Abstract
To investigate the microevolution of Helicobacter bizzozeronii in the human stomach, comparative genomics of antrum-derived populations, obtained 3 months before (T0) and 6 months after (T1) an unsuccessful eradication treatment, was performed. For each time point, the DNA of bacterial mass, representing the population diversity in three biopsies, was mixed in equal amounts and sequenced using Illumina technology. Polymorphic sites (PSs) were detected by mapping the reads against an isogenic reference genome, derived from a corpus isolate obtained at T0. The total numbers of PSs detected in the H. bizzozeronii population at T0 and T1 were 128 and 223, affecting 81 and 134 coding sequences, respectively. At T0 in 91.4% of the PSs the mutation appeared at a frequency of 50% or less. On the contrary, in the majority of the PSs observed in T1 (71.3%) the mutation had a frequency >75%. Although only a minority of mutations were fixed in the antrum-derived population at T0, a certain level of allelic variability, compared with the corpus-derived reference genome, was present and most likely arose as consequence of the long-term colonization of the patient. The treatment probably induced a sudden decrease of population size, selecting a subpopulation, which acted as founder for the new population at T1 characterized by a higher number of fixed mutations. These data demonstrate that genome plasticity is an important common prerequisite among gastric Helicobacter species for adaptation to the stomach environment allowing the bacterium to evolve rapidly once a selective pressure is applied.
Keywords: Helicobacter heilmannii sensu lato, Helicobacter bizzozeronii, resistance mutations, compensatory mutations
Introduction
The human-adapted pathogen Helicobacter pylori is one of the most common causes of bacterial infections worldwide and is recognized as an etiologic agent of chronic gastritis, peptic ulcers, gastric adenocarcinoma, and MALT lymphoma (Suerbaum and Michetti 2002). H. pylori colonization generally initiates in early childhood and persists throughout the lifetime of its host (Suerbaum and Michetti 2002). In addition to H. pylori, humans can be sporadically infected by other Helicobacter species that are also able to cause gastritis (Haesebrouck et al. 2009). These non-H. pylori gastric Helicobacter species, referred to as H. heilmannii sensu lato, appear as long, tightly coiled spiral rods that are easily differentiated from H. pylori by histological examination of gastric biopsies (Haesebrouck et al. 2011). H. heilmannii s.l. comprises several Helicobacter species, including H. bizzozeronii, H. felis, H. suis, and H. heilmannii sensu stricto, which are all known to colonize the gastric mucosa of different animal species. Because their highly fastidious nature, these zoonotic microorganisms have been cultivated from the gastric mucosa of only three human patients (Andersen et al. 1999; Kivistö et al. 2010; Wuppenhorst et al. 2012).
Several studies have revealed that the H. pylori population in an individual human host shows extensive diversity (Morelli et al. 2010; Kennemann et al. 2011). This diversity could be due to either the presence of multiple strains or the accumulation of variants within individual strains, which arise during long periods of persistence as a result of genetic drift and intra-strain recombination (Kang and Blaser 2006). It has been proposed that the balance between genomic integrity and diversification in H. pylori creates a dynamic pool of genetic variants with sufficient diversity to occupy all potential niches in the stomach (Kang and Blaser 2006). Because the hyper-variability observed in an H. pylori population from a single stomach most likely reflects particular selection pressures which drove the co-evolution of this bacterium and humans (Linz et al. 2007), what happens when a different Helicobacter species colonizes the human stomach? How is the equilibrium between genome integrity and diversification modulated during the adaptation process in a new host?
It has been suggested that a wider metabolic flexibility, compared with what has been observed for H. pylori, confers to the bacterial species belonging to H. heilmannii s.l. the ability to jump from their natural hosts to humans (Schott et al. 2011). Thus, it is possible that after initial rapid adaptation, the human gastric environment constrains the evolution of the microorganism, as described for certain opportunistic bacterial pathogens (Yang et al. 2011). However, like H. pylori, this group of micro-organisms is highly specialized for colonization of the stomach. In theory, species that are closely related from the evolutionary point of view and colonize in the same ecological niche, develop similar mechanisms to cope with the same environment. Therefore, we hypothesize that to successfully colonize the stomach, all gastric Helicobacter species need to undergo extensive genome diversification.
To verify our hypothesis, we analysed the microevolution of a human-derived H. bizzozeronii (Kivistö et al. 2010). In March 2008 (Time 0, T0), H. bizzozeronii was isolated from corpus and antral biopsies obtained from a woman with severe gastric symptoms. The symptoms were subsided after triple therapy consisting in the administration of metronidazole, tetracycline, and lansoprazole for a week. However, the patient continued to suffer from mild nausea associated with eating warm foods and in a follow-up examination in November 2008 (Time 1, T1) H. bizzozeronii was re-isolated from three antrum biposies. The antimicrobial susceptibility of the antrum-derived H. bizzozeronii population at T0 and T1 was evaluated by agar dilution method using bacterial mass obtained from each biopsy. In addition, the susceptibility of the corpus H. bizzozeronii population CIII-1ORG and of its derived clone CIII-1GEN, from which the isogenic reference genome was sequenced, was studied. All the samples were resistant to tetracycline, according to the EUCAST clinical breakpoint for H. pylori (>1 µg/mL) (EUCAST 2012). For metronidazole, H. bizzozeronii obtained from antrum biopsies at T0 as well as the corpus-derived clone CIII-1GEN were sensitive (MIC = 4 µg/mL; clinical breakpoint > 8 µg/mL), whereas H. bizzozeronii isolated after the treatment (T1) were resistant (MIC = 32 µg/mL). Interestingly, H. bizzozeronii CIII-1ORG showed an MIC of metronidazole equal to 32 µg/mL, indicating the simultaneous presence of metronidazole susceptible and resistant H. bizzozeronii variants before the treatment. The occurrence of heteroresistant phenotypes obtained from a single biopsy was an indication of the presence of certain level of genetic heterogeneity in the H. bizzozeronii population colonizing the stomach of the patient. Therefore, to predict the variability existing in the bacterial population and to analyse the microevolution of H. bizzozeronii in the human stomach, a sequence library representing genetic diversity among antrum-derived H. bizzozeronii at T0 and T1 was generated using Illumina technology and was mapped against the isogenic reference genome CIII-1GEN. As we were interested in identifying polymorphic sites (PSs) where the mutations could occur in only a fraction of H. bizzozeronii population, we applied a model using the Breseq computational pipeline (Barrick et al. 2009), which allowed the population to have an arbitrary mixture of the two most frequent bases at each position in the genome (Jerome et al. 2011). This approach allowed the identification of not only the mutations but also their frequencies (reported as the percentage of reads containing the mutation relative to the total number of reads mapping to the position), yielding an estimation of the heterogeneity displayed in the antrum-derived H. bizzozeronii population at both time points.
Heterogeneity of Antrum-Derived H. bizzozeronii Population in a Human Host before and after Treatment
At T0, the total number of PSs detected in the H. bizzozeronii population was 128 (table 1 and supplementary tables S1–S4, Supplementary Material online), of which 85.9% (110) were characterized by single nucleotide polymorphisms (SNPs) and 14.1% (18) were characterized by insertions or deletions (indels). A total of 91 SNPs affected coding sequences (CDSs); among these, 18 SNPs were synonymous substitutions, whereas 73 SNPs were nonsynonymous. We further divided the results into four groups, depending on the percentage of reads presenting the mutation at each position (table 1). For the majority of PSs (91.4%), the mutations appeared at frequencies of 50% or less, indicating that the majority of the reads belonging to the antrum-derived H. bizzozeronii population at T0 did not differ from the isogenic reference genome. In fact, applying a consensus approach (which considers only the mutations present in more than 50% of the sequence reads) the population at T0 differed from the isogenic reference genome at only 12 positions. Although these data indicate that only a minority of mutations were fixed in the antrum-derived H. bizzozeronii population at T0, a certain level of allelic variability, which probably arose as consequence of the long-term colonization of the patient (Kivistö et al. 2010), was present. Moreover, it has been shown that during an early phase of H. pylori colonization, antral and corpus environments favored the growth of different variants (Kennemann et al. 2011). The fixation of some mutations in the antrum-derived H. bizzozeronii population compared with the corpus isolate suggests that, as described for H. pylori, different regions of the stomach may promote the adaptation of specific Helicobacter sub-populations (Kang and Blaser 2006; Suerbaum and Josenhans 2007).
Table 1.
PSs Detected in the Antrum-Derived Helicobacter bizzozeronii Population at T0 and T1
| Events | Total |
<25% |
25–50% |
50–75% |
>75% |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| T0 | T1 | T0 | T1 | T0 | T1 | T0 | T1 | T0 | T1 | |
| SNPs | 110 | 203 | 13 | 52 | 86 | 4 | 4 | 3 | 7 | 144 |
| Synonymous substitutions | 18 | 35 | 6 | 6 | 8 | 3 | 1 | 2 | 3 | 24 |
| Nonsynonymous substitutions | 73 | 136 | 4 | 39 | 62 | 0 | 3 | 0 | 4 | 97 |
| Intergenic regions or noncoding DNAs | 19 | 32 | 3 | 7 | 16 | 1 | 0 | 1 | 0 | 23 |
| DIPs | 18 | 20 | 6 | 5 | 11 | 0 | 0 | 0 | 1 | 15 |
| Total | 128 | 223 | 20 | 57 | 97 | 4 | 4 | 3 | 8 | 159 |
Note.—The results were divided in four groups depending of the percentage of the reads presenting the mutation in each position.
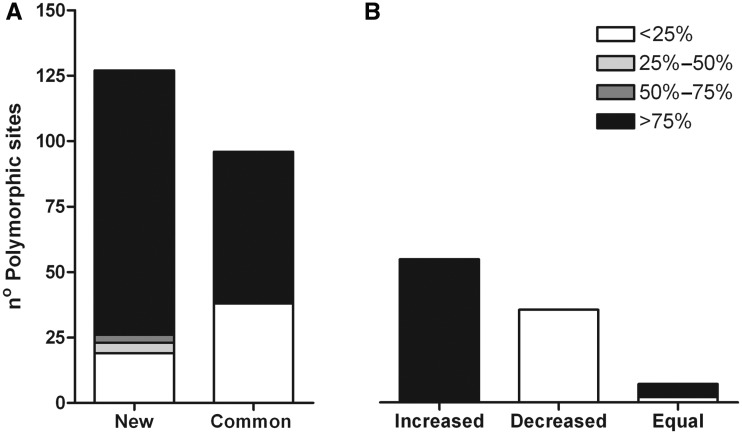
Six months after the failed treatment (T1), the number of PSs detected in the H. bizzozeronii population increased extensively. At T1, a total of 223 PSs were identified, of which 91.1% (203) were characterized by SNPs and 8.9% (20) were characterized by indels (table 1 and supplementary tables S5–S8, Supplementary Material online). In contrast to T0, the majority of PSs (71.3%) at T1 contained mutations at a frequency of 75% or more of the sequence reads. Moreover, among the 223 PSs detected at T1, 127 positions were not present at T0, 54 positions displayed the same mutation as detected at T0 but with a >1.5-fold increase in mutation frequency and 35 positions displayed a >1.5-fold decrease in mutation frequency compared with observations at T0 (fig. 1). Interestingly, new mutations at T1 occurred more readily at a high frequency (>75%, Pearson’s χ2 test, P = 0.003571). Moreover, among the common PSs, the mutations occurring with frequencies of 75% or more showed a >1.5-fold increase compared with those observed at T0, whereas those occurring with frequencies of 25% or less at T1 had a higher frequency at T0.
Fig. 1.—
PSs detected in the antrum-derived Helicobacter bizzozeronii population at T1 in comparison with those detected at T0 divided into four groups depending on the percentage of the reads presenting the mutation in each position. (A) New: number of PSs absent in T0; Common: number of PSs present in both sampling points (B) Changes in frequency of the reads presenting the mutation in T1 compared with T0: Increased: 1.5-fold increase; Decreased: 1.5-fold decrease; Equal: no differences.
To summarize, during 9 months of chronic infection and after the treatment, the H. bizzozeronii population showed an increased accumulation of sequence diversity. We observed both acquisition and loss of PSs as well as increased and decreased frequencies of existing mutations. We detected a high number of new polymorphisms in the population at T1, the majority of which appear to be under stabilizing selection. However, the computational pipeline used in this study did not allow us to distinguish whether the polymorphisms observed were a consequence of mutation or recombination events. Neither can we exclude the possibility that new polymorphisms observed in T1 were already present in the population at T0 but were not revealed in our analysis because either they were not sampled or they were present but at levels below the limit of detection (<5% of the reads).
Several studies suggest that intra/interstrain recombination plays an important role in the generation of genetic differences in H. pylori during long-term colonization (Kennemann et al. 2011; Salama et al. 2007). Nevertheless, genome-wide analysis between pairs of sequential H. pylori isolates taken at 3-year intervals from chronically infected individuals showed a high mutation rate (2.5 × 10−5 mutations per site and year) underlining the role of genetic drift in the accumulation of sequence diversity (Kennemann et al. 2011). Supposing that all new polymorphism fixed in the H. bizzozeronii population at T1 (104, applying the consensus approach, as described earlier) originated from mutation events, the number of mutations per site and year can be predicted to be approximately 7.9 × 10−5: a value comparable with the rate previously observed for H. pylori (Kennemann et al. 2011). Thus, genetic drift may explain the increased number of new PSs in T1 compared with T0. However, the increased and decreased frequencies of existing mutations at T1 are most likely a consequence of the treatment. The therapy probably induced a sudden decrease of population size by selecting resistant individuals which acted as founders for the new population at T1. Therefore, several mutations were stochastically co-selected with the resistant-associated mutations. Some of these mutations probably contributed to decrease the fitness cost of the antibiotic resistance and were fixed during the following expansion of the population.
Effect of the Mutations on the Coding Regions of the H. bizzozeronii Genome and Their Association with Resistance to Metronidazole
Nonsynonymous substitutions or indels affected a total of 80 and 133 CDSs in the antrum-derived H. bizzozeronii population at T0 and T1, respectively. We were able to functionally classify approximately 66% of the mutated CDSs (table 2). Several genes involved in motility and 13 of 20 methyl-accepting chemotaxis proteins described in H. bizzozeronii were affected by mutations in the antrum-derived isolates at both time-points, suggesting the importance of these genes in the adaptation of non-H. pylori Helicobacter species to the human stomach (Schott et al. 2011). However, because the majority of affected genes showed single amino acid substitutions, it is hard to deduce whether each mutation influences the function of the protein, and further studies are warranted to address this point.
Table 2.
Functional Classification of the Mutated CDSs in the Antrum-Derived Helicobacter bizzozeronii Population Isolated at Both Time Points
| Function Categories | Mutated CDSs at T0 | Mutated CDSs at T1 |
|---|---|---|
| Unknown function | 28 | 47 |
| Predicted function | 53 | 87 |
| Motility and chemotaxis | 15 | 19 |
| Respiratory chain | 6 | 12 |
| Cell wall biosynthesis | 5 | 11 |
| Proteolytic enzymes | 2 | 5 |
| Gene regulation | 2 | 4 |
| Protein synthesis | 3 | 4 |
| Amino acid metabolism | 2 | 3 |
| DNA replication | 3 | 3 |
| Glycan biosynthesis | 1 | 3 |
| Central metabolism | 1 | 3 |
| Purine metabolism | 2 | 3 |
| ABC transporters | 1 | 2 |
| Bacterial replication | 1 | 2 |
| Lipid metabolism | 3 | 2 |
| Mobile elements | 2 | 2 |
| Transporter | 1 | 2 |
| Virulence-associated genes | 1 | 2 |
| Acid acclimation | 0 | 1 |
| Cofactors | 0 | 1 |
| DNA repair system | 1 | 1 |
| Metals omeostasis | 1 | 1 |
| RNA degradation | 0 | 1 |
Point mutations or indels affected the frame of several CDSs in the populations from both T0 and T1 (table 3). In these cases, the effect associated with the frameshifts can be predicted by loss of protein integrity due to formation of a premature stop codon. This phenomenon is typically related to variability in nucleotide number of intragenic simple sequence repeats (SSRs) due to slipped-strand mispairing (Coenye and Vandamme 2005; Schott et al. 2011), resulting in an on–off switching of the associated gene products (phase variation). Previously, when considering a minimum of 9 nucleotides to identify SSRs, we predicted 43 loci in the genome of H. bizzozeronii to be potentially affected by phase variation (Schott et al. 2011). However, 14 of these SSRs did not show any changes in repeat length in the antrum-derived H. bizzozeronii population (supplementary table S9, Supplementary Material online). On the contrary, 10 intragenic homopolymeric tracts with repeat numbers between 6 and 8 nucleotides showed variation between the reads from populations at T0 or at T1 (table 3). The 10 genes that showed instable, short SSRs encode outer membrane proteins and proteins involved in the central metabolism, respiratory chain, cell motility, and chemotaxis. With the exception of two genes, we observed a trend toward an in-frame state in the antrum-derived H. bizzozeronii population at T1.
Table 3.
List of the 10 Genes Which Showed Unstable Short-SSRs in the Antrum-Derived Helicobacter bizzozeronii Population
| Position (bp) | Locus_tag | Description | Variation Detected | In-Frame ORF Tract Lengtha | Frequency of In-Frame ORF State (%) |
||
|---|---|---|---|---|---|---|---|
| T0 | T1 | ||||||
| Central metabolism and respiratory chain | |||||||
| 98,504 | HBZC1_00960 | Oxygen-insensitive NAD(P)H nitroreductase (rdxA) | C8–C9 | C9 | 0 | 95.8 | ↑ |
| 1,144,741 | HBZC1_12300 | l-2-hydroxygluturate oxidase (lhgO) | G8–G9 | G8 | 73 | 88 | ↑ |
| Motility and chemotaxis | |||||||
| 325,928 | HBZC1_03490 | Flagellar protein (flbB) | C8–C9 | C8 | 0 | 100 | ↑ |
| 896,590 | HBZC1_09500 | Putative methyl-accepting chemotaxis protein | G7–G6 | G7 | 68 | 93 | ↑ |
| Outer membrane proteins | |||||||
| 809,831 | HBZC1_08590 | Putative outer membrane protein | G8–G7 | G7 (merged with HBZC1_08580) | 0 | 78.9 | ↑ |
| 903,088 | HBZC1_09570 | Putative outer membrane protein | T8–T7 | T8 | 100 | 0 | ↓ |
| 939,820 | HBZC1_10100 | Putative outer membrane protein | T6–T7 | T7 (merged with HBZC1_10110) | 33 | 100 | ↑ |
| Other proteins | |||||||
| 128,638 | HBZC1_01400 | Hypothetical protein | G–G8 | G8 | 10 | 0 | ↓ |
| 549,532 | HBZC1_05780 | Hypothetical protein | T8–T7 | T7 (merged with HBZC1_05780) | 30.5 | 100 | ↑ |
| 1,661,749 | HBZC1_17960 | Replicative DNA helicase | C7–C6 | C6 | 6.5 | 12 | ↑ |
aThe “In-frame ORF tract length” was defined as the homopolymeric tract length that generated the longest potential open-reading frame.
Interestingly, the frame of the nitroreductase HBZC1_00960, showing 47% identity with H. pylori RdxA (HP0954), was affected only in the population isolated after treatment. It has been described that inactivation of the nitroreductases rdxA and frxA is associated with the acquisition of resistance to metronidazole in H. pylori (Olekhnovich et al. 2009). We observed a frame length extension of the H. bizzozeronii rdxA homolog associated with the disruption of the C-terminal cysteine-containing conserved region IACLXAL, which most likely affects protein function (Olekhnovich et al. 2009). RdxA is the only nitroreductase showing changes in H. bizzozeronii isolated after the treatment, suggesting its implication in the metronidazole resistance of this bacterial species.
Resistance mutations often confer a fitness cost to the bacterium, making it less competitive than a susceptible organism in an antibiotic-free environment (Engstrand 2009). It has been described that RdxA is the primary protein responsible for intracellular NAD(P)H-oxidase activity (Olekhnovich et al. 2009) in H. pylori. Therefore, inactivation of this gene may decrease the fitness of Helicobacter spp. Because more than 90% of the reads showed the insertion, even 6 months after the treatment, we can suppose that H. bizzozeronii developed compensatory mutations to restore fitness to levels comparable with the susceptible strains.
It is well known that the mechanisms leading to antibiotic resistance are highly pleiotropic (Davies and Davies 2010). For example, modifications in regulatory proteins have been associated with acquired antibiotic resistance in H. pylori (Tsugawa et al. 2011). Similar amino acid substitutions in two major Helicobacter global regulatory proteins, Fur and NikR, were detected in the H. bizzozeronii population at both time points. These data suggest that pleiotropic effects may drive the adaptation of H. bizzozeronii in different parts of the stomach. However, after the treatment at T1, the frequency of mutations observed in Fur and NikR changed (supplementary table S5, Supplementary Material online). In addition, at T1 amino acid substitutions in two other putative regulatory proteins, carbon starvation protein A (CstA, HBZC1_06990), and carbon starvation induced protein (CsiD, HBZC1_12310), were observed. Therefore, pleiotropy may also contribute to the compensation of fitness decrease of the bacterial population due to the acquisition of resistance mutations. Another possible example of a compensatory mutation is the increased frequency of HBZC1_12300 being in an active state at T1. HBZC1_12300 is a homolog of E. coli lhgO, which is involved in the recovery of α-ketoglutarate following reduction by other enzymes (Schott et al. 2011). The higher frequency of in-frame lhgO in the resistant population could be indicative of increased needs to supply the citric acid cycle, either to comply an increased energy requirements or to compensate for the inactivation of other pathways.
Conclusions
In conclusion, our study showed that to successfully colonize the human stomach, H. bizzozeronii undergoes extensive genome diversification. We observed that H. bizzozeronii generates remarkable genetic diversity, allowing the bacterium to evolve rapidly once a selective pressure is applied. Antimicrobial treatment drove the microevolution of the antrum-derived population of H. bizzozeronii, selecting a subpopulation which acted as founder for the new population at T1. These data indicate that the enormous genome plasticity observed in H. pylori is not a consequence of co-evolution within a single host but rather is a fundamental prerequisite for adaptation to the stomach environment, which is common among all gastric Helicobacter species.
Materials and Methods
For this study, human-derived H. bizzozeronii was isolated in March 2008 from a patient with severe gastric symptoms and in November 2008 after a failed treatment (Kivistö et al. 2010). H. bizzozeronii was cultured as previously described (Schott et al. 2011). DNA was extracted using a ZR Fungal/Bacterial RNA MiniPrep kit (Zymo Research Co, Irvine, CA). Based on previous AFLP results, which showed no differences among H. bizzozeronii isolated from antrum (Kivistö et al. 2010), we randomly selected three out of seven biopsies obtained in March 2008 (AII-2, AI-3, and AII-4) and all three obtained in November 2008 (IA-1, IA-2, and IA-3). To generate the sequence libraries, equal amounts of DNA from bacterial mass isolated from each biopsy were pooled for each time point, here referred to as Antrum T0 and Antrum T1. Sequences were obtained using Illumina sequencing technology (BaseClear BV, Leiden, The Netherlands) with 50 (Antrum T0) and 75 cycles (Antrum T1) yielding approximately 12 million reads for each pool of strains.
The Illumina reads were trimmed using Condetri perl script (Smeds and Kunstner 2011) with default settings. Only reads from pairs for which both reads passed the quality threshold were aligned to the isogenic reference genome CIII-1GEN using SSAHA2 (Ning et al. 2001) as part of the Breseq pipeline v0.17 (Barrick et al. 2009; http://barricklab.org/breseq, last accessed December 7, 2012). The isogenic reference genome was obtained from a single colony of a corpus-derived population, CIII-1ORG, isolated at T0 (Schott et al. 2011). Polymorphisms were predicted by applying the model implemented in the breseq pipeline that allowed the population to be, in each position, an arbitrary mixture of the two most frequent bases. Only polymorphisms where both bases were predicted to be present at frequencies ≥5% in the population were reported (Jerome et al. 2011). The mapping results were manually checked for misalignments. Polymorphic positions associated with misalignments or ambiguous bases were deleted. Finally, due to their intrinsic hyper variability, the polymorphic positions related to known SSRs were excluded by comparative analysis.
Supplementary Material
Supplementary tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Academy of Finland (FCoE MiFoSa, no. 118602 and 141140). M.R. was supported by an Academy of Finland Postdoctoral Fellowship (no. 132940).
Literature Cited
- Andersen LP, et al. Characterization of a culturable “Gastrospirillum hominis” (Helicobacter heilmannii) strain isolated from human gastric mucosa. J Clin Microbiol. 1999;37:1069–1076. doi: 10.1128/jcm.37.4.1069-1076.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Coenye T, Vandamme P. Characterization of mononucleotide repeats in sequenced prokaryotic genomes. DNA Res. 2005;12:221–233. doi: 10.1093/dnares/dsi009. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrand L. How will next-generation sequencing contribute to the knowledge concerning Helicobacter pylori? Clin Microbiol Infect. 2009;15:823–828. doi: 10.1111/j.1469-0691.2009.02962.x. [DOI] [PubMed] [Google Scholar]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. 2012 Clinical Breakpoints. Available from: www.eucast.org/clinical_breakpoints/ (last accessed December 7, 2012) [Google Scholar]
- Haesebrouck F, et al. Non-Helicobacter pylori helicobacter species in the human gastric mucosa: a proposal to introduce the terms H. heilmannii sensu lato and sensu stricto. Helicobacter. 2011;16:339–340. doi: 10.1111/j.1523-5378.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- Haesebrouck F, et al. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin Microbiol Rev. 2009;22:202–223. doi: 10.1128/CMR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome JP, et al. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One. 2011;6:e16399. doi: 10.1371/journal.pone.0016399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Blaser MJ. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat Rev Microbiol. 2006;4:826–836. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- Kennemann L, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci U S A. 2011;108:5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivistö R, Linros J, Rossi M, Rautelin H, Hanninen ML. Characterization of multiple Helicobacter bizzozeronii isolates from a Finnish patient with severe dyspeptic symptoms and chronic active gastritis. Helicobacter. 2010;15:58–66. doi: 10.1111/j.1523-5378.2009.00730.x. [DOI] [PubMed] [Google Scholar]
- Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli G, et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010;6:e1001036. doi: 10.1371/journal.pgen.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olekhnovich IN, Goodwin A, Hoffman PS. Characterization of the NAD(P)H oxidase and metronidazole reductase activities of the RdxA nitroreductase of Helicobacter pylori. FEBS J. 2009;276:3354–3364. doi: 10.1111/j.1742-4658.2009.07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, et al. Genetic analysis of Helicobacter pylori strain populations colonizing the stomach at different times postinfection. J Bacteriol. 2007;189:3834–3845. doi: 10.1128/JB.01696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott T, Kondadi PK, Hanninen ML, Rossi M. Comparative genomics of Helicobacter pylori and the human-derived Helicobacter bizzozeronii CIII-1 strain reveal the molecular basis of the zoonotic nature of non-pylori gastric Helicobacter infections in humans. BMC Genomics. 2011;12:534. doi: 10.1186/1471-2164-12-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Kunstner A. ConDeTri—a content dependent read trimmer for illumina data. PLoS One. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- Tsugawa, et al. Two amino acids mutation of ferric uptake regulator determines Helicobacter pylori resistance to metronidazole. Antioxid Redox Signal. 2011;14:15–23. doi: 10.1089/ars.2010.3146. [DOI] [PubMed] [Google Scholar]
- Wuppenhorst N, et al. Culture of a gastric non-Helicobacter pylori Helicobacter from the stomach of a 14-year-old girl. Helicobacter. 2012 doi: 10.1111/j.1523-5378.2012.00990.x. Advance Access published August 21, 2012, doi:10.1111/j.1523-5378.2012.00990.x. [DOI] [PubMed] [Google Scholar]
- Yang L, et al. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A. 2011;108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]