Abstract
A characteristic feature of eukaryote and prokaryote genomes is the co-occurrence of nucleotide substitution and insertion/deletion (indel) mutations. Although similar observations have also been made for chloroplast DNA, genome-wide associations have not been reported. We determined the chloroplast genome sequences for two morphotypes of taro (Colocasia esculenta; family Araceae) and compared these with four publicly available aroid chloroplast genomes. Here, we report the extent of genome-wide association between direct and inverted repeats, indels, and substitutions in these aroid chloroplast genomes. We suggest that alternative but not mutually exclusive hypotheses explain the mutational dynamics of chloroplast genome evolution.
Keywords: Araceae, indels, phylogeny, repeats, substitution mutations, taro
Introduction
Comparative studies of chloroplast genome sequences have investigated divergences spanning an enormous range of evolutionary times. These have included studies of intraspecific variation in domesticated plants (Yamane et al. 2003), studies of early land plant evolution (Kugita et al. 2003) and also the earliest events of oxygenic photosynthesis (Martin et al. 2002). This range of comparisons has been possible because of the conservative nature of chloroplast (cp) genome evolution (Palmer 1985), which involves relatively slow rates of sequence evolution in some parts of the cp genome (Sammut and Huttley 2011) and elevated rates in other parts (Magee et al. 2010; Sammut and Huttley 2011).
Molecular evolution of the cp genome sequences is typically modeled as a time reversible substitution process, in which changes at any one site are independent of changes at any other site (Liò and Goldman 1998; Drouin et al. 2008). However, observations have suggested more complex processes of evolution in which both lineage-specific and nonrandom spatial patterns of substitution occur (Liò and Goldman 1998; Lee et al. 2007; Gruenheit et al. 2008; Magee et al. 2010; Wu et al. 2011; Zhong et al. 2011). Such observations have practical significance for understanding the limitations of cp genomes in phylogenetic analyses of highly diverged lineages (Gruenheit et al. 2008), and for understanding the mutational dynamics of “hotspot” regions studied in comparisons of closely related taxa (Shaw et al. 2007; Worberg et al. 2007).
In prokaryotes and eukaryotes, analyses of DNA sequence alignments show that indels commonly occur in regions that are hotspots for nucleotide substitutions. Alternative hypotheses have been proposed to explain this co-occurrence. It has been suggested that certain genome regions are predisposed to mutational events such as substitutions and insertion/deletions—“the regional difference hypothesis” (Silva and Kondrashov 2002; Hardison et al. 2003). A second hypothesis explaining the association between indels and substitutions is that certain (large) indels act to induce substitutions through a DNA repair process that recruits error-prone DNA polymerases—“the indel-induced mutation hypothesis” (Tian et al. 2008; Zhu et al. 2009). A third and related hypothesis is that it is the presence of repeat sequences rather than indels per se, that actually promotes replication fork arrest, causing the recruitment of the error-prone DNA polymerases, and in doing so generates nucleotide substitutions (McDonald et al. 2011).
These hypotheses have not been explicitly investigated in cp genomes yet these genomes are known to contain very high densities of direct and inverted oligonucleotide repeats. Associations between repeats, indels, and substitutions have also been reported in cp genomes (McLenachan et al. 2000; Lockhart et al. 2001 and references cited therein). Cp genome repeats include simple sequence repeats (SSRs, also known as microsatellites) and other moderate to long (8–48 bp) repeats. Contraction and expansion of the SSR units, caused by slipped strand mispairing during DNA replication (Levinson and Gutman 1987), frequently produces short indels at these SSR loci (Masood et al. 2004). The moderate-to-long repeats have also been suggested to cause indels (Kawata et al. 1997) and inversions (Kim and Lee 2005; Whitlock et al. 2010). Most angiosperms also contain two large inverted repeat (IR) regions, commonly known as IRa and IRb (5–76 kb; Palmer 1991).
Here, we report the cp genome sequences of two morphotypes of taro (Colocasia esculenta; var. RR and var. GP; Matthews 1985) and examine the genome wide association of repeats (excluding IRa and IRb), indels and substitutions in the cp genomes of these taro morphotypes and four other distantly related aroids in the duckweed (Lemnoideae) subfamily.
The Colocasia esculenta cp Genome
Colocasia esculenta (L.) Schott, commonly known as taro, is an ancient root crop in subfamily Aroideae of the monocot family Araceae. This species is distributed in the tropical to subtropical and some temperate regions of the world (Bown 1988).
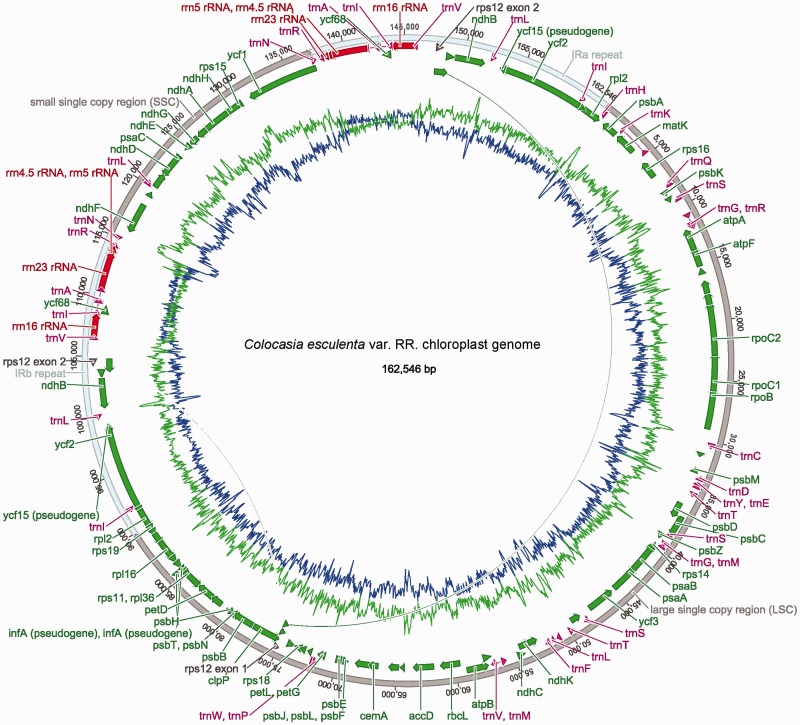
Gene arrangement and other features of the C. esculenta cp genome are shown in figure 1. Size of the cp genome was 162,546 bp (GC content: 36.1%) in var. RR, and 162,424 bp (GC content: 36.2%) in var. GP. The GC content varied from 42.4% in IRs to 34.4% in the large single copy (LSC) and only 28.4% in the small single copy (SSC) regions of the taro cp genomes. Higher GC content in the IR regions corresponded to the presence of the ribosomal DNA locus. Pair-wise sequence alignment between the taro cp genomes revealed 99.5% identical sequence, 241 substitutions, and 92 indels. The LSC region contained 141 (58.6%) substitutions and 65 (71%) indels, the SSC region contained 83 (34.4%) substitutions and 25 (27%) indels, whereas the IRa and IRb regions collectively contained only 17 (7%) substitutions and 2 (2%) indels, indicating that the IR was the most evolutionarily stable region. Prominent differences between the two taro cp genomes were found at the IRb–SSC boundary (numerous indels making up a 91 bp difference in size), and at the SSC–IRa boundary (a shift of 64 bp in the repeat boundary without causing indels). Thus, the IR boundaries at both ends of the SSC region were polymorphic at intraspecific level in taro. Polymorphism between the two taro cp genomes included 59 substitutions in 29 protein coding genes. Among these, the most polymorphic gene was ycf1 even when normalized for its size, showing 16 substitutions between the two genomes. Some protein coding genes (including atpH, psbM, and psbZ) and tRNA genes (including trnH, trnG, and trnW) in particular showed a relatively high density of substitutions and indels within 20 bp upstream of their respective coding regions. Whether this observation has functional significance needs to be further explored. A set of 30 functional tRNA genes covering all 20 amino acids required for protein synthesis was present in the taro cp genome.
Fig. 1.—
Colocasia esculenta var. RR chloroplast genome (GenBank accession: JN105690). Brown lines in the outer circle represent the LSC and SSC regions, cyan lines represent the IRs, whereas inner green lines show AT and blue lines show GC percentage throughout the cp genome.
The overall gene arrangement was similar between taro (C. esculenta) and the duckweed (Lemna minor; Mardanov et al. 2008) cp genomes. However, notable differences were as follows:
trnH gene is reported in the LSC region in duckweed, whereas the 5′-end of this gene extended into the IRa region in taro.
infA gene is completely missing in duckweed, but a pseudo-copy of this gene with internal stop codons was observed in taro.
A single functional rpl2 gene spanning the IRb–LSC boundary is reported in duckweed, whereas two functional copies of this gene were found in taro, one in each of the IR regions.
A pseudo-copy of ycf68 gene is reported in duckweed; however, a functional copy of this gene was observed in each IR region in taro.
Duckweed has ycf1 and rps15 genes within its IR regions, whereas these genes were placed within the SSC region in taro.
The infA gene is considered to be among the most mobile cp genes. Multiple independent gene transfers from cp to nuclear genomes are thought to have occurred during angiosperm evolution (Millen et al. 2001). The ycf68 gene is present in a range of plant families as a functional or a pseudo-gene, and may have functional significance even in its noncoding form (Raubeson et al. 2007). Other genes showing variation in comparison with L. minor include trnH, rpl2, ycf1, and rps15. These are located at or near the boundaries of IRs with single copy regions. These boundaries are well known to exhibit expansion and contraction in angiosperms (Whitlock et al. 2010) as well as in gymnosperms (Lin et al. 2012). A comparison of the size and percentage proportions of LSC, SSC, and IR regions in taro and other aroid cp genomes is given in table 1. Characterization of these boundaries is likely to provide useful insights into the dynamics of single copy—IR boundary shifts in Colocasia and other aroid cp genomes.
Table 1.
Comparison among Total Size (bp) and Sizes of the LSC, SSC, and Two IR Regions in Taro and Other Aroid Chloroplast Genomes
| Species | GenBank ID | Genome Size | LSC | SSC | IR |
|---|---|---|---|---|---|
| Colocasia esculenta var. GP | JN105689 | 162,424 | 89,670 (55.21) | 22,208 (13.67) | 25,273 (31.12) |
| C. esculenta var. RR | JN105690 | 162,546 | 89,817 (55.26) | 22,075 (13.58) | 25,327 (31.16) |
| Lemna minor | NC010109 | 165,955 | 89,906 (54.17) | 13,603 (8.20) | 31,223 (37.63) |
| Spirodela polyrhiza | JN160603 | 168,788 | 91,222 (54.04) | 14,056 (8.33) | 31,755 (37.63) |
| Wolffiella lingulata | JN160604 | 169,337 | 92,015 (54.34) | 13,956 (8.24) | 31,683 (37.42) |
| Wolffia australiana | JN160605 | 168,704 | 91,454 (54.21) | 13,394 (7.94) | 31,930 (37.85) |
Note.—Percentage proportions of the LSC, SSC, and IRs are given in parenthesis.
Correlations among Repeats, Indels, and Substitutions in Aroid cp Genomes
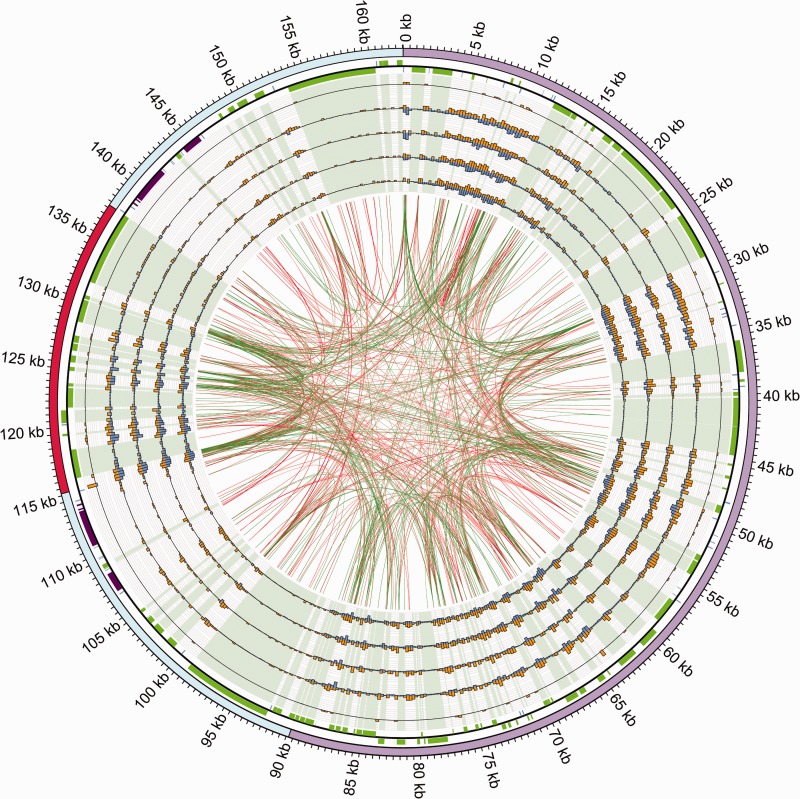
We have visualized the extent to which indel and substitution mutations are nonrandomly distributed between taro and other aroid cp genomes, using a Circos (Krzywinski et al. 2009) plot as given in figure 2. This plot shows that substitutions are very closely correlated in their distribution with moderate (15 bp) to long (48 bp) repeat sequences mainly found in noncoding regions. Correlation (r) and related values for these data are given in table 2. Correlations were highly significant in comparisons of three types of mutations, including 1) repeats and substitutions, 2) substitutions and indels, and 3) repeats and indels. In a pairwise comparison of the two closely related taro genomes, the strength of correlations was greatest for “repeats and indels” followed by “substitutions and indels” and then “repeats and substitutions.” In contrast, when pairwise comparison was made between a taro genome and a more distantly related aroid genome, the strength of correlations reversed. The strongest correlation was for “repeats and substitutions” followed by “substitutions and indels” and then “repeats and indels” (table 2). The strongest correlation value observed was for “repeats and indels” in comparison of the two taro genomes. Similar observations have previously been reported in prokaryotes and eukaryotes (Kawata et al. 1997; McDonald et al. 2011) and have led to a hypothesis that repeat sequences play a pivotal role in generation of indel and substitution mutations (McDonald et al. 2011).
Fig. 2.—
Circos plot of taro (Colocasia esculenta) var. RR showing the relationship between short repeats within the chloroplast genome and distribution of indels and substitutions in pairwise comparisons of taro var. RR cp genome with other aroid cp genomes. All data in the histogram tracks are shown in nonoverlapping 250 bp bins, with the taro var. RR genome taken as a reference for the coordinate space. Tracks from the outermost to innermost show: taro var. RR chloroplast ideogram (LSC in purple, SSC in red, and IRs in cyan); genome annotation on the positive and negative strand (genes in green; tRNAs in blue and rRNAs in purple); five circular plots showing comparisons between C. esculenta var. RR with 1) C. esculenta var. GP, 2) Lemna minor, 3) Spirodela polyrhiza, 4) Wolffia australiana, and 5) Wolffiella lingulata. For each genome comparison, the number of indels in each 250 bp bin is shown in orange (scale of 0–10), and the number of substitutions is shown in blue (scale of 0–160). Across these five plots, the light green coloring indicates the coding regions. The figure center shows the results of Reputer mapping using the taro var. RR chloroplast genome. Two ends of a red line mark the two locations of the forward (direct) repeats, whereas those of a green line mark the two locations of the reverse (inverted) repeats on the genome. In this part of the figure, the large IRs are not plotted, as they would obscure a large part of the figure. Number of repeats shown in the diagram is 667, with a size range from 15 to 48 bp (average repeat size: 16 bp).
Table 2.
Comparisons among the Pairwise Alignments (Colocasia esculenta var. RR taken as a Reference) to Calculate the Correlations between 1) Repeats and Substitutions, 2) Insertion-Deletions (indels) and Substitutions, and 3) Repeats and Indels
| Comparison | C. esculenta var. GP | Wolffiella lingulata | Wolffia australiana | Lemna minor | Spirodela polyrhiza |
|---|---|---|---|---|---|
| Repeats and substitutions | |||||
| Correlation between repeats and substitutions (r) | 0.245 | 0.391 | 0.416 | 0.424 | 0.491 |
| Significance of correlation (t) | 6.44*** | 10.81*** | 11.657*** | 11.92*** | 14.37*** |
| Coefficient of determination (r2) | 0.060 | 0.152 | 0.173 | 0.180 | 0.241 |
| Insertion–deletions (indels) and substitutions | |||||
| Correlation between indels and substitutions (r) | 0.391 | 0.220 | 0.245 | 0.323 | 0.387 |
| Significance of correlation (t) | 10.82*** | 5.75*** | 6.43*** | 8.71*** | 10.69*** |
| Coefficient of determination (r2) | 0.153 | 0.048 | 0.060 | 0.105 | 0.150 |
| Repeats and indels | |||||
| Correlation between repeats and indels (r) | 0.640 | 0.168 | 0.178 | 0.224 | 0.212 |
| Significance of correlation (t) | 21.20*** | 4.33*** | 4.59*** | 5.87*** | 5.51*** |
| Coefficient of determination (r2) | 0.409 | 0.028 | 0.032 | 0.050 | 0.045 |
Note.—The alignments compared closely related (var. RR to var. GP) and distantly related (var. RR to W. lingulata, W. australiana, L. minor, and S. polyrhiza) aroid chloroplast genomes. The alignments were partitioned into 651 nonoverlapping bins of 250 bp size each to calculate these correlations.
***All correlations were highly significant at 0.001α and 649 degree of freedom.
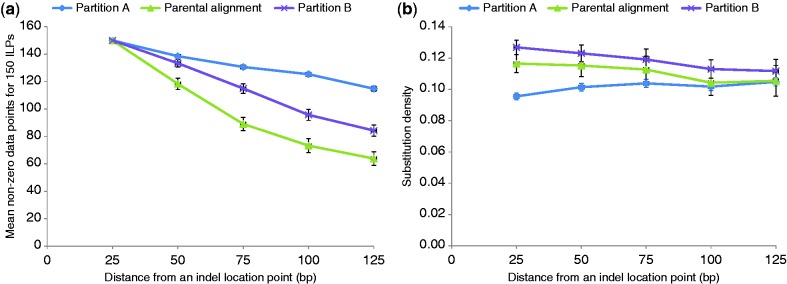
Since Tian et al. (2008) proposed that moderate-to-large–sized indels induce substitutions in their surrounding sequences, we also investigated this relationship in a multiple sequence alignment (parental alignment) of all six aroid cp genomes. From the this parental alignment, we extracted data partitions containing distinct indel location points (ILPs) to make mutually exclusive partitions with respect to locations of the ILPs. Partition A contained ILPs associated with SSR indels in both coding and noncoding regions. Partition B contained ILPs associated with large (oligonucleotide long, non-SSR) indels in both coding and noncoding regions. Partition C contained ILPs in noncoding regions, associated with both SSR indels and large indels. Partition D contained ILPs in coding regions, associated with both SSR and large indels. The density of substitutions in all partitions was highly dependent upon inverse of distance from the ILPs (r2 ranged from 0.85 to 0.97 for all bin sizes; supplementary fig. S1, Supplementary Material online). Higher substitution density in bins closer to the ILPs was a general trend in all five comparisons above, including the partition in which coding regions were removed (partition C); however, in this case, distance from the ILPs was relatively shorter than in the other four comparisons. The indel-induced mutation hypothesis was further explored in a comparison including the parental alignment and partitions A and B, as shown in figure 3. From this comparison, it is evident that the partition B (containing ILPs associated with large indels) displayed a higher density of substitutions closer to ILPs, and the density of substitutions decreased with an increase in distance from the ILPs. In contrast, the partition A (containing ILPs associated with SSRs) exhibited a low density of substitutions close to ILPs, and the density of substitutions showed a net increase with increase in distance from the ILPs. These observations are consistent with the indel-induced mutation hypothesis suggested for diploid eukaryote (Tian et al. 2008) as well as bacterial genomes (Zhu et al. 2009).
Fig. 3.—
Results showing (a) the number of mean non-zero data points used to calculate the substitution density and (b) the values of substitution density in 125 bp sequence adjacent to the ILPs for the parental alignment as well as two of its partitions, A and B (Partition A contains ILPs associated with SSR indels in coding and noncoding regions, while partition B contains ILPs associated with large indels in coding and noncoding regions). Lower than 150 values for non-zero data points at >25 bp distance in (a) represents that taking an average for 1,000 random iterations, lesser than 150 ILPs are 125 bp apart from each other in all three types of comparisons.
It is well known that certain regions of the chloroplast genome show different rates of mutations (Lee et al. 2007; Gruenheit et al. 2008; CBOL Plant Working Group 2009; Zhong et al. 2011). These are observations consistent with a regional difference hypothesis (Silva and Kondrashov 2002; Hardison et al. 2003) and the suggestion that purifying selection operates at both coding and noncoding regions (Petersen et al. 2011). However, these explanations are alone insufficient to explain substitution and indel patterns of the chloroplast genome. The extent of genome wide correlations reported here for indels, repeats, and substitution provides further support for the hypothesis by McDonald et al. (2011), which emphasizes the evolutionary importance of the repeats in causing mutations. In addition, our observations on substitution densities also provide support for an indel-induced mutation hypothesis (Tian et al. 2008; Zhu et al. 2009) and further our understanding for the sometimes poor fit between time reversible substitution models and chloroplast sequence data. Perhaps, most interestingly, the relationship between repeats, substitutions, and indels implies that, if the distribution of repeat sequences in a chloroplast genome is determined, there is a possibility to predict the mutational hotspot regions and other sequences that are most appropriate for population genetic, phylogeographic, and phylogenetic analyses.
Materials and Methods
Taro plants (C. esculenta var RR; voucher number MPN:46548, and var GP; voucher number MPN:46549 in the Dame Ella Campbell Herbarium, Massey University, New Zealand) were obtained from the University of Auckland campus. Chloroplasts were enriched following procedure given in Atherton et al. (2010). DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, USA) and quantified using a Qubit Fluorometer (Invitrogen) and Quant-iT-ds DNA HS Assay kit (Invitrogen). Illumina sequence reads were generated using the GAIIx platform at the Massey Genome Service, Massey University, New Zealand. IIlumina sequencing produced 33 million reads of 75 base long (16.5 million paired-end reads) for var. RR, and 26.4 million reads of 75 base long (13.2 million paired-end reads) for var. GP. The reads were mapped to the duckweed cp genome (L. minor; Mardanov et al. 2008) using BWA mapping tool (Li and Durbin 2009). Mapping results were visualized using Tablet (Milne et al. 2010). The reads from var. RR were de novo assembled into contiguous sequences (“contigs”) of variable lengths using Velvet (v.0.7.60; Zerbino and Birney 2008), as described elsewhere (Collins et al. 2008). These contigs were BLAST-searched (Altschul et al. 1997) to determine homology to the duckweed cp genome. The contigs of cp origin were assembled in Geneious Pro (Drummond et al. 2009) to deduce the cp genome of the taro var. RR morphotype. The two IRs were distinguished by visual inspection of the boundaries between the repeat and single copy regions. Genome annotation was carried out using Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. 2004) and also by direct comparison with the duckweed cp genome. Contigs were generated similarly for the var. GP morphotype. The completed var. RR cp genome was then used as our reference genome to help assemble the var. GP cp genome. To verify integrity of the de novo assembly process, the original 75 base long reads from both taro samples were mapped back to their respective, assembled cp genomes. Summary statistics for the BWA mapping of 75 base long reads to the L. minor cp genome, as well as to their respective assembled var. RR and var. GP genomes are given in table 3.
Table 3.
Summary Statistics for BWA Mapping of 75 Base, Paired-End Reads Obtained from the Colocasia esculenta var. RR and var. GP Morphotypes to the Lemna minor Chloroplast Genome and to Their Assembled Chloroplast Genomes
| Parameter |
L. minor |
C. esculenta var. RR |
C. esculenta var. GP |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR1 | RR2 | RPE | GR1 | GR2 | GPE | RR1 | RR2 | RPE | GR1 | GR2 | GPE | |
| Genome coverage (%) | 68.5 | 68.1 | 85 | 68.5 | 68.7 | 85.6 | 99.99 | 100 | 100 | 100 | 100 | 100 |
| Average coverage depth | 129 | 128 | 337 | 319 | 317 | 825 | 296 | 294 | 593 | 665 | 659 | 1,338 |
| Maximum coverage depth | 674 | 623 | 1,531 | 1,984 | 1,853 | 5,194 | 641 | 656 | 1,020 | 1,940 | 2,021 | 3,304 |
Note.—The acronyms RR1, RR2, and RPE represent mapping with the read 1, read 2, and paired-end (reads 1 and 2 taken together) reads obtained from the var. RR morphotype. Similarly, GR1, GR2, and GPE represent mapping with the read 1, read 2, and paired-end reads obtained from the var. GP morphotype.
The var. RR cp genome was pairwise aligned to the var. GP cp genome, as well as to four aroid cp genomes from the Lemnoideae subfamily, using DIALIGN alignment (Morgenstern 2004). The four aroid cp genomes included L. minor (GenBank ID: NC010109; Mardanov et al. 2008), Spirodela polyrhiza (GenBank ID: JN160603), Wolffiella lingulata (GenBank ID: JN160604), and Wolffia australiana (GenBank ID: JN160605; Wang and Messing 2011). Selecting C. esculenta var. RR cp genome as a reference for the coordinate positions, indels, and substitutions were counted in pairwise comparisons in nonoverlapping bins of 250 bp through the entire length of the aligned cp genomes (partitioning each of the five alignments into 651 bins). For the substitution count, indels in the var. RR cp genome were deleted from the alignments to preserve the coordinate positions. Similar patterns of indel and substitution counts were obtained using a MAFFT alignment (Katoh et al. 2005; results not shown). A total of 5,000 forward (direct) and reverse (inverted) repeats with a minimum size of 14 bp, a maximum size of 48 bp, and a maximum of three nucleotide mismatch between the two repeat copies in the taro var. RR cp genome were calculated using Reputer (Kurtz et al. 2001). Of these 5,000 repeats, 667 locations of the forward and reverse in var. RR (minimum size: 15 bp; zero mismatch between the two copies), as well as polymorphic sites (indels and substitutions) in all five pairwise comparisons with respect to the var. RR cp genome were plotted as a circular diagram using Circos (Krzywinski et al. 2009). Correlations (r) were calculated between numbers of 1) repeats and substitutions, 2) substitutions and indels, and 3) repeats and indels. This was done for comparisons of closely related (two taro genomes) and distantly related (taro with other Lemnoideae) cp genomes. The correlation values (r) were used to determine the significance of correlation (t) and the coefficient of determination (r2), according to Lowry (2012).
To further investigate the relationships between substitutions and indels, a multiple sequence alignment of all six aroid cp genomes was generated using DIALIGN alignment (Morgenstern 2004). Hyper variable regions causing problems in the alignment were removed to ensure conservative estimates. This 122-kb long parental alignment contained 457 ILPs. This parental alignment was used to generate mutually exclusive alignment combinations with respect to locations of the ILPs, to include ILPs associated with coding and noncoding regions and SSR indels (171 ILPs; partition A) and coding and noncoding regions and large indels (286 ILPs; partition B). The parental alignment was also used to generate two further mutually exclusive alignment combinations to include ILPs associated with SSR indels and large indels in noncoding regions (376 ILPs; partition C) and SSR indels and large indels in coding regions (81 ILPs; partition D). Using a Perl script, we counted the number and positions of substitutions with respect to the ILPs, and plotted the substitution density as a function of distance from the ILPs in nonoverlapping bins of 50, 100, 150, 200, and 250 bp each for the parental alignment as well as partitions A, B, and D; and 10, 20, 30, 40, and 50 bp for the partition C. The effect of large indels in causing substitutions was further explored by comparing first three alignment combinations (parental alignment along with partitions A and B) and plotting the substitution density as a function of distance from the ILPs in 125 bp sequence adjacent to the ILPs. For this purpose, a jacknifing approach was used to randomly select 150 ILPs from each of these three partitions with 1,000 random iterations to count substitutions within the 125 bp distance, divided into five nonoverlapping bins of 25 bp in size. Plots showing the relationship between substitutions and ILPs were generated using MS Excel 2010 worksheets.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge the financial assistance of the New Zealand Royal Society (The New Zealand Marsden Fund and James Cook Fellowship scheme) and the Higher Education Commission, Government of Pakistan.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton RA, et al. Whole genome sequencing of enriched chloroplast DNA using the Illumina GAII platform. Plant Methods. 2010;6:22. doi: 10.1186/1746-4811-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown D. London: Century Hutchinson; 1988. Aroids: plants of the arum family. [Google Scholar]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LJ, Biggs PJ, Voelckel C. An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Inform. 2008;21:3–14. [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast, and nuclear genomes of seed plants. Mol Phylogenet Evol. 2008;49:827–831. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, et al. Geneious. 2009 Available from: http://www.geneious.com/ (last accessed December 10, 2012) [Google Scholar]
- Gruenheit N, Lockhart PJ, Steel M, Martin W. Difficulties in testing for covarion-like properties of sequences under the confounding influence of changing proportions of variable sites. Mol Biol Evol. 2008;25:1512–1520. doi: 10.1093/molbev/msn098. [DOI] [PubMed] [Google Scholar]
- Hardison RC, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata M, Harada T, Shimamoto Y, Oono K, Takaiwa F. Short inverted repeats function as hotspots of intermolecular recombination giving rise to oligomers of deleted plastid DNAs (ptDNAs) Curr Genet. 1997;31:179–184. doi: 10.1007/s002940050193. [DOI] [PubMed] [Google Scholar]
- Kim K-J, Lee H-L. Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cells. 2005;19:104–113. [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugita M, et al. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res. 2003;31:716–721. doi: 10.1093/nar/gkg155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-L, Jansen RK, Chumley TW, Kim K-J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman G. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Wu CS, Huang YY, Chaw SM. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 2012;4:374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liò P, Goldman N. Models of molecular evolution and phylogeny. Genome Res. 1998;8:1233–1244. doi: 10.1101/gr.8.12.1233. [DOI] [PubMed] [Google Scholar]
- Lockhart PJ, et al. Phylogeny, radiation, and transoceanic dispersal of New Zealand alpine buttercups: molecular evidence under split decomposition. Ann MO Bot Gard. 2001;88:458–477. [Google Scholar]
- Lowry R. Concepts & applications of inferential statistics. 2012 Available from: http://vassarstats.net/textbook/index.html (last accessed August 14, 2012) [Google Scholar]
- Magee AM, et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010;20:1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov AV, et al. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 2008;66:555–564. doi: 10.1007/s00239-008-9091-7. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masood MS, et al. The complete nucleotide sequence of wild rice (Oryza nivara) chloroplast genome: first genome wide comparative sequence analysis of wild and cultivated rice. Gene. 2004;340:133–139. doi: 10.1016/j.gene.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Matthews PJ. Nga taro o Aotearoa. J Polynesian Soc. 1985;94:253–272. [Google Scholar]
- McDonald MJ, Wang W-C, Huang H-D, Leu J-Y. Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 2011;9:e1000622. doi: 10.1371/journal.pbio.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLenachan PA, et al. Markers derived from amplified fragment length polymorphism gels for plant ecology and evolution studies. Mol Ecol. 2000;9:1899–1903. doi: 10.1046/j.1365-294x.2000.01075.x. [DOI] [PubMed] [Google Scholar]
- Millen RS, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. Chloroplast DNA and molecular phylogeny. Bioessays. 1985;2:263–267. [Google Scholar]
- Palmer JD. Plastid chromosomes: structure and evolution. In: Vasil IK, Bogorad L, editors. Cell culture and somatic cell genetics in plants. Vol. 7A: The molecular biology of plastids. San Diego (CA): Academic Press; 1991. pp. 5–53. [Google Scholar]
- Petersen K, Schöttler MA, Karcher D, Thiele W, Bock R. Elimination of a group II intron from a plastid gene causes a mutant phenotype. Nucleic Acids Res. 2011;39:5181–5192. doi: 10.1093/nar/gkr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raubeson LA, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammut R, Huttley G. Regional context in the alignment of biological sequence pairs. J Mol Evol. 2011;72:147–159. doi: 10.1007/s00239-010-9409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Silva JC, Kondrashov AS. Patterns in spontaneous mutation revealed by human-baboon sequence comparison. Trends Genet. 2002;18:544–547. doi: 10.1016/s0168-9525(02)02757-9. [DOI] [PubMed] [Google Scholar]
- Tian D, et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- Wang W, Messing J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One. 2011;6:e24670. doi: 10.1371/journal.pone.0024670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock BA, Hale AM, Groff PA. Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLoS One. 2010;5:e11533. doi: 10.1371/journal.pone.0011533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worberg A, et al. Phylogeny of basal eudicots: insights from non-coding and rapidly evolving DNA. Organisms Divers Evol. 2007;7:55–77. [Google Scholar]
- Wu C-S, Wang Y-N, Hsu C-Y, Lin C-P, Chaw S-M. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and Cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 2011;3:1284–1295. doi: 10.1093/gbe/evr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yamane K, Yasui Y, Ohnishi O. Intraspecific cpDNA variations of diploid and tetraploid perennial buckwheat, Fagopyrum cymosum (Polygonaceae) Am J Bot. 2003;90:339–346. doi: 10.3732/ajb.90.3.339. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, et al. Systematic error in seed plant phylogenomics. Genome Biol Evol. 2011;3:1340–1348. doi: 10.1093/gbe/evr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang Q, Tang P, Araki H, Tian D. Genomewide association between insertions/deletions and the nucleotide diversity in bacteria. Mol Biol Evol. 2009;26:2353–2361. doi: 10.1093/molbev/msp144. [DOI] [PubMed] [Google Scholar]