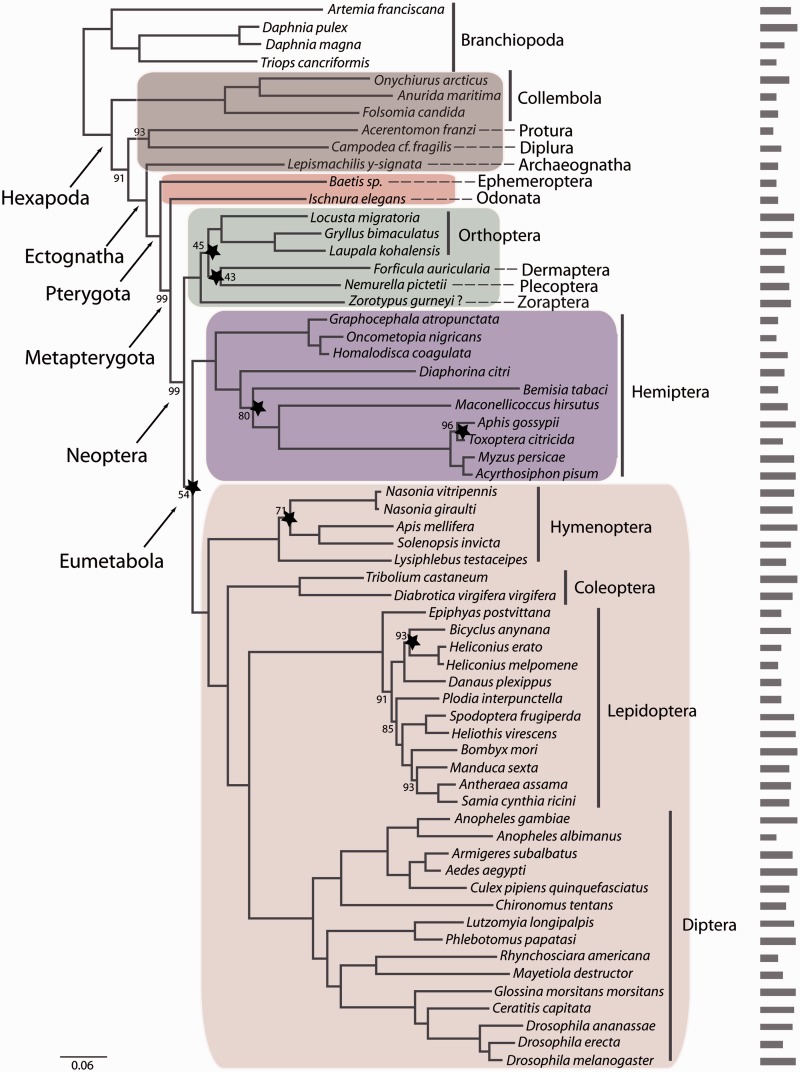
Fig. 2.—
RAxML topology derived from data matrix P_matrix_g (62 species, 285 genes, 79506 amino acid positions), PROTCATWAGF. Support values are derived from 1,000 bootstrap replicates. Bootstrap values are only given for nodes that lack maximum support. Stars indicate nodes for which at least 90% of data (=256 genes) have to be concatenated to recover this specific node based on the RADICAL analyses. Also shown is a barplot indicating the number of putative orthologous genes for each of the taxa in this data set. Color code: primarily wingless hexapods, gray; “palaeopteran” insects, red; polyneopteran insects, green, paraneopteran insects, purple; and holometabolan insects, pink.
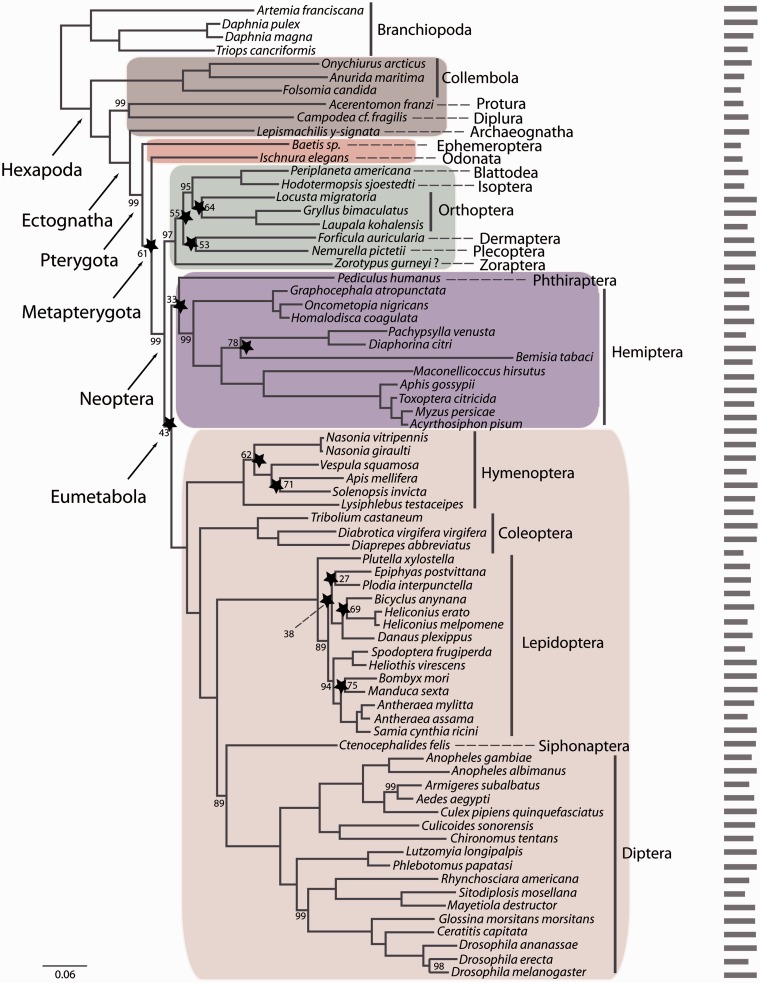
Fig. 3.—
RAxML topology derived from data matrix P_matrix_s (73 species, 102 genes, 24507 amino acid positions), PROTCATWAGF model. Support values are derived from 1,000 bootstrap replicates. Bootstrap values are only given for nodes that lack maximum support. Stars indicate nodes for which at least 90% of data (=92 genes) have to be concatenated to recover this specific node based on the RADICAL analyses. Also shown is a barplot indicating the number of putative orthologous genes for each of the taxa in this data set. Color code: primarily wingless hexapods, gray; “palaeopteran” insects, red; polyneopteran insects, green, paraneopteran insects, purple; and holometabolan insects, pink.
The Eumetabola hypothesis (Paraneoptera + Holometabola) remains inconclusive in all analyses (39–54% bootstrap support). In fact, this group shares several synapomorphies (Beutel and Pohl 2006) but most topologies derived from molecular sequence data alone do not recover this clade at all (Whiting et al. 1997; Wheeler et al. 2001; Misof et al. 2007; von Reumont et al. 2009; Meusemann et al. 2010) or only with low support (Kjer 2004; Ishiwata et al. 2010; Simon et al. 2010).
In addition, we evaluated the concatenation patterns of the data sets with RADICAL (Narechania et al. 2012). The outcome of a RADICAL analysis is a characterization of the strength of support for the concatenated phylogenetic hypothesis over the course of a concatenation chain. The approach allows for the identification of problematic nodes in a phylogenetic hypothesis through the concatenation process, even when the support for a particular node appears to be robust given high bootstrap or Bayes posterior support. The RADICAL curves for the data sets in this analysis highlight that topologies for any combination of genes quickly approach the concatenated tree topologies (figs. 1–3) during concatenation (supplementary fig. S3, Supplementary Material online). However, the RADICAL curves for the three data sets also indicate that the fixation point (Consensus Fork Index [CFI] = N, where N is equal to the number of nodes in the concatenated tree or when all nodes are identical to the concatenated tree) is only reached after concatenation of nearly all genes due to incongruence of partitions along the concatenation path. For example, based on the M_matrix data set, RADICAL identified five nodes (indicated by a star in fig. 1) as problematic. For these nodes, 90% of all genes (=300 genes) are required to recover the total evidence topology. Also for the P_matrix_g RADICAL identified seven nodes as problematic and 13 nodes for the P_matrix_s data set. In all three data sets, RADICAL identified 1) the node supporting Eumetabola (=Paraneoptera + Holometabola), 2) the node supporting the sistergroup relationship of Plecoptera and Dermaptera, and 3) the node supporting the sistergroup of Plecoptera + Dermaptera to remaining Polyneoptera (except Zoraptera) as problematic (table 2).
Table 2.
Phylogenetic Hypotheses within the Insect Tree Addressed in This Study
| Data set | ML Analyses |
RADICAL Analyses |
KOGs (RADICAL Analyses Based on P_matrix_g) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M_matrix | P_matrix_g | P_matrix_s | M_matrix | P_matrix_g | P_matrix_s | Cell | Info | Meta | Poorly | |
| No. of species | 53 | 62 | 73 | 53 | 62 | 73 | 62 | 62 | 62 | 62 |
| No. of genes | 335 | 285 | 102 | 335 | 285 | 102 | 85 | 80 | 78 | 42 |
| Positions (aa) | 71369 | 79509 | 24507 | 71369 | 79509 | 24507 | 24133 | 19709 | 23964 | 11700 |
| Density (%) | 70 | 75 | 85 | 70 | 75 | 85 | ||||
| Clade support | ||||||||||
| Hexapoda | 100 | 100 | 100 | 15 | 10 | 1 | 20 | 10 | 10 | 10 |
| Ectognatha | n.a. | 100 | 100 | n.a. | 50 | 20 | 85 | 30 | 75* | 40 |
| Pterygota | 100 | 100 | 99 | 15 | 55 | 45 | 45 | 55 | 40 | 42* |
| Metapterygota (=Odonata + Neoptera) | 100 | 99 | 61 | 130 | 110 | 95 | 85 | 35 | 35*A | 42*B |
| Neoptera | 77 | 99 | 99 | 310 | 150 | 55 | 50* | 55 | 75 | 1* |
| Polyneoptera | 100 | 100 | 97 | 160 | 115 | 40 | 85* | 30 | 75 | 42* |
| Orthoptera, (Blattodea, Isoptera) | 100 | n.a. | 95 | 240 | n.a. | 70 | n.a. | n.a. | n.a. | n.a. |
| (Dermaptera, Plecoptera) | 48 | 43 | 53 | 325 | 275 | 95 | 75* | 65* | 75 | 42* |
| Zoraptera, (remaining Polyneoptera) | 100 | 100 | 97 | 160 | 115 | 40 | 85* | 30 | 75 | 42* |
| (Dermaptera, Plecoptera), (Orthoptera, (Blattodea, Isoptera)) | 67 | 45 | 55 | 300 | 275 | 95 | 40* | 65* | 78* | 42* |
| Eumetabola (=Paraneoptera + Holometabola) | 39 | 54 | 43 | 330 | 275 | 100 | 20* | 30*C | 55 | 1* |
| Paraneoptera | 100 | 100 | 33 | 115 | 50 | 100 | 80 | 55 | 20 | 35* |
| Holometabola | 100 | 100 | 100 | 50 | 35 | 30 | 40 | 25 | 55 | 40 |
| Hymenoptera, remaining Holometabla | 100 | 100 | 100 | 50 | 35 | 30 | 40 | 25 | 55 | 40 |
| Coleoptera, (Lepidoptera, Diptera) | 100 | 100 | 100 | 20 | 35 | 30 | 35 | 25 | 35 | 20 |
| (Lepidoptera, Diptera) | 100 | 100 | 100 | 35 | 35 | n.a. | 35 | 20 | 20 | 15 |
Note.—For selected clades (1st column), the bootstrap support values based on the RAxML analyses (2nd column) and the number of genes required for fixation based on the RADICAL analyses (3rd column) for all three matrices are given. In the 4th column, the number of the major KOG genes required for fixation based on the P_matrix_g data set and the RADICAL analyses are given.
* indicates the number of genes for which a node no longer occurs in any randomized concatenation set of that size or larger. For two nodes (Metapterygota and Eumetabola), alternative hypotheses were tested using the P_matrix_g data set and RADICAL based on the major KOG categories (4th column). A: concatenation of 75 meta genes support the Palaeoptera hypothesis; B: concatenation of 15 poorly genes support the Palaeoptera hypothesis. C: concatenation of 75 info genes support a sistergroup relationship of Polyneoptera and Holometabola.
The Polyneopteran Relationships and the Phylogenetic Position of Zoraptera
The interrelationships of the 11 polyneopteran orders are far from resolved and even the monophyly of this neopteran infraclass is disputed. Within Polyneoptera only two clades, Dictyoptera (Blattodea, Isoptera, and Mantodea) and Xenonomia (Grylloblattodea + Mantophasmatodea) have become better resolved (table 1). Other proposed groups within Polyneoptera are not widely accepted due to the lack of convincing morphological synapomorphies and contradictory or only poorly resolved relationships based on molecular data sets, for example, Orthopterida (=Orthoptera + Phasmatodea) and Eukinolabia (=Phasmatodea + Embioptera) (but see Letsch et al. 2012).
The phylogenetic position of the three remaining polyneopteran orders (Dermaptera, Plecoptera, and Zoraptera) is even more unclear. Here, the placement of Plecoptera and Zoraptera within Polyneoptera has even been questioned; Zoraptera + Paraneoptera (Beutel and Weide 2005) and Plecoptera + remaining Neoptera (Beutel and Gorb 2006). In addition, these three orders have been mostly neglected in molecular studies. Consequently, this study provides one of the most comprehensive molecular data sets for these enigmatic orders and advances us toward the resolution of the Polyneoptera.
Dermaptera is a key order for resolving the phylogenetic position of Plecoptera and Zoraptera, due to their inferred sistergroup relationships to both. Here, two hypotheses are debated: Haplocerata (=Dermaptera + Zoraptera) or Dermaptera + Plecoptera (table 1). Zoraptera is indeed the most enigmatic insect lineage with respect to its evolutionary history, with more than 10 discussed positions within Polyneoptera as well as Paraneoptera (Yoshizawa 2007). The term “Zoraptera-problem” (Beutel and Weide 2005) is as well deserved as the “Strepsiptera-problem” (Kristensen 1981). Indeed, molecular sequence data for Zoraptera are still rare (19 sequences, 13 of them rRNA genes http://www.ncbi.nlm.nih.gov/nuccore?term=zoraptera; last accessed April 2012). In contrast, the sequence information available for Strepsiptera including several nuclear coding genes, EST projects, a complete mitochondrial genome as well as a recently published genome-project has greatly improved the phylogenetic position of this previously phylogenetically ambiguous insect order (McMahon et al. 2009; Wiegmann et al. 2009; Longhorn et al. 2010; McKenna and Farrell 2010; Talavera and Vila 2011; Niehuis et al. 2012). However, based on the molecular data and/or morphological characters available for Zoraptera 4 of the 10 discussed phylogenetic positions of Zoraptera have gained increased support: 1) Zoraptera + Dictyoptera; 2) Zoraptera + Dermaptera (=Haplocerata); 3) Zoraptera + Paraneoptera; and 4) Zoraptera + Embioptera (=Mystroptera) (table 1).
Using the first transcriptomic data for the three discussed orders (Dermaptera, Plecoptera, and Zoraptera), our analyses provide conclusive support for monophyletic Polyneoptera (100–97%), contesting the hypothesis of Zoraptera + Paraneoptera and Plecoptera + remaining Neoptera. Zoraptera splits off first within Polyneoptera followed by the clade (Plecoptera + Dermaptera) + remaining Polyneoptera. In addition, no support for the hypothesis Zoraptera + Dermaptera (=Haplocerata) or Zoraptera + Dictyoptera is found. Still, we have to consider that important polyneopteran orders are missing to fully explore the phylogenetic position of these three orders (but see Letsch et al. 2012). Especially, the exact position of Plecoptera and Dermaptera within Polyneoptera remains problematic even and despite using extensive molecular data sets. Although a sistergroup relationship of Plecoptera and Dermaptera is recovered in all of our three analyses, the bootstrap values are overall weak (53–43%). In addition, the node supporting this group (Plecoptera + Dermaptera) has been identified as a problematic node in all three data sets by the RADICAL analyses.
The lack of genomic information from all polyneopteran orders might be also the reason why the exact phylogenetic position of the three orders is still inconclusive.
In sum, based on this first phylogenomic approach to infer the phylogenetic position of Zoraptera, we contest three of the four hypotheses concerning the position of Zoraptera: 1) Zoraptera + Dictyoptera, 2) Zoraptera + Dermaptera (=Haplocerata), and 3) Zoraptera + Paraneoptera.
The Impact of Matrix Selection to Infer Insect Evolution
Controversies about the effects of missing data on phylogenomic studies still exist (Wiens 2003; Philippe et al. 2004, 2005; de Queiroz and Gatesy 2007; Hartmann and Vision 2008; Lemmon et al. 2009). Although it has been suggested that the low number of informative or overlapping characters cause the inaccurate placement of incomplete taxa, there is also evidence that missing data might enhance tree reconstruction artifacts (Wiens and Moen 2008; Lemmon et al. 2009). Consequently, several studies consider including/excluding taxa and characters to avoid a high percentage of missing data (Philippe et al. 2007; Dunn et al. 2008; Simon et al. 2009; Meusemann et al. 2010; von Reumont et al. 2012) but automated methods to create a matrix for the phylogenetic analyses based on explicit criteria are still rare.
To further address this issue, we compared different approaches to reduce overall missing data, first applying an automated method, MARE (Meyer et al. April 2011) (M_matrix), which aims to increase the number of taxa with potentially informative genes by excluding genes that have lower tree-likeness scores, and second applying a PERL script that selects taxa/genes based on presence/absence (P_matrix_g and P_matrix_s). All three matrices have 96 genes in common (supplementary fig. S2, Supplementary Material online) and none of the matrices exhibited superior performance over the others. Recently, von Reumont et al. (2012) proposed that MARE might introduce potential artifacts especially among deep nodes due to removal of genes with older and distorted phylogenetic signal. This assumption could not be confirmed by our results. Indeed, the inferred interrelationships of the insect orders in all three topologies were essentially the same with comparable bootstrap supports. However, the phylogenetic signal of each gene in the matrices and especially the interactions of these signals (the ratio of phylogenetic-to-nonphylogenetic signal) are unknown. Based on this and a previous study, we propose that reducing missing data have a positive effect on the inferred relationships within the Insect Tree (see supplementary figure 6 in Meusemann et al. 2010), but there is no difference in selecting taxa/genes based on information content or simple presence/absence, for the insect data set used in this study.
Another major point in phylogenomic and phylogenetic studies in general is taxon sampling, as it is one potential source of long-branch attraction (LBA) artifacts (Hillis et al. 2003; Brinkmann et al. 2005). We have addressed this issue in the P_matrix_s analyses. In this data set, the taxon sampling was increased (73 species of initial 78 species included) and mainly underrepresented genes were excluded. The inferred insect relationships based on this approach are in agreement with the M_matrix and P_matrix_g analyses. This indicates that our results are robust with respect to the number of selected species and genes based on our original matrix.
The Influence of Gene and/or Taxon Sampling on the Insect Tree: A Never Ending “Palaeoptera Problem”
The transition from nonwinged to winged insects still represents one of the major obstacles for insect systematics—the so-called “Palaeoptera Problem” (see Simon et al. 2009; Trautwein et al. 2012; Yeates et al. 2012). Based on our analyses, strong support for the clade Metapterygota (Odonata + Neoptera) is provided (bootstrap support: 100–99%) (table 2). Only in the P_matrix_s analyses does the clade receive weak support (61%). However, if we compare the inferred insect relationships with Meusemann et al. (2010) and von Reumont et al. (2012), both of which use wide taxon sampling across arthropod lineages, there is strong conflict in the support for relationships among the “palaeopterous” orders. In the study of Meusemann et al. (2010), the ML analyses are inconclusive, but “Palaeoptera” (Odonata + Ephemeroptera) is strongly supported in Bayesian analyses. von Reumont et al. (2012) provide strong support for “Palaeoptera” in the reduced ML analyses (100–91%), whereas the unreduced ML analyses are inconclusive. In contrast, Simon et al. (2009) using a smaller taxon sampling across insects support the clade Chiastomyaria (Ephemeroptera + Neoptera). Hence, all three possible sistergroup relationships of Ephemeroptera, Odonata, and Neoptera are supported by using the same EST/transcriptome data and the same ortholog prediction approach but different matrix composition, making the “Palaeoptera Problem” more enigmatic than before.
To further evaluate whether the support for the clade Metapterygota in this study is only a result of taxon sampling or if the phylogenetic signals of the genes represented in the different matrices have an influence, we searched for genes in our original orthologs data set that are also represented in the SOS data set of Meusemann et al. (2010). Of the 129 genes represented in the SOS data set of Meusemann et al. (2010), 85 genes were identified in our original orthologs data set. Based on these 85 genes and a taxon sampling identical to the P_matrix_g analyses, ML analyses were performed (-f a; 1,000 bootstrap replicates). Again the clade Metapterygota (Odonata + Neoptera) received support, although all relatively weak (64%) (supplementary fig. S4, Supplementary Material online).
Removing distantly related taxa from the outgroup sampling (e.g., several crustacean taxa, myriapods, and chelicerates) and increasing the in-group sampling have a major impact on the basal insect relationships—the relative placement of the “palaeopterous” orders Ephemeroptera and Odonata. These circumstances lead us to propose that not only exploring systematic bias and impact of missing data but also the effect of a priori defined taxon sampling for the inferred relationships is an important issue for future work on phylogenetically ambiguous regions of the Insect Tree. The right way to increase the accuracy of a phylogenomic tree remains an open question, as there is a trade-off between sampling size and computation time.
The Influence of Gene Function to Infer the Evolutionary History of Insects
Another key question in phylogenomic studies is the selection of a core set of genes for analysis. What genes should be used to recover the “true” species tree? Naturally, the selected genes should have orthologs across as many of the taxa sampled as possible, but the challenge is to evaluate which genes harbor the phylogenetic signal to resolve a phylogenetic question. Ideally independent molecular loci should reflect the same evolutionary history to make the results robust, but different genomic regions can have different evolutionary histories along the branches of a species tree (Degnan and Rosenberg 2006).
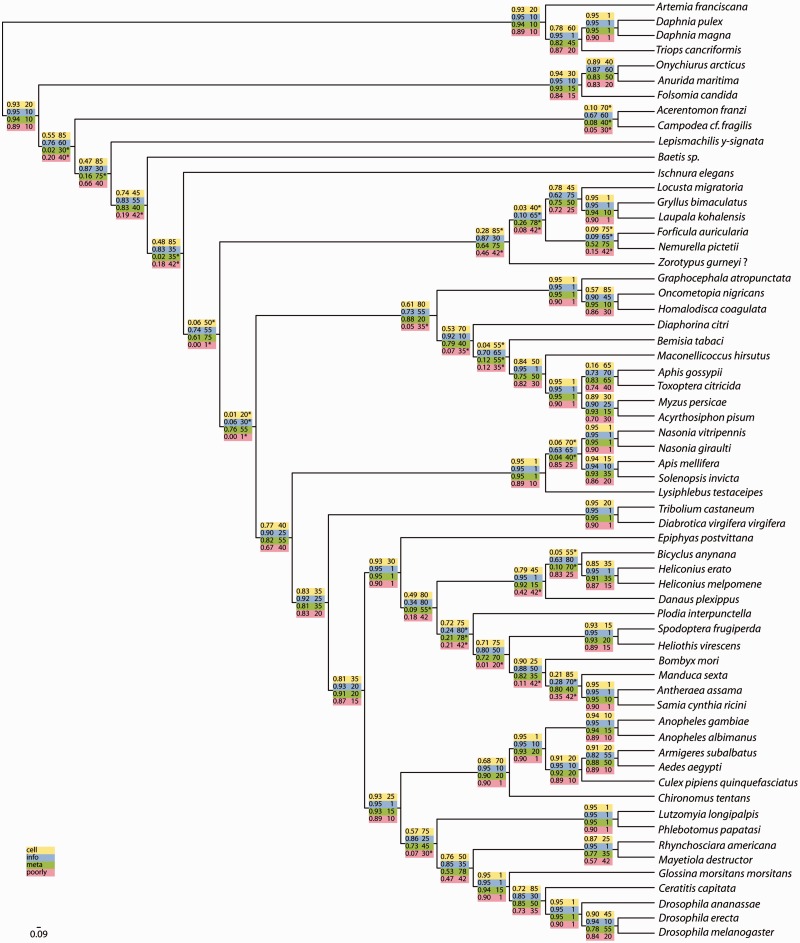
To address the assumption that the phylogenetic signal of a gene depends on functional constraints and evolutionary history (Philippe et al. 2011), we performed additional analyses. The P_matrix_g data set was used to evaluate the source of incongruence for partitions and gene categories based on their function to infer insect relationships. Therefore, the biological function of the represented genes was assigned through Blast against the eukaryotic orthologous groups (KOGs) database. The genes were concatenated according to their major functional classification: 1) cellular processes and signaling (cell = 85 genes), 2) information storage and processing (info = 80 genes), 3) metabolism (meta = 78 genes), and 4) poorly recognized (poorly = 42 genes).
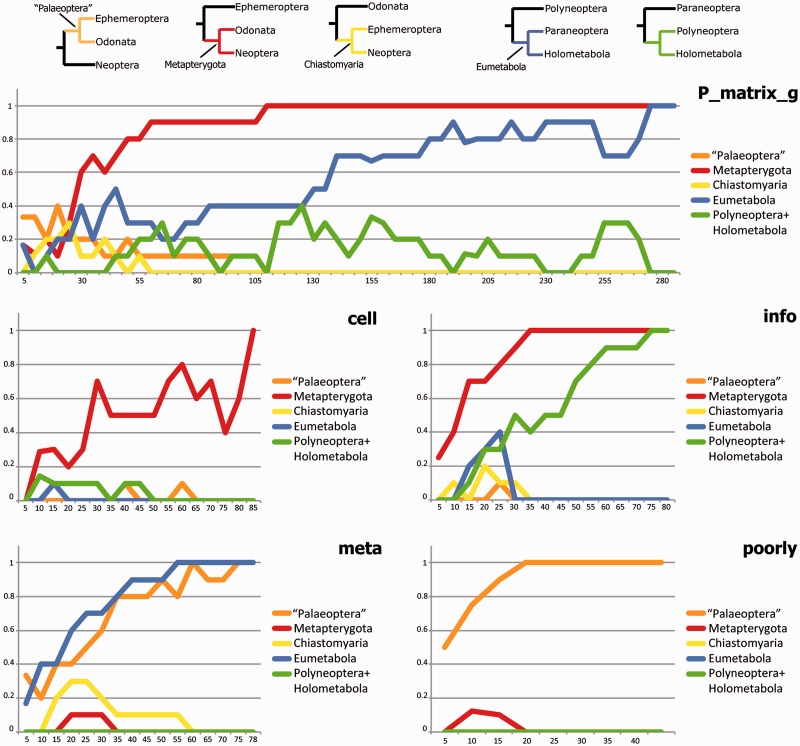
To evaluate whether these four categories exhibit strong agreement with the total evidence topology based on the P_matrix_g analyses (fig. 2), we applied RADICAL. These analyses highlight that for most deep nodes 1) nearly all genes of each major KOG are required to recover the total evidence topology and 2) the KOG categories have a substantial proportion of genes that disagree with the total evidence topology (fig. 4 and table 2). For example, the node supporting Metapterygota (Odonata + Neoptera) is stabilized when all cell genes are concatenated (85) and also when 35 info genes are concatenated. However, this node disappear in any concatenation set larger than 35 for meta genes and in the concatenation set larger than 42 for poorly genes. Also for the node supporting the Eumetabola hypothesis (Paraneoptera + Holometabola), the functional subgroups harbors conflicting signal. This node is recovered after concatenation of 55 meta genes but disappear in any concatenation set larger than 20 for cell genes, 30 for info genes and 1 for poorly genes. In contrast, the nodes supporting Holometabola, the first-branching of Hymenoptera within Holometabola or the inter- and intra-relationships for holometabolous orders are nearly all well recovered by all functional subgroups (fig. 4 and table 2).
Fig. 4.—
RADICAL analysis of functional subgroups. AUC values (left column) and fixation points are provided across all total evidence nodes for the functional groups 1) cellular processes and signaling (cell = 85 genes); 2) information storage and processing (info = 80 genes); 3) metabolism (meta = 78 genes); and 4) poorly recognized (poorly = 42 genes). AUC values indicate the proportion of total concatenation space occupied by that node and the fixation point indicates the number of genes required before the node appears in all concatenation sets of that size. The star indicates the number of genes for which a node no longer occurs in any randomized concatenation set of that size or larger.
These results demonstrate that for some short ancient internodes, for example, the basal pterygote divergence or the neopteran lineage divergence, some functional subgroups disagree with the total evidence topology and might harbor phylogenetic signal for alternative phylogenetic relationships. To further evaluate this assumption, we selected two controversially discussed relationships of insect lineages and assessed nodal support within the total evidence tree and their alternatives: 1) basal pterygote divergence: “Palaeoptera,” Metapterygota, or Chiastomyaria and 2) Eumetabola (=Paraneoptera + Holometabola) vs. Polyneoptera + Holometabola (fig. 5 for hypotheses). RADICAL was used to assign the support for these five nodes for the P_matrix_g data set as well as for the four functional subgroups based on this data set (fig. 5 and table 2). For the basal pterygote divergence, the Metapterygota hypothesis is recovered by concatenation of approximately 110 genes. The support for this hypothesis stems from the cell genes and mainly the info genes, whereas the meta genes and the poorly genes support the alternative “Palaeoptera” hypothesis. The Eumetabola hypothesis is generally only recovered after concatenation of nearly the complete data set (280 genes). Indeed, the analysis based on the functional subgroups show that only the meta genes recover this hypothesis, whereas the info genes support the alternative (Polyneoptera + Holometabola).
Fig. 5.—
RADICAL analysis of nodal support. Shown are the RADICAL curves for each of the five phylogenetic hypotheses (at the top) for the complete P_matrix_g data set and the four functional subgroups based on this data set. The y axis shows the CFI and the x axis the concatenation size (number of genes).
The analyses show that the phylogenomic matrices have more complex phylogenetic signal and that the functional subgroups recover different scenarios of ancient rapid insect evolution, for example, the basal pterygote or the neopteran lineage divergence. Horizontal transfer, gene duplication or incomplete lineage sorting can lead to this incongruence in the evolutionary history of the functional subgroups (Kubatko and Degnan 2007). Another explanation would be that the different evolutionary signals are a result of the different evolutionary processes that act upon the functional subgroups and that the functional role of these genes in the cell is important for the phylogenetic signal they carry (Graur and Li 2000). These issues might become more obvious when whole genomes are available for the diverse insect lineages. Using complete taxa with high number of overlapping characters could then provide the opportunity to find genes and/or functional subgroups that harbors the same evolutionary history along the branches as the species under investigation. In addition, eventually comparative research on regulatory genes may become also helpful for deep phylogenetic studies and might bridge some gaps between description and causal explanations (Hadrys et al. 2012).
Conclusion
In this study, we provide the first transcriptomic data for three enigmatic polyneopteran orders Dermaptera, Plecoptera, and Zoraptera. Based on comprehensive phylogenomic analyses, we provide conclusive support for monophyletic Polyneoptera. Although the interaction of gene choice and taxon-sampling still remains unknown, we could not identify any influence of different approaches to reduce the missing data in inferring insect relationships.
In contrast, our additional analyses highlight that especially for the ancient rapid radiation of the insects, for example, basal pterygote divergence or split of neopteran infraclasses, the taxon-sampling and gene function have a huge impact on the inferred relationships. Consequently, further extended analyses (in terms of data quantity as well as quality) are necessary to finally confirm the inferred phylogenetic relationships of the most critical groups presented in this study (e.g., Metapterygota and Eumetabola). Currently, it seems that the available molecular data for insects is insufficient to recover some ancient splits within insect evolution and that large phylogenomic matrices harbor a high percent of conflicting phylogenetic signal for these short internodes. As long as we do not have independent alternative characters, for example, genetic characters such as gene order, genome rearrangements, intron and transposon positions, which might provide a greater understanding of insect evolution, we can only suggest which taxa/genes and/or which functional subgroups might reflect the “true evolutionary history” of insects. In sum, inferring insect relationships offers a great opportunity to explore the extent and source of biases and how the resolution of ancient rapid radiations might be influenced by the choice of taxa and genes.
Supplementary Material
Supplementary figures S1–S4 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft [DFG]) special priority program “Deep Metazoan Phylogeny” SPP1174 grant to H.H. (DFG HA 1947/5). S.S. acknowledges funding by the DFG grant (DFG HA 1947/5) and a fellowship within the Postdoc-Program of the German Academic Exchange Service (DAAD). The authors thank Leonardo Calderon Obaldia for collecting the specimens of Zorotypus gurneyi?. They express their gratitude to Ryuichiro Machida and especially Michael Engel for their effort in determining Zorotypus gurneyi?. They also thank Associate Editor Günter Wagner and two anonymous reviewers for providing constructive comments which greatly improved this manuscript.
Literature Cited
- Baurain D, Brinkmann H, Philippe H. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Mol Biol Evol. 2007;24:6–9. doi: 10.1093/molbev/msl137. [DOI] [PubMed] [Google Scholar]
- Beutel RG, Gorb S. A revised interpretation of the evolution of attachment structures in hexapoda with special emphasis on Mantophasmatodea. Arthropod System Phylogeny. 2006;64:3–25. [Google Scholar]
- Beutel RG, Pohl H. Endopterygote systematics—where do we stand and what is the goal (Hexapoda, Arthropoda)? System Entomol. 2006;31:202–219. [Google Scholar]
- Beutel R, Weide D. Cephalic anatomy of Zorotypus hubbardi (Hexapoda: Zoraptera): new evidence for a relationship with Acercaria. Zoomorphology. 2005;124:121–136. [Google Scholar]
- Brinkmann H, et al. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol. 2005;54:743–757. doi: 10.1080/10635150500234609. [DOI] [PubMed] [Google Scholar]
- Boudreaux HB. Arthropod phylogeny with special reference to insects. New York: Wiley; 1979. [Google Scholar]
- de Queiroz A, Gatesy J. The supermatrix approach to systematics. Trends Ecol Evol. 2007;22:34–41. doi: 10.1016/j.tree.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 2009;9:157. doi: 10.1186/1471-2148-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel MS, Grimaldi DA. A winged Zorotypus in Miocene amber from the Dominican Republic (Zoraptera: Zorotypidae), with discussion on relationships of and within the order. Acta Geológica Hispánica. 2000;35:149–164. [Google Scholar]
- Foottit RG, Adler PH. Insect biodiversity: science and society. Oxford (UK): Wiley-Blackwell; 2009. [Google Scholar]
- Friedemann K, Wipfler B, Bradler S, Beutel RG. On the head morphology of Phyllium and the phylogenetic relationships of Phasmatodea (Insecta) Acta Zoologica. 2012;93:184–199. [Google Scholar]
- Graur D, Li WH. Fundamentals of molecular evolution. Sunderland (MA): Sinauer Associates; 2000. [Google Scholar]
- Grimaldi DA, Engel MS. Evolution of the insects. New York: Cambridge University Press; 2005. [Google Scholar]
- Hadrys H, et al. Isolation of hox cluster genes from insects reveals an accelerated sequence evolution rate. PLoS One. 2012;7:e34682. doi: 10.1371/journal.pone.0034682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann S, Vision TJ. Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol Biol. 2008;8:95. doi: 10.1186/1471-2148-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Is sparse taxon sampling a problem for phylogenetic inference? Syst Biol. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, et al. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles) Mol Biol Evol. 2006;23:268–278. doi: 10.1093/molbev/msj041. [DOI] [PubMed] [Google Scholar]
- Inward D, Beccaloni G, Eggleton P. Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol Lett. 2007;3:331–335. doi: 10.1098/rsbl.2007.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata K, et al. Phylogenetic relationships among insect orders based on three nuclear protein-coding gene sequences. Mol Phylogenet Evol. 2010;58:169–180. doi: 10.1016/j.ympev.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Jarvis KJ, Haas F, Whiting MF. Phylogeny of earwigs (Insecta: Dermaptera) based on molecular and morphological evidence: reconsidering the classification of Dermaptera. Systematic Entomology. 2005;30:442–453. [Google Scholar]
- Jintsu Y, Uchifune T, Machida R. Structural features of eggs of the basal phasmatodean Timema monikensis Vickery and Sandoval, 1998 (Insecta: Phasmatodea: Timematidae) Arthropod System Phylogeny. 2010;68:71–78. [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kjer KM. Aligned 18S and insect phylogeny. Syst Biol. 2004;53:506–514. doi: 10.1080/10635150490445922. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Carle FL, Litman J, Ware J. A molecular phylogeny of Hexapoda. Arthropod System Phylogeny. 2006;65:35–44. [Google Scholar]
- Klass KD, Zompro O, Kristensen NP, Adis J. Mantophasmatodea: a new insect order with extant members in the Afrotropics. Science. 2002;296:1456–1459. doi: 10.1126/science.1069397. [DOI] [PubMed] [Google Scholar]
- Kristensen NP. Phylogeny of insect orders. Annu Rev Entomol. 1981;26:135–157. [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Kück P, Meusemann K. FASconCAT: convenient handling of data matrices. Mol Phylogenet Evol. 2010;56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Brown JM, Stanger-Hall K, Lemmon EM. The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Syst Biol. 2009;58:130–145. doi: 10.1093/sysbio/syp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsch HO, et al. Insect phylogenomics: results, problems and the impact of matrix composition. Proc Biol Sci. 2012;279:3282–3290. doi: 10.1098/rspb.2012.0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhorn SJ, Pohl HW, Vogler AP. Ribosomal protein genes of holometabolan insects reject the Halteria, instead revealing a close affinity of Strepsiptera with Coleoptera. Mol Phylogenet Evol. 2010;55:846–859. doi: 10.1016/j.ympev.2010.03.024. [DOI] [PubMed] [Google Scholar]
- McKenna DD, Farrell BD. 9-genes reinforce the phylogeny of holometabola and yield alternate views on the phylogenetic placement of Strepsiptera. PLoS One. 2010;5:e11887. doi: 10.1371/journal.pone.0011887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon DP, Hayward A, Kathirithamby J. The mitochondrial genome of the 'twisted-wing parasite' Mengenilla australiensis (Insecta, Strepsiptera): a comparative study. BMC Genomics. 2009;10:603. doi: 10.1186/1471-2164-10-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusemann K, et al. A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol. 2010;27:2451–2464. doi: 10.1093/molbev/msq130. [DOI] [PubMed] [Google Scholar]
- Meyer B, Meusemann K, Misof B. MARE: MAtrix REduction—a tool to select optimized data subsets from supermatrices for phylogenetic inference. Bonn (Germany): Zentrum fuür molekulare Biodiversitätsforschung (zmb) am ZFMK; 2011. [Google Scholar]
- Misof B, Misof K. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. System Biol. 2009;58:21–34. doi: 10.1093/sysbio/syp006. [DOI] [PubMed] [Google Scholar]
- Misof B, et al. Towards an 18S phylogeny of hexapods: accounting for group-specific character covariance in optimized mixed nucleotide/doublet models. Zoology (Jena) 2007;110:409–429. doi: 10.1016/j.zool.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Narechania A, et al. Random addition concatenation analysis: a novel approach to the exploration of phylogenomic signal reveals strong agreement between core and shell genomic partitions in the cyanobacteria. Genome Biol Evol. 2012;4:30–43. doi: 10.1093/gbe/evr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, et al. Genomic and morphological evidence converge to resolve the enigma of strepsiptera. Curr Biol. 2012;22:1309–1313. doi: 10.1016/j.cub.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Ott M, Zola J, Stamatakis A, Aluru S. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. Proceedings of the 2007 ACM/IEEE conference on Supercomputing. Reno (NV) 2007 ACM. p. 1–11. [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N. Phylogenomics. Annu Rev Ecol Evol System. 2005;36:541–562. [Google Scholar]
- Philippe H, et al. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Philippe H, et al. Acoel flatworms are not platyhelminthes: evidence from phylogenomics. PLoS One. 2007;2:e717. doi: 10.1371/journal.pone.0000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael JA, Engel MS. A new species of Zorotypus from Central Amazonia, Brazil (Zoraptera: Zorotypidae) American Museum Novitates. 2006:1–11. [Google Scholar]
- Regier JC, et al. Resolving arthropod phylogeny: exploring phylogenetic signal within 41 kb of protein-coding nuclear gene sequence. Syst Biol. 2008;57:920–938. doi: 10.1080/10635150802570791. [DOI] [PubMed] [Google Scholar]
- Regier JC, et al. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Savard J, et al. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber F, et al. OrthoSelect: a protocol for selecting orthologous groups in phylogenomics. BMC Bioinformatics. 2009;10:219. doi: 10.1186/1471-2105-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharanowski BJ, et al. Expressed sequence tags reveal Proctotrupomorpha (minus Chalcidoidea) as sister to Aculeata (Hymenoptera: Insecta) Mol Phylogenet Evol. 2010;57:101–112. doi: 10.1016/j.ympev.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Simon S, Schierwater B, Hadrys H. On the value of elongation factor-1alpha for reconstructing pterygote insect phylogeny. Mol Phylogenet Evol. 2010;54:651–656. doi: 10.1016/j.ympev.2009.09.029. [DOI] [PubMed] [Google Scholar]
- Simon S, Strauss S, von Haeseler A, Hadrys H. A phylogenomic approach to resolve the basal pterygote divergence. Mol Biol Evol. 2009;26:2719–2730. doi: 10.1093/molbev/msp191. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Talavera G, Vila R. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol. 2011;11:315. doi: 10.1186/1471-2148-11-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MD, Whiting MF. Comparison of two alignment techniques within a single complex data set: POY versus Clustal. Cladistics. 2005;21:272–281. [Google Scholar]
- Trautwein MD, et al. Advances in insect phylogeny at the dawn of the postgenomic era. Annu Rev Entomol. 2012;57:449–468. doi: 10.1146/annurev-ento-120710-100538. [DOI] [PubMed] [Google Scholar]
- von Reumont BM, et al. Can comprehensive background knowledge be incorporated into substitution models to improve phylogenetic analyses? A case study on major arthropod relationships. BMC Evol Biol. 2009;9:119. doi: 10.1186/1471-2148-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reumont BM, et al. Pancrustacean phylogeny in the light of new phylogenomic data: support for remipedia as the possible sister group of hexapoda. Mol Biol Evol. 2012;29:1031–1045. doi: 10.1093/molbev/msr270. [DOI] [PubMed] [Google Scholar]
- Wheeler WC, Whiting MF, Wheeler QD, Carpenter JM. The phylogeny of the extant hexapod orders. Cladistics. 2001;17:113–169. doi: 10.1111/j.1096-0031.2001.tb00115.x. [DOI] [PubMed] [Google Scholar]
- Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC. The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol. 1997;46:1–68. doi: 10.1093/sysbio/46.1.1. [DOI] [PubMed] [Google Scholar]
- Whiting MF, Bradler S, Maxwell T. Loss and recovery of wings in stick insects. Nature. 2003;421:264–267. doi: 10.1038/nature01313. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, et al. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biol. 2009;7:34. doi: 10.1186/1741-7007-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ. Missing data, incomplete taxa, and phylogenetic accuracy. Syst Biol. 2003;52:528–538. doi: 10.1080/10635150390218330. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Moen DS. Missing data and the accuracy of Bayesian phylogenetics. J Syst Evol. 2008;46:307–314. [Google Scholar]
- Wipfler B, Machida R, Müller M, Beutel RG. On the head morphology of Grylloblattodea (Insecta) and the systematic position of the order, with a new nomenclature for the head muscles of Dicondylia. Systematic Entomology. 2011;36:241–266. [Google Scholar]
- Xie Q, Tian X, Qin Y, Bu W. Phylogenetic comparison of local length plasticity of the small subunit of nuclear rDNAs among all Hexapoda orders and the impact of hyper-length-variation on alignment. Mol Phylogenet Evol. 2009;50:310–316. doi: 10.1016/j.ympev.2008.10.025. [DOI] [PubMed] [Google Scholar]
- Yeates DK, Cameron SL, Trautwein M. A view from the edge of the forest: recent progress in understanding the relationships of the insect orders. Austr J Entomol. 2012;51:79–87. [Google Scholar]
- Yoshizawa K. The Zoraptera problem: evidence for Zoraptera + Embiodea from the wing base. System Entomol. 2007;32:197–204. [Google Scholar]
- Yoshizawa K, Johnson KP. Aligned 18S for Zoraptera (Insecta): phylogenetic position and molecular evolution. Mol Phylogenet Evol. 2005;37:572–580. doi: 10.1016/j.ympev.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Yoshizawa K. Monophyletic Polyneoptera recovered by wing base structure. System Entomol. 2011;36:377–394. [Google Scholar]
- Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zhang YY, et al. The complete mitochondrial genome of the cockroach Eupolyphaga sinensis (Blattaria: Polyphagidae) and the phylogenetic relationships within the Dictyoptera. Mol Biol Rep. 2010;37:3509–3516. doi: 10.1007/s11033-009-9944-1. [DOI] [PubMed] [Google Scholar]
- Zwick P. The Plecoptera—who are they? The problematic placement of stoneflies in the phylogenetic system of insects. Aquatic Insects. 2009;31:181–194. [Google Scholar]