Abstract
Staphylococcus aureus is a frequent cause of serious infections and also a human commensal. The emergence of community-associated methicillin-resistant S. aureus led to a dramatic increase in skin and soft tissue infections worldwide. This epidemic has been driven by a limited number of clones, such as USA300 in the United States. To better understand the extent of USA300 evolution and diversification within communities, we performed comparative whole-genome sequencing of three clinical and five colonizing USA300 isolates collected longitudinally from three unrelated households over a 15-month period. Phylogenetic analysis that incorporated additional geographically diverse USA300 isolates indicated that all but one likely arose from a common recent ancestor. Although limited genetic adaptation occurred over the study period, the greatest genetic heterogeneity occurred between isolates from different households and within one heavily colonized household. This diversity allowed for a more accurate tracking of interpersonal USA300 transmission. Sequencing of persisting USA300 isolates revealed mutations in genes involved in major aspects of S. aureus function: adhesion, cell wall biosynthesis, virulence, and carbohydrate metabolism. Genetic variations also included accumulation of multiple polymorphisms within select genes of two multigene operons, suggestive of small genome rearrangements rather than de novo single point mutations. Such rearrangements have been underappreciated in S. aureus and may represent novel means of strain variation. Subtle genetic changes may contribute to USA300 fitness and persistence. Elucidation of small genome rearrangements reveals a potentially new and intriguing mechanism of directed S. aureus genome diversification in environmental niches and during pathogen–host interactions.
Keywords: evolution, genome rearrangement, repeat deletions
Introduction
Community-associated Staphylococcus aureus methicillin-resistant (CA-MRSA) infections first emerged in the United States over a decade ago and have since become epidemic (Adcock et al. 1998; Herold et al. 1998; Naimi et al. 2001). Most of these infections affect skin and soft tissues, but 5–10% are invasive with potentially fatal outcomes (Kaplan et al. 2005). This CA-MRSA epidemic has been driven by a limited number of clones worldwide (DeLeo et al. 2010), such as pulsed-field gel electrophoresis (PFGE)-type USA300 in the United States. Since the first isolation of USA300 in California in 2000, the pathogen rapidly spread across the United States and by 2004 accounted for the majority of skin and soft tissue infections presenting to Emergency Departments in urban centers (Moran et al. 2006). USA300 remains the leading cause of community-associated bacterial infections in the United States (Talan et al. 2011).
The sequential acquisition of mobile genetic elements (MGEs), such as Panton–Valentine leukocidin (PVL) and arginine-catabolic mobile element (ACME), has been considered as an essential step in the evolution of USA300 (Diep and Otto 2008). A previous genome-wide comparison of 10 clinically and geographically diverse USA300 clinical isolates suggested that the majority of these isolates were closely related and had undergone recent clonal expansion and diversification (Kennedy et al. 2008). This study also demonstrated that a discrete number of single-nucleotide polymorphisms (SNPs) in the core genome of USA300 may equally alter the virulence of a given isolate in a murine sepsis model (Kennedy et al. 2008). Although some MGEs in USA300 carry classic determinants of virulence, such as toxins or immune evasion molecules (Diep et al. 2006), the molecular factors underlying the fitness and adaptability of these strains remain incompletely determined.
Many studies of the evolution of the S. aureus genome relate to anti-infective chemotherapy, where most of the accumulated mutations were likely the result of strong selective pressure of the therapeutic agents. These studies specifically documented the evolution of resistance to vancomycin (Mwangi et al. 2007; Howden et al. 2008, 2011) or linezolid during chemotherapy (Gao et al. 2010). Moreover, whole-genome sequencing of a global collection of sequence type 239 (ST239) S. aureus isolates indicated that over a quarter of the detected homoplasies were directly related to evolution of resistance to antibiotics (Harris et al. 2010). This study also estimated that the core genome divergence was approximately 1 SNP per ∼6 weeks. Sequence analysis of three longitudinally sampled S. aureus ST30 isolates collected from a single patient with cystic fibrosis over a 26-month period documented the accumulation of 23 SNPs and 15 insertions/deletions (InDels), mainly in genes involved in virulence, global regulation, metabolism, and antibiotic resistance (McAdam et al. 2011).
Although progress has been made, our understanding of the evolution and dissemination of CA-MRSA as an endemic pathogen within communities is limited. Epidemiological data suggest that some of the unique characteristics of CA-MRSA include the ability to persist, cause recurrent infections, and transmit (or spread) among household members (Wagenvoort et al. 1997; Cook et al. 2007). The ability to adapt during persistent colonization may be a potential determinant of increased fitness and transmissibility of bacterial pathogens (Zdziarski et al. 2010). Here, we performed comparative whole-genome sequence analysis of eight USA300 isolates collected from three households over a 15-month period to investigate evolution of the epidemic USA300 genome in vivo.
Materials and Methods
Ethics
We obtained written informed consent from each individual before conducting an interview or obtaining samples. Parental consent was required for the participation of children younger than 18 years, and pediatric assent was obtained from those capable of providing it. Index participants were compensated $10 for their time. The Institutional Review Board of Columbia University Medical Center, New York, approved this study.
Sample Selection and Molecular Genotyping
Between 2004 and 2007, a population-based longitudinal study, funded by the Centers for Disease Control and Prevention, was performed to determine the spread of CA-MRSA in households with an MRSA index case in Northern Manhattan. This is home to the largest Dominican community in the country (approximately 222,000). Potential study subjects were identified by a positive clinical culture with either a CA-MRSA or a hospital-associated (HA)-MRSA isolate. Patients were not eligible to participate if they were living in a nursing home or shelter or if they already had a prior positive culture for MRSA. A total of 114 eligible index cases were enrolled. The index cases and their household members were interviewed and samples (nares or open wounds) collected upon recruitment and at 4-month intervals for up to 16 months. The clinical isolate was also available for further genotyping in 67% (76) of cases. A total of 403 positive S. aureus samples were identified over the study period. USA300 was the predominant strain and identified at least once in 38/114 (33.3%) households. Of these, USA300 colonized or infected more than two members in seven (18.4%) households and was detectable on two or more occasions in 11/38 households. Three of these households were remarkable in that they had persistent colonization of different family members with USA300 for more than 1 year.
Molecular strain typing was carried out using PFGE, multilocus sequence typing (MLST), and S. aureus protein A (spa) sequencing (spa-typing) as described (Enright et al. 2000; Harmsen et al. 2003). In brief, SmaI-digested samples were subjected to PFGE (Chung et al. 2000; Bhat et al. 2009). The resulting band patterns were analyzed by Bionumerics software (version 4.0, Applied Maths, Ghent, Belgium) to determine relatedness between strains (Tenover et al. 1995). Profiles with >80% similarity were considered closely related. MLST was carried out as described (Enright et al. 2000), and ST were assigned using saureus.mlst.net, accessed November 30, 2012. Spa-typing was carried out using polymerase chain reaction sequencing, and spa-types were automatically assigned using Ridom Staph-Type software (version 1.5) and compared with http://spaserver2.ridom.de, accessed November 30, 2012.
Further screening for the presence of PVL (Kaneko et al. 1997), the ACME (Diep et al. 2008), and for the type of staphylococcal chromosomal cassette (SCC) mec was determined by PCR as described (Milheirico et al. 2007).
Growth Assays and Antimicrobial Susceptibility Testing
Bacteria were grown in trypticase soy broth (TSB, Becton Dickenson) overnight and incubated after a 1:100 dilution in microtiter plates at 37°C with shaking. Optical density (OD) measurements were automatically recorded every 2 min in a Victor3 microplate reader (Perkin Elmer) and used for calculation of the doubling time for each strain. Drug susceptibilities of all S. aureus isolates were determined by Kirby Bauer standard disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines (Rosenthal et al. 2010). The following antibiotics were tested: penicillin, cefoxitin, oxacillin, erythromycin, tetracycline, levofloxacin, gentamicin, clindamycin, trimethoprim-sulfamethoxazole, and rifampin. The mupirocin E-test was performed after inoculation of a single colony in TSB followed by incubation at 37°C. At a cell density equivalent to a 0.5 McFarland standard, bacteria were streaked evenly onto Muller Hinton agar (MHA)/5% sheep blood plates (BD). Plates were incubated at 37°C for 24 h.
Whole-Genome Comparative Sequencing and Phylogenetic Analysis
Total bacterial DNA was isolated from overnight cultures using a DNAeasy Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions, except lysostaphin was added to samples, and they were incubated for at least 3 h at 37°C. Genome sequencing was performed using a SOLiD 3 System (Applied Biosystems). Mate-pair libraries were prepared according to the manufacturer’s recommendations. Raw colorspace reads generated by the SOLiD sequencer were mapped to the USA300 FPR3757 strain using Corona-Lite (Applied Biosystems) and ZOOM (Bioinformatics Solutions, Inc.). This approach gave on average 112× sequence coverage and identified all SNPs, small insertions/deletions (InDels, insertions up to 4 bp and deletions up to 11 bp), and large deletions using the USA300 FPR3757 strain as the reference strain. InDels and MGEs were verified by PCR and/or PCR-directed capillary DNA sequencing using oligonucleotide primers (Sigma Genosys, The Woodlands, TX) designed with Vector NTI software (Invitrogen Corp., Carlsbad, CA). Purified PCR products were sequenced using a 3730XL DNA analyzer at the Genomics Core Facility, Research Technologies Section, Rocky Mountain Laboratories as described previously (Kennedy et al. 2008). Sequences were initially aligned to strain FPR3757 (Diep et al. 2006), and SNP and non-SNP insertions, deletions and transpositions in genomic and plasmid DNA were obtained. Meta data were deposited in the NCBI short read archive (http://www.ncbi.nlm.nih.gov/sra, accessed November 30, 2012) under the accession number SRA059180.
Phylogenetic analyses were performed using 777 concatenated SNP nucleotides (or 806 if HOU-MR is included) in the core genome of all eight isolates compared with the FPR3757 reference strain. Alternatively, phylogenetic analyses that included strain COL and previously published USA300 isolates (Kennedy et al. 2008) were performed using 1,577 concatenated SNP nucleotides. Phylogenetic trees were generated by the maximum likelihood method using 1,000 bootstraps using the PhyML plugin (Guindon and Gascuel 2003) in the Geneious software package (Biomatters Ltd., New Zealand).
Analysis of Small Insertion and Deletions
For analysis of small insertion and deletions, we determined a quality cutoff for InDels. We graphed the total coverage for every InDel called for each individual strain that showed a biasymptotic line and then identified the “break” in the line for each isolate. From this analysis, we compiled a list of the high coverage calls for InDels. For most of the isolates, this was at least a ∼10 fold coverage, but in some instances, this was lower (6-fold coverage in one case).
Results
Persistence of USA300 After Infection in Households in Northern Manhattan
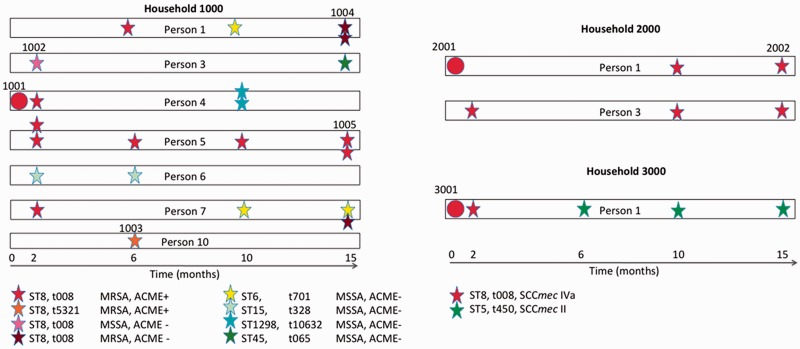
To examine the evolution of USA300 strains within community households, we selected isolates from the two households (HH1000 and HH2000) containing members that had persistent colonization with USA300 for more than 1 year (fig. 1). In addition, we selected isolates from a household (HH3000) with nonpersistent colonization (fig. 1 and table 1). Notably, these isolates were selected after an initial USA300 skin infection within each household (index case) (table 1).
Fig. 1.—
Timeline of Staphylococcus aureus colonization in three community households and sample selection for whole-genome sequence comparisons. Shown are two households with persistent USA300 colonization (HH1000, left panel, and HH2000, top right panel) and one household (HH3000, lower right panel) without persistent USA300 colonization. Note that for HH1000, only the 7 colonized members of the 12 total members sharing the household are indicated. Circles indicate index infection and stars symbolize S. aureus colonization of household members at 2, 6, 10, or 15 months, respectively. Multiple stars indicate colonization of more than one body site. Color of stars specifies clonal type (red star, USA300 t008; orange star, USA300 spa-variant; pink, USA300 MSSA; and dark red, USA300 ACME negative). Other colors (green, blue, and yellow) symbolize non-USA300 isolates. Isolate numbers are shown for sequenced samples only.
Table 1.
Phenotypic and Genotypic Characterization of Samplesa
| Isolate | Source | Doubling Time (min) | Hemolysins | spa | PFGE rel. to USA300-0114 (%) | SCCmec | ACME | PVL | Ox | LVX | T/S | Ery |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HH1000 | ||||||||||||
| 1001 | Wound | 58:30 | α, δ | t008 | 96 | IVa | + | + | R | R | S | R |
| 1002 | Colonizer | 56:55 | α, δ | t008 | 85 | MSSA | − | − | S | R | R | S |
| 1003 | Colonizer | 58:49 | α, δ | t5321 | 96 | IVa | + | + | R | R | S | R |
| 1004 | Colonizer | 59:06 | α, δ | t008 | 82 | IVa | − | + | R | S | S | R |
| 1005 | Colonizer | 62:22 | α, δ | t008 | 96 | IVa | + | + | R | R | S | R |
| HH2000 | ||||||||||||
| 2001 | Abscess | 61:50 | α, δ | t008 | 100 | IVa | + | + | R | R | S | R |
| 2002 | Colonizer | 66:09 | α, δ | t008 | 100 | IVa | + | + | R | R | S | R |
| HH3000 | ||||||||||||
| 3001 | Abscess | 71:51 | α++, δ | t008 | 100 | IVa | + | + | R | S | S | R |
Note.—Ox, oxacillin; LVX, levofloxacin; T/S, trimethoprim/sulfamethoxazole; Ery, erythromycin.
aAll isolates were resistant to penicillin but sensitive to vancomycin, linezolid, tetracycline, and rifampin. All isolates were ST8 by MLST.
The first household (HH1000) was shared by 12 members of an extended family, of which four were colonized with S. aureus at the time of enrollment (fig. 1). Over the course of the study, 7/12 individuals were colonized with S. aureus, and USA300 was the predominant pulsed-field type (fig. 1). Although most USA300 isolates from this household were MRSA, spa-type t008, and ACME and PVL positive, there was also an methicillin susceptible S. aureus (MSSA) isolate (isolate 1002), a spa-type variant (isolate 1003), and an ACME-negative isolate (isolate 1004; table 1). Members of the second household (HH2000) were also colonized persistently with USA300, and all isolates were indistinguishable by conventional molecular genotyping (table 1). In the household containing members that were not persistently colonized by S. aureus (HH3000), the USA300 isolate from the index case (3001) was replaced by an unrelated MRSA strain (fig. 1). Therefore, this sample selection represents a cross-sectional comparison of S. aureus isolates from index infections (1001, 2001, and 3001) as well as longitudinal pairs (2001 and 2002; 1001 and 1002, 1003, 1004, or 1005, respectively; fig. 1).
These eight isolates differed minimally in their drug resistance profiles, hemolysis pattern, and growth characteristics, although 3001 had an increased doubling time compared with the other isolates (table 1).
SNPs, InDels, and Evidence of Small-Genome Rearrangements among Closely Related USA300 Isolates
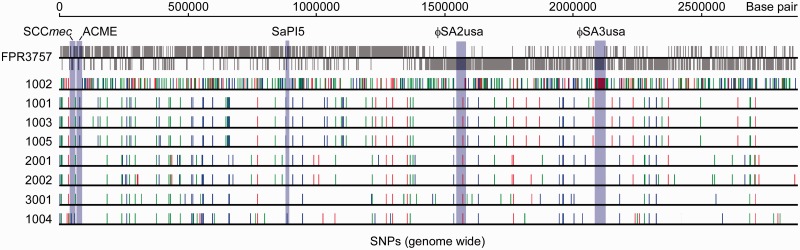
Using comparative whole-genome sequencing, a total of 1,365 SNPs were detected in the core genome and MGEs of the eight sequenced strains compared with the genome of the USA300 reference strain (FPR3757) (Diep et al. 2006). Of these SNPs, 293 were intergenic, 429 were nonsynonymous, and 643 were synonymous. There were a combined 747 SNPs among the eight New York USA300 isolates and 777 SNPs compared with the reference genome FPR3757. Of these, 209 (26.9%) SNPs were intergenic, 319 (41.1%) were nonsynonymous, and 249 (32.0%) were synonymous. The majority of mutations (n = 561) were only detected in the MSSA isolate, 1002 (fig. 2), 30 SNPs were found in all sequenced isolates and a combined 186 SNPs were present in the seven MRSA isolates relative to the reference strain (range was 63–113 SNPs for individual isolates) (fig. 3). These SNPs clustered into 101 open-reading frames, and multiple SNPs were detected in 15 genes.
Fig. 2.—
Distribution of SNPs in the core genome of the 8 USA300 household isolates. Blue, SNP in an intergenic region; red, nonsynonymous SNP; and green, synonymous SNP. The light-purple-shaded areas indicate the position of selected MGEs in the FPR3757 reference strain. Forward (above the line) and reverse (below the line) strand open-reading frames of FPR3757 are shown in gray at the top of the aligned sequences.
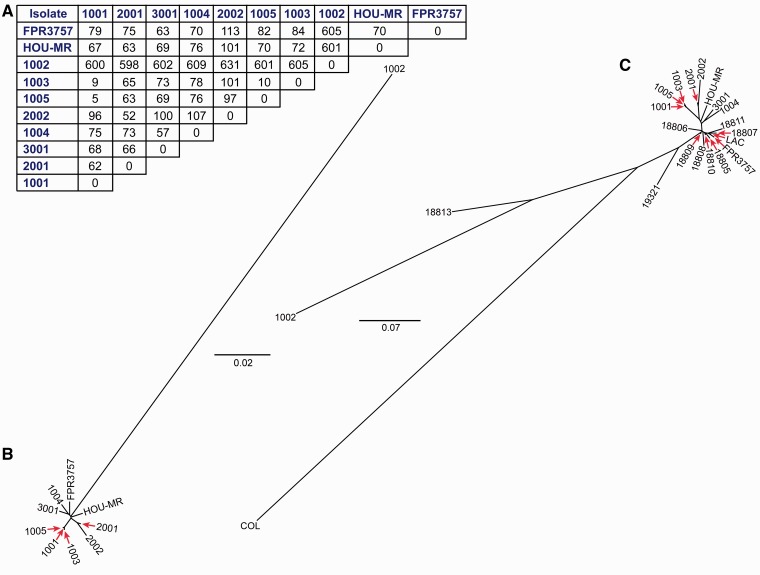
Fig. 3.—
SNP matrix and SNP-based phylogenetic analysis. (A) SNP matrix. (B) Phylogenetic analysis based on 806 concatenated core genomic SNP nucleotides in the eight household USA300 isolates and strain HOU-MR relative to the FPR3757 USA300 reference strain. (C) Phylogenetic analysis of household isolates, geographically diverse USA300 isolates, and USA300 reference strains, based on 1,577 concatenated core genomic SNP nucleotides.
We detected similar ratios of nonsynonymous to synonymous SNPs in all but two isolates (table 2). The highest ratio was found in the nonpersisting isolate 3001 (3.4:1), which may indicate this isolate has undergone recent diversification (Castillo-Ramirez et al. 2011).
Table 2.
Summary of Mutations among MRSA Isolates
| House | Strain | SNPs | IG | NS | S | Ratio |
|---|---|---|---|---|---|---|
| 1000 | 1001 | 79 | 31 | 33 | 14 | 2.4:1 |
| 1003 | 84 | 33 | 35 | 15 | 2.3:1 | |
| 1004 | 70 | 28 | 28 | 14 | 2:1 | |
| 1005 | 82 | 33 | 34 | 14 | 2.4:1 | |
| 2000 | 2001 | 75 | 34 | 30 | 11 | 2.7:1 |
| 2002 | 113 | 34 | 37 | 42 | 0.9:1 | |
| 3000 | 3001 | 63 | 28 | 27 | 8 | 3.4:1 |
Note.—IG, intergenic; NS, nonsynonymous SNP; S, synonymous SNP.
Compared with the FPR3757 reference genome, 24 genes in the eight query isolates had regions of large deletions. These deletions were mainly located in fibronectin-binding proteins, hypothetical cytosolic proteins, or proteins of unknown function and were located in large repeat regions.
We note that ACME was absent in isolates 1004 and 1002 (MSSA), consistent with prior PCR-genotyping (table 2). This MSSA isolate also lacked other MGEs commonly associated with USA300, including SaPI5, ΦSA2usa, and ΦSA3usa (fig. 2).
Phylogeny
To determine the extent of diversity among the core genomes of the eight household isolates relative to the USA300 reference genome FPR3757, we performed phylogenetic analysis using concatenated SNP nucleotides identified in the core genome of isolates from Northern Manhattan households (fig. 3A–C). This analysis indicates that MSSA isolate 1002 likely diverged earlier from a common ancestor and is not immediately related to the other USA300 isolates from HH1000 (fig. 3B), whereas all seven USA300 MRSA isolates and HOU-MR, another reference USA300 strain (Highlander et al. 2007), were more closely related. When SNPs from the core genomes of 10 previously sequenced and geographically diverse USA300 isolates were included in the phylogenetic analyses (Highlander et al. 2007; Kennedy et al. 2008), the Northern Manhattan MRSA isolates clustered closely together and suggest a relatively recent common ancestor. Overall, these isolates were closely related to the main cluster that represents the epidemic USA300 clone (fig. 3C).
Furthermore, isolates 2001 and 2002 (HH2000) clustered together as did most MRSA isolates from HH1000 (1001, 1005, and 1003, representing the initial clinical isolate, late colonization isolate, and spa-variant, respectively). However, one of the ACME-negative isolates, 1004, was less closely related to this branch and thus likely did not recently arise in HH1000. Rather, isolate 1004 was more closely related to the nonpersisting isolate (3001) from HH3000. Based on the high-resolution data obtained from whole-genome comparison and phylogenetic analysis, three distinct USA300 subclones (1001, 1002, and 1004) were likely acquired independently and introduced into HH1000. This idea contrasts with results from conventional genotyping (PFGE, spa-typing, and PCR typing for MGEs), which, in light of the epidemiology of the strains collected in one household, suggested immediate common ancestry of these three USA300 isolates. On the other hand, the spa-clustering algorithm BURP (Mellmann et al. 2007) predicted that isolate 1003 (spa-type t5321) was more distantly related to the other USA300 household isolates, but based on whole-genome sequence comparison, it is very closely related to 1001 and only differs from this isolate by two additional SNPs other than those in spa.
Cross-Sectional Comparison of the Three Infectious Isolates
Although index isolates 1001 and 2001 from the “persisting” households 1000 and 2000 each contained 10 or 11 unique nonsynonymous SNPs, respectively (table 3), they also shared seven unique nonsynonymous mutations that were not present in the “non-persisting” household (HH3000). These mutations resided within genes encoding proteins involved in replication and recombination repair, coenzyme transport, carbohydrate transport and metabolism, and genes encoding proteins of unknown function (table 3).
Table 3.
Summary of Nonsynonymous SNPs and Small Indels
| Household/isolate | Gene | COG | Mutation |
|---|---|---|---|
| HH1000/1005 | ileS | J | I473N |
| Zinc protease | R | P52S | |
| Nicotinate phosphoribosyl transferase | H | G340A | |
| sdrC | None | Δ75 bp SD region | |
| HH1000/1003 | spa | JM | E393A, Δ24 bp |
| gid | J | A133V | |
| Nramp | P | Y304F | |
| HH2000/2002 | CDP-ribitol-phosphotransferase | M | S400N |
| lacC | G | W218a | |
| moeA | H | M25I | |
| hlgA | None | D130E | |
| hlgC | None | L2I, L79F, L126F | |
| Conserved hypothetical protein | None | 9 bp insertion | |
| HH1000 and HH2000 combined, “Persisting strains” | gyrA | L | L84S |
| tRNA-Arg | S | E17D | |
| Hypothetical membrane-span protein | S | D71N | |
| parC | L | Y80S | |
| O2-independ.coprophyrinogen-oxidase | H | T145A | |
| Transaldolase | G | I6V | |
| Transporter, MMPL family | R | K795Q | |
| HH3000/3001, “Non-persister” | Cytosolic protein | C | G26D |
| yqiG | C | A219E | |
| Cell division protein ftsH | O | D336G | |
| Hypothetical membrane-span protein | S | P220L | |
| Transcriptional regulator, MarR | K | W48a | |
| Hypothetical membrane-span protein | TK | W199R | |
| Arogenate dehydrogenase | E | G336V, Y335a | |
| Erythrocyte membrane-span protein | LVDSRM | T8428S | |
| proC | E | D152N | |
| kipl | E | E60G | |
| rpmA | J | Y54H | |
| icaA | Δ5 bp (239a) | ||
| lukE | None | Δ1 bp (6a) | |
| Hypothetical cytosolic protein | J | N721K |
aMissense mutation.
The nonpersisting isolate 3001 contained 12 unique nonsynonymous SNPs in genes involved in transcription, translation, post-translational modification (cell division protein ftsh), energy production and conversion, protein turnover, amino acid transport, and metabolism (table 3). In addition, this isolate had premature stop codons in marR (transcriptional regulator) and lukE (encoding a leukocidin subunit), and a frame shift mutation in icaA (table 3).
Longitudinal Evolution of Mutations in HH1000
Only three nonsynonymous SNPs and one deletion were identified in each, the spa-variant (1003), and the late colonizing isolate (1005) compared with the index isolate (1001) (table 3). For 1003, SNPs and a repeat deletion were detected in spa, consistent with the known evolution of that spa-type. Two additional SNPs were found in genes encoding proteins involved in translation (glucose-inhibited division protein, gid) or inorganic ion transport and metabolism (a homolog of natural resistance associated macrophage protein, Nramp) (table 3).
Relative to 1001, isolate 1005, a late colonizing isolate, harbored mutations in genes encoding proteins involved in translation (IleS), coenzyme transport and metabolism, and a zinc protease and a 75-bp repeat deletion in an adhesion molecule known as SdrC (table 3). Sequencing of additional USA300 isolates collected in household HH1000 (table 4) indicates co-occurrence and a possible genetic linkage of the ileS mutation and a 75-bp deletion within the region encoding for serine-aspartate repeats in sdrC. IleS is a class I tRHA synthetase and the molecular target of mupirocin. Low-level resistance (MIC = 8–256 µg/ml) has been linked to a number of mutations in ileS that result in changes in amino acids, such as V588F and V631F (Henkel and Finlay 1999; Harbarth et al. 2000; Antonio et al. 2002), affecting the interaction of mupirocin with the Rossman fold (Harbarth et al. 2000). Our observed I473N mutation is predicted to fall within the IleS core domain, which is contained within the Rossman fold. However, isolate 1005 is not resistant to mupirocin and the MIC (0.4 µg/ml) was identical to that of the index clinical isolate (1001) by E-test. This finding is consistent with the reported lack of exposure of family members to mupirocin during the study period.
Table 4.
Summary of Samples and Mutations in Household 1000
| Sample ID | Timepoint (months) | Person | Source | SdrC, SAUSA300_0546 | IleS, SAUSA300_1087 | Zn prot, SAUSA300_1172 | Nicotinate PT, SAUSA300_1894 |
|---|---|---|---|---|---|---|---|
| 1001 | 0 | 4 | Clinical | Wt | I473 | P52 | G340A |
| 1010 | 2 | 4 | Buttocks | Wt | I473 | P52 | G340 |
| 1011 | 2 | 5 | Nasal | Wt | I473 | P52 | G340 |
| 1012 | 2 | 5 | Axilla | Wt | I473 | P52 | G340 |
| 1013 | 2 | 7 | Nasal | Wt | I473 | P52 | G340 |
| 1014 | 6 | 1 | Nasal | Wt | I473 | P52 | G340A |
| 1015 | 6 | 5 | Nasal | Δ834-875 | I473N | P52 | G340 |
| 1016 | 10 | 5 | Nasal | Δ834-875 | I473N | P52S | G340 |
| 1017 | 15 | 5 | Nasal | Δ834-875 | I473N | P52S | G340 |
| 1005 | 15 | 5 | Axilla | Δ834-875 | I473N | P52S | G340 |
USA300 Mutations Unique to HH2000 and Evolution
In household HH2000, the index (2001) and colonizing (2002) isolates harbored mutations in five genes involved in cell wall and membrane biogenesis, carbohydrate and coenzyme transport and metabolism, and S. aureus virulence (table 3). These mutations included a missense mutation W218* in the tagatose-6-phosphatate kinase, which is encoded by the lacC gene. Staphylococcus aureus is one of the few organisms that uses the d-tagatose-6-phosphate pathway exclusively to metabolize lactose and d-galactose. This pathway is essential for S. aureus survival in lactose-rich media such as milk. In its functional absence, d-galactose accumulates, and bacterial growth is inhibited (Bettenbrock et al. 1999).
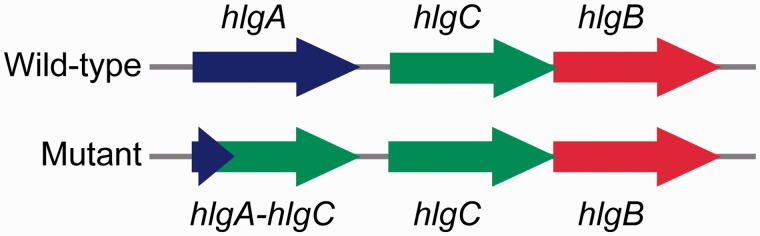
We noted an overlap between regions of large deletions and accumulation of synonymous SNPs in two clusters of genes (hlgA and hlgC, encoding gamma-hemolysin subunits, and tarL, encoding CDP-ribitol ribitolphosphotransferase) in isolate 2002 from household 2000. Resequencing of these genes revealed additional nonsynonymous and synonymous SNPs, which all clustered within the 5’-region of each gene. Blast searches of these selected regions revealed a chimeric gene, in which the first 372 bp were consistent with the hlgA sequence, and the remainder of the gene (573 bp) was replaced by hlgC sequence (fig. 4). This orientation was further verified by locus-spanning PCR. However, no additional sequence changes were observed in the adjacent hlgC or hlgB genes. The putative breakpoint followed a region of 17-bp homology containing two long A stretches positioned at the 3’-end of a 200-bp region of ∼80% nucleotide identity.
Fig. 4.—
Small region genome rearrangement. Schematic representation of small genome rearrangement within the hlgACB locus.
Isolate 2002 harbored an additional putative gene rearrangement in a tandem gene cluster. Similar to hlgA, the tarL gene (encoding CDP-ribitol ribophosphatase, SAUSA300_0251) had 456 bp at the 3’-end of the gene replaced by sequence from the upstream-located paralog tarB (SAUSA300_0247) in isolate 2002. No additional changes were noted within this locus that also contains other enzymes of the teichoic cell wall synthesis machinery (Qian et al. 2006).
All the SNPs and areas of genome rearrangements were also present in all additional USA300 isolates from this household from timepoint 3 onward (fig. 1) but not at the beginning of the study period, suggesting that isolate 2002 had replaced 2001 among household members (not shown).
Discussion
Using genome-wide analysis of sequentially sampled USA300 isolates from three community households, we discovered that there was limited evolution of these subclones over a ∼15-month period. Despite this finding, there were noted changes in the DNA of these isolates, and they mainly occurred in genes that encoded for major aspects of S. aureus function, such as cell adhesion, cell wall biosynthesis, virulence, and carbohydrate metabolism. On the basis of the close epidemiological relationship of the sequenced household isolates, we were also able to identify regions of small genome rearrangements within two multigene operons. Although large genome replacements have been recognized as a means of major clone evolution, such as for ST239 (Robinson et al. 2005), small genome rearrangements within operons may represent a more common and previously underappreciated mechanism of S. aureus evolution and adaptation.
Most of the biological and genome differences were observed between the persisting and nonpersisting USA300 isolates. Although almost all isolates harbored a relatively high number of nonsynonymous mutations relative to synonymous mutations, the strain with the highest nonsynonymous-to-synonymous SNP ratio did not persist. This preferential accumulation of nonsynonymous SNPs may indicate the recent emergence of slightly deleterious mutations, and thus, purifying selection has not yet occurred (Castillo-Ramirez et al. 2011). In addition, this isolate was noted to have a decreased doubling time and harbor a premature stop codon in the transcriptional regulator marR, consistent.
Although this study did not allow for a calculation of the USA300 mutation rate, it suggests a limited accumulation of novel SNPs (only three SNPs in each, HH1000 and HH2000) over the 15-month study period appears. These numbers appear in contrast to the hospital-associated S. aureus isolates of the ST239 lineage, in which the estimated rate of mutation is one SNP every 6 weeks (Harris et al. 2010). This may indicate that the household isolates studied here already had reached an equilibrium in host adaptation to genetically closely related individuals. As a parallel, host imprinting and divergent adaptation of bacteria to individual patients has been suggested in asymptomatic Escherichia coli bacteriuria patients (Zdziarski et al. 2010). Here, the repeated exposure of patients to the identical E. coli isolate led to the repeated individual-specific evolution of almost all genomic changes. Alternatively, the difference in SNP frequency between the household and hospital-associated isolates emphasizes the potential role of high-level antibiotic pressure in driving evolution of S. aureus strains. It should be noted, however, that approximately 50% of healthy individuals residing in this study community report some antibiotic use over a 6-month period (Uhlemann et al. 2011).
Although the majority of longitudinally evolved SNPs were isolate specific, the long-term persisting isolates from both households underwent adaptation in molecules implicated in bacterial adhesion to human host cells, namely a truncation of SD repeats in sdrC in isolate 1005 and a small gene rearrangement in the tarL gene in isolate 2002. SdrC belongs to the multigene family of surface receptors that promote attachment to host extracellular matrix proteins (Josefsson et al. 1998). SdrC is believed to play a role in the multifactorial process of binding to desquamated nasal epithelial cells (Weidenmaier et al. 2005; Corrigan et al. 2009) and to the neuronal cell adhesion molecule β-neurexin (Barbu et al. 2010). Although much of the biology of SdrC remains unknown, it shares most of its structural organization with clumping factor A (ClfA), a closely related member of the MSCRAMM family (Josefsson et al. 1998). A previous study provided evidence that the number of SD repeats in ClfA plays an important role in modulating the proper display of domain A, thereby allowing subsequent binding of ClfA to fibrinogen (Hartford et al. 1997). This finding suggests an important regulatory role for variable SD repeat units in other members of this gene family such as SdrC. Deletions or insertions of SD repeats in SdrC may provide a rapid mechanism of optimized binding to host cells and colonization of a particular human host. The ability to adhere to human cells may have further been altered by the gene rearrangement in tarL. This gene is thought to encode for the CDP-ribitol ribitolphospho-transferase (EC 2.7.8.14), which is involved in wall teichoic acid (WTA) synthesis (Qian et al. 2006). A S. aureus mutant devoid of WTA showed a complete lack of nasal colonization and endovascular infections and was more resistant to human β-defensin 3 (Weidenmaier et al. 2003, 2005; Koprivnjak et al. 2008). The observed reassembly of one of the enzymes in the WTA machinery may allow for adaptive colonization and persistence in a particular human host. We also observed an apparently genetically linked second small genome rearrangement in the gamma-hemolysin operon in the same isolate. Gamma-hemolysin is a bicomponent pore-forming leukotoxin encoded by three genes, hlgA, hlgB, and hlgC, all within the same operon. hlgA and hlgC encode LukS subunits, and hlgB encodes a LukF subunit. These subunits assemble as either HlgAB or HlgCB, and both forms have potent hemolytic and leukotoxic activity with differing cell tropism (Supersac et al. 1998). Gamma-hemolysin subunits are highly upregulated by USA300 in blood, although the toxin may only have a modest contribution to virulence (Malachowa et al. 2011). Variation of the composition of the LukS subunit may potentially allow a subtle adaptation of S. aureus for host cell tropism or manifest loss of virulence factors as has been suggested for bacteria during their chronic colonization state (Zdziarski et al. 2010; McAdam et al. 2011). In addition, leukotoxins are now known to enhance or directly trigger the host inflammatory response (Zivkovic et al. 2011; Graves et al. 2012; Malachowa et al. 2012; Yoong and Pier 2012), which would suggest an important role of the rearrangements in antigenic diversification and environmental host adaptation.
Rapid diversification of surface proteins encoded by paralogous tandem gene clusters genes have been recognized in many different bacterial species (van der Woude and Baumler 2004) via multiple different molecular mechanisms such as inversion, deletion, gene conversion via recombination, or slipped strand mispairing during DNA replication. Our findings here are consistent with sequential homologous recombination, gene duplication, and generation of chimeric gene sequences in the tarLB and hlgAC loci. However, the small sample size precludes the exact mapping of the putative breakpoints. Additional evidence for gene rearrangements in S. aureus has also been detected in the ssl and lpl clusters, likely due to multiple rounds of unequal crossing-over events between sister chromosomes around centrally conserved sequences (Tsuru et al. 2006; Tsuru and Kobayashi 2008).
Limitations of this study include the relatively small number of sequenced isolates. However, using this high-resolution sequence analysis, we were also able to reclassify three possible transmission events within one household when compared with a combination of conventional typing techniques, including spa, MLST, and PFGE. The direct implication of this refined typing technique is that USA300 was introduced at least three times into a single community household (HH1000). This suggests that members of this household either had continuous exposure to one or several unidentified high-risk reservoirs in their social networks or potentially harbored a genetic predisposition for colonization with ST8 S. aureus. As whole-genome sequencing becomes more affordable and sequence analysis more efficient, these tools will greatly aid in refining epidemiological investigations.
The phylogenetic analysis incorporating geographically diverse USA300 isolates (Kennedy et al. 2008) suggested that all USA300 MRSA household isolates have evolved as a single clade in the Northern Manhattan community from a relatively recent common ancestor within the epidemic USA300 cluster. This further illustrates the importance of local communities as reservoirs for transmission and spread of endemic S. aureus clones such as USA300.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health and by National Institutes of Health grants (K08 AI090013 to A.-C.U. and R01 AI077690 and R01 AI077690-S1 to F.D.L.); the Centers for Diseases Control (to F.L.); and the Paul A. Marks scholarship (to A.-C.U.).
Literature Cited
- Adcock PM, Pastor P, Medley F, Patterson JE, Murphy TV. Methicillin-resistant Staphylococcus aureus in two child care centers. J Infect Dis. 1998;178:577–580. doi: 10.1086/517478. [DOI] [PubMed] [Google Scholar]
- Antonio M, McFerran N, Pallen MJ. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:438–442. doi: 10.1128/AAC.46.2.438-442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbu EM, et al. beta-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS Pathog. 2010;6:e1000726. doi: 10.1371/journal.ppat.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenbrock K, Siebers U, Ehrenreich P, Alpert CA. Lactobacillus casei 64H contains a phosphoenolpyruvate-dependent phosphotransferase system for uptake of galactose, as confirmed by analysis of ptsH and different gal mutants. J Bacteriol. 1999;181:225–230. doi: 10.1128/jb.181.1.225-230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M, et al. Staphylococcus aureus ST398, New York city and Dominican Republic. Emerg Infect Dis. 2009;15:285–287. doi: 10.3201/eid1502.080609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramirez S, et al. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog. 2011;7:e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M, et al. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6:189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- Cook HA, Furuya EY, Larson E, Vasquez G, Lowy FD. Heterosexual transmission of community-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2007;44:410–413. doi: 10.1086/510681. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Diep BA, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves SF, et al. Sublytic concentrations of Staphylococcus aureus Panton-Valentine leukocidin alter human PMN gene expression and enhance bactericidal capacity. J Leukoc Biol. 2012;92:361–374. doi: 10.1189/jlb.1111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harbarth S, et al. Risk factors for persistent carriage of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2000;31:1380–1385. doi: 10.1086/317484. [DOI] [PubMed] [Google Scholar]
- Harmsen D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartford O, Francois P, Vaudaux P, Foster TJ. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol Microbiol. 1997;25:1065–1076. doi: 10.1046/j.1365-2958.1997.5291896.x. [DOI] [PubMed] [Google Scholar]
- Henkel T, Finlay J. Emergence of resistance during mupirocin treatment: is it a problem in clinical practice? J Chemother. 1999;11:331–337. doi: 10.1179/joc.1999.11.5.331. [DOI] [PubMed] [Google Scholar]
- Herold BC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- Highlander SK, et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden BP, et al. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3755–3762. doi: 10.1128/AAC.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden BP, et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 2011;7:e1002359. doi: 10.1371/journal.ppat.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson E, et al. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology. 1998;144:3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- Kaneko J, Muramoto K, Kamio Y. Gene of LukF-PV-like component of Panton-Valentine leukocidin in Staphylococcus aureus P83 is linked with lukM. Biosci Biotechnol Biochem. 1997;61:541–544. doi: 10.1271/bbb.61.541. [DOI] [PubMed] [Google Scholar]
- Kaplan SL, et al. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin Infect Dis. 2005;40:1785–1791. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1227–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivnjak T, Weidenmaier C, Peschel A, Weiss JP. Wall teichoic acid deficiency in Staphylococcus aureus confers selective resistance to mammalian group IIA phospholipase A(2) and human beta-defensin 3. Infect Immun. 2008;76:2169–2176. doi: 10.1128/IAI.01705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malachowa N, et al. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One. 2011;6:e18617. doi: 10.1371/journal.pone.0018617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malachowa N, et al. Staphylococcus aureus leukotoxin GH promotes inflammation. J Infect Dis. 2012;206:1185–1193. doi: 10.1093/infdis/jis495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One. 2011;6:e24301. doi: 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A, et al. Based upon repeat pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 2007;7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran GJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- Mwangi MM, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimi TS, et al. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996–1998. Clin Infect Dis. 2001;33:990–996. doi: 10.1086/322693. [DOI] [PubMed] [Google Scholar]
- Qian Z, et al. Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: genes, genomic organization and gene duplication. BMC Genomics. 2006;7:74. doi: 10.1186/1471-2164-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DA, et al. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet. 2005;365:1256–1258. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- Rosenthal VD, et al. International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003-2008, issued June 2009. Am J Infect Control. 2010;38:95–104. doi: 10.1016/j.ajic.2009.12.004. e102. [DOI] [PubMed] [Google Scholar]
- Supersac G, Piemont Y, Kubina M, Prevost G, Foster TJ. Assessment of the role of gamma-toxin in experimental endophthalmitis using a hlg-deficient mutant of Staphylococcus aureus. Microb Pathog. 1998;24:241–251. doi: 10.1006/mpat.1997.0192. [DOI] [PubMed] [Google Scholar]
- Talan DA, et al. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis. 2011;53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- Tenover FC, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuru T, Kawai M, Mizutani-Ui Y, Uchiyama I, Kobayashi I. Evolution of paralogous genes: reconstruction of genome rearrangements through comparison of multiple genomes within Staphylococcus aureus. Mol Biol Evol. 2006;23:1269–1285. doi: 10.1093/molbev/msk013. [DOI] [PubMed] [Google Scholar]
- Tsuru T, Kobayashi I. Multiple genome comparison within a bacterial species reveals a unit of evolution spanning two adjacent genes in a tandem paralog cluster. Mol Biol Evol. 2008;25:2457–2473. doi: 10.1093/molbev/msn192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann AC, et al. The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One. 2011;6:e22407. doi: 10.1371/journal.pone.0022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenvoort JH, Toenbreker HM, Nurmohamed A, Davies BI. Transmission of methicillin-resistant Staphylococcus aureus within a household. Eur J Clin Microbiol Infect Dis. 1997;16:399–400. doi: 10.1007/BF01726373. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, Kristian SA, Peschel A. Bacterial resistance to antimicrobial host defenses—an emerging target for novel antiinfective strategies? Curr Drug Targets. 2003;4:643–649. doi: 10.2174/1389450033490731. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, et al. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis. 2005;191:1771–1777. doi: 10.1086/429692. [DOI] [PubMed] [Google Scholar]
- Yoong P, Pier GB. Immune-activating properties of Panton-Valentine leukocidin improve the outcome in a model of methicillin-resistant Staphylococcus aureus pneumonia. Infect Immun. 2012;80:2894–2904. doi: 10.1128/IAI.06360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdziarski J, et al. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog. 2010;6:e1001078. doi: 10.1371/journal.ppat.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic A, et al. TLR 2 and CD14 mediate innate immunity and lung inflammation to staphylococcal Panton-Valentine leukocidin in vivo. J Immunol. 2011;186:1608–1617. doi: 10.4049/jimmunol.1001665. [DOI] [PubMed] [Google Scholar]