Abstract
Anopheles gambiae M and S are thought to be undergoing ecological speciation by adapting to different larval habitats. Toward an improved understanding of the genetic determinants and evolutionary processes shaping their divergence, we used a 400,000 single-nucleotide polymorphism (SNP) genotyping array to characterize patterns of genomic differentiation between four geographically paired M and S population samples from West and Central Africa. In keeping with recent studies based on more limited genomic or geographic sampling, divergence was not confined to a few isolated “speciation islands.” Divergence was both widespread across the genome and heterogeneous. Moreover, we find consistent patterns of genomic divergence across sampling sites and mutually exclusive clustering of M and S populations using genetic distances based on all 400,000 SNPs, implying that M and S are evolving collectively across the study area. Nevertheless, the clustering of local M and S populations using genetic distances based on SNPs from genomic regions of low differentiation is consistent with recent gene flow and introgression. To account for these data and reconcile apparent paradoxes in reported patterns of M–S genomic divergence and hybridization, we propose that extrinsic ecologically based postmating barriers vary in strength as environmental conditions fluctuate or change.
Keywords: divergent selection, genome scan, introgression, population genomics, SNP genotyping, speciation islands
Introduction
A fundamental problem in evolutionary biology is to understand the genomic architecture associated with adaptive population divergence and reproductive isolation. Scans of genome-wide patterns of differentiation between populations can be used to map candidate regions contributing to isolation, especially when diversification is occurring despite some ongoing gene flow (Via 2009; Nosil and Feder 2012). In the presence of gene flow, only genome regions under strong divergent natural selection are expected to resist introgression, resulting in a heterogeneous pattern of differentiated and homogeneous regions (Wu 2001). Genome-wide scans of differentiation between incompletely reproductively isolated populations have largely revealed the predicted heterogeneous pattern (Nosil et al. 2009; Nosil and Feder 2012). However, interpretation of genome scans is fraught with difficulties (Noor and Bennett 2009; Turner and Hahn 2010), and unresolved questions remain concerning the impact of gene flow on the number, size, and genomic distribution of exceptionally diverged regions during the speciation process.
At the heart of the debate is the role that genome structure plays in facilitating population divergence in the presence of gene flow. Two central issues concern: 1) the importance of physical features of the genome that reduce recombination (e.g., chromosomal rearrangements and centromeres) in fostering divergence and 2) whether divergence is predicated on a small number of genes clustered in a few genomic “islands of speciation” (Turner et al. 2005) or on a larger number of genes arrayed across the genome. Empirical studies have revealed conflicting patterns (Nosil et al. 2009). It is unclear whether these differing outcomes are the result of: 1) truly different evolutionary mechanisms acting among taxa, 2) comparisons involving disparate stages along the divergence continuum, 3) studies employing different sampling schemes with varying levels of resolution and sensitivity for detecting divergence, or 4) some combination of the above.
The M and S forms of the African malaria mosquito Anopheles gambiae (Diptera: Culicidae) (della Torre et al. 2001) represent a model system for the study of ecological speciation (Rundle and Nosil 2005; Schluter 2009). These mosquitoes are thought to be diverging despite incomplete reproductive isolation due to differential ecological adaptation to alternative larval habitats (Diabate et al. 2008; Gimonneau et al. 2012a; Kamdem et al. 2012; Lehmann et al. 1997) in different eco-geographical settings (Caputo et al. 2008; Costantini et al. 2009; Simard et al. 2009). The M form mainly exploits more temporally stable larval habitats containing relatively high levels of environmental stressors; in the savannas of West Africa, M larval habitats tend to be rice fields with high predator density, whereas in the Central African rainforest, these may be polluted urban dumping grounds. Conversely, the S form exploits ephemeral habitats associated with seasonal rainfall that are largely unpolluted and predator free. Despite the difference in larval habitat, M and S overlap extensively and apparently continuously across much of West and Central Africa, where they may be strictly sympatric and synchronously breeding (della Torre et al. 2005). Premating reproductive barriers (Diabate et al. 2009; Pennetier et al. 2010; Sanford et al. 2011) generally limit heterotypic matings between local M and S populations to ∼1% across most of their range of overlap (Tripet et al. 2001; della Torre et al. 2005). The exception is in the western extreme of their West African range, where hybridization rates as high as 20% have been recorded (Caputo et al. 2008, 2011; Oliveira et al. 2008). No intrinsic postmating barriers have been detected between M and S (Diabate et al. 2007; Hahn et al. 2012). However, extrinsic, environment-based postmating barriers are presumed to be strong and to act against maladapted hybrids in the alternate M versus S larval habitats (Lehmann and Diabate 2008; Turner and Hahn 2010). Although pre- and postmating reproductive isolation may reduce effective gene flow significantly through most of West and Central Africa, neither barrier is absolute. The realized rate of gene flow between M and S is not known (Turner and Hahn 2010; White et al. 2010; Hahn et al. 2012), but several lines of evidence indicate that at least some contemporary gene flow is occurring. Single-nucleotide polymorphism (SNP) genotyping of individual mosquitoes from Guinea Bissau and Ghana has identified individuals of mixed ancestry (Marsden et al. 2011; Weetman et al. 2012) and genes conferring resistance to insecticides and/or pathogens, as well as other apparently neutral sequences, have introgressed between M and S (Djogbenou et al. 2008; Etang et al. 2009; White et al. 2011; Choi and Townson 2012). However, as we discuss later, the extent of introgression is still an open question at the center of ongoing debate that has important ramifications for understanding M–S divergence.
The M and S forms of A. gambiae have played a key role in the development of current concepts about speciation genomics. The metaphor of “speciation islands” was originally developed by Turner et al. (2005) to describe the pattern of genomic differentiation detected in the first microarray-based genome scan of M versus S divergence. Interrogating the M and S genomes with ∼142,000 unique probes from predicted protein-coding genes, only three small regions of significant M–S differentiation were observed. Two of these regions were adjacent to centromeres on chromosomes 2 and X, and the third was located on chromosome 2R. Collectively, the differentiated regions represented <2.8 Mb of the total 260 Mb A. gambiae genome (∼1%) and contained only 67 of the 12,670 annotated protein coding genes (0.5%). Interpreted under the prevailing assumption of substantial introgressive hybridization between M and S, these results suggested that the genes responsible for ecological and reproductive isolation were few in number and mainly confined to three isolated “speciation islands,” the largest two of which coincided with low recombination regions near centromeres on chromosome 2 and the X. Low differentiation across the remainder of the genome was attributed to the homogenizing effects of gene flow.
Subsequent studies cast doubt on the widely held assumption of substantial introgressive hybridization between M and S. Following the microarray-based approach of Turner et al. (2005), White et al. (2010) documented three regions of elevated M–S divergence adjacent to the centromeres on all three chromosomes comprising the A. gambiae complement (centromeric divergence on chromosome 3 had been obscured in the original study due to incomplete genome assembly in that region). White et al. (2010) also found near-complete association of form-specific alleles in these unlinked centromeric regions within both forms, demonstrating strong linkage disequilibrium between M genotypes (and S genotypes) across a study area including Mali, Burkina Faso, and Cameroon. Hahn et al. (2012) argued that in the absence of evidence for biased cotransmission of form-specific allelic combinations (e.g., centromeric drive) on these otherwise independently assorting chromosomes, such strong linkage disequilibrium would be unlikely if levels of introgression were qualitatively similar to the ∼1% hybridization rate (Turner and Hahn 2010). They, therefore, posed an alternative minimal gene flow hypothesis to explain the heterogeneous pattern of M–S genome divergence. Under this hypothesis, low divergence is largely due to shared ancestral polymorphism resulting from incomplete lineage sorting. Moreover, the high divergence centromere-proximal regions are not necessarily “speciation islands.” Rather, they may contain advantageous alleles that arose and swept to high frequency separately in M and S populations but need not be responsible for ecological or reproductive isolation between the two taxa (Noor and Bennett 2009; Turner and Hahn 2010; White et al. 2010; Hahn et al. 2012). Resolving the extent of introgression between M and S is therefore critical for interpreting the meaning of patterns of genomic divergence between these two mosquitoes.
Questions have also arisen concerning the original characterizations of genomic islands of speciation in A. gambiae. The apparent concentration of M–S divergence in isolated regions of reduced recombination could be an artifact of: 1) the relatively low-resolution gene-based approach used in initial mosquito genome scan studies and/or 2) inadequate sampling designs in which only five individuals of each taxon were hybridized to arrays, effectively constraining detectable divergence to regions of fixed or highly skewed frequency differences. More recent higher resolution genomic scans of M–S divergence in Mali based on genome resequencing (Lawniczak et al. 2010) and genotyping of more than 400,000 SNPs (Neafsey et al. 2010) have revealed additional regions of divergence dispersed along all chromosome arms in the genome that were missed by the original gene-based arrays. These studies and other lower density SNP genotyping surveys of natural M and S populations from Ghana, Cameroon, and Guinea Bissau (Weetman et al. 2010, 2012) demonstrate that divergence is more widespread and extensive than previously appreciated.
The changing view of the structure of genomic differentiation between M and S calls for more detailed and standardized studies to firmly resolve the nature, as well as consistency, of divergence across the range of overlap of these mosquitoes and an assessment of the extent to which the pattern has been affected by gene flow. The former task has been primarily limited by methodology and adequate sampling, which can be rectified. However, assessing the impact of gene flow is more difficult. One approach adopted by two recent studies has been to investigate population genomic divergence in an area of unusually high hybridization rates (∼20%) in Guinea Bissau (Oliveira et al. 2008). Based on detection of admixed individuals, these studies showed that the level of introgression in Guinea Bissau was qualitatively consistent with the high rate of hybridization, suggesting that hybrids might not be as strongly selected against at this locality as they are presumed to be elsewhere in Africa (Marsden et al. 2011; Weetman et al. 2012). However, the apparently high level of introgression was asymmetric (largely from M to S), and mating was still not panmictic in Guinea Bissau (Caputo et al. 2011; Marsden et al. 2011; Weetman et al. 2012). Moreover, 15 outlier SNPs (mostly near centromeres but one on 3R in an area of normal recombination) were found to display consistent M–S divergence in Guinea Bissau and other locations in West and Central Africa, suggesting that these regions are being maintained by form-related divergent selection countering gene flow (Weetman et al. 2012).
Guinea Bissau, however, may represent an anomalous area where M and S have recently come into secondary contact (Caputo et al. 2011), and it is possible that changing environmental conditions have reduced the strength of ecologically based postmating barriers. If so, studying M–S divergence in Guinea Bissau and other areas of atypically high hybridization where ecological and reproductive isolation may be breaking down (Caputo et al. 2011; Weetman et al. 2012) is unlikely to be broadly instructive about genome regions that contribute to M–S isolation under conditions more typical of the range of overlap between these two mosquitoes in West and Central Africa.
Accordingly, here we characterize M–S divergence in multiple geographic regions where recorded hybridization rates are more typical (0–1%; della Torre et al. 2005; Costantini et al. 2009; Simard et al. 2009). Although the relative rarity of M–S hybridization compared with Guinea Bissau can make it difficult to sample F1 hybrids in these areas, a 1% rate of interbreeding is not trivial in population genetic terms and would be sufficient to homogenize between-form variation unless countered by strong selection (Slatkin 1987). The only previous study to have mapped genomic divergence between natural population samples of M and S at very high resolution (more than 400,000 markers) studied only one locality in Mali (Neafsey et al. 2010). We extend that study by assessing the nature and consistency of patterns of M–S divergence and testing for evidence of gene flow through genome-wide scans of four replicated pairs of local M and S populations from West and Central Africa, using the same custom 400K SNP genotyping array (Neafsey et al. 2010). We report results confirming widespread and heterogeneous yet consistent genomic differentiation between M and S mosquitoes across West and Central Africa. We also found evidence for recent introgression between local M and S demes. Toward a reconciliation of the apparent paradox between strong genetic associations of form-specific alleles across the genome and credible evidence of ongoing introgressive hybridization, we propose that extrinsic ecologically based postmating barriers vary in strength as a function of fluctuating or changing environmental conditions.
Materials and Methods
Mosquito Sampling and Identification
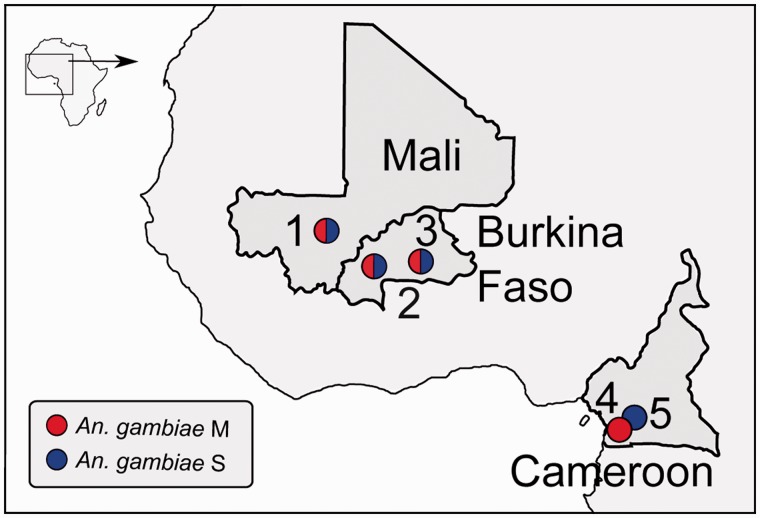
Mosquitoes were collected as indoor resting or host-seeking adults from the five localities numbered in figure 1. Those localities are 1) the southern Malian villages given in Neafsey et al. (2010); the Burkina Faso villages of 2) Samandeni (11°27′N, 04°27′W), and 3) Monemtenga (12°06′N, 01°17′W), sampled in 2005; and the Cameroonian villages of 4) Campo (02°22′N, 09°49′E), and 5) Bibouleman (02°52′N, 11°15′E), sampled in 2005. The first three localities were sites where M and S mosquitoes were collected together, whereas sites 4 and 5 represented localities where M and S, respectively, were collected in geographic proximity (distance of 169 km separating populations). Morphological analysis was first performed to identify collected mosquitoes as belonging to the A. gambiae sibling species complex; only female mosquitoes were analyzed further. Molecular determination of A. gambiae M or S form was based on a diagnostic polymerase chain reaction-RFLP molecular assay targeting the ribosomal DNA repeat on the X chromosome (Santolamazza et al. 2004). Karyotyping of chromosome 2 inversions was not performed for these samples, but the known physical location of chromosomal rearrangements that commonly segregate in both forms (all but one are shared polymorphisms; della Torre et al. 2005) is indicated in figure 2.
Fig. 1.—
Sampling locations in Africa. Localities sampled are represented by numbered circles. Corresponding village names and geographic coordinates are given in Materials and Methods.
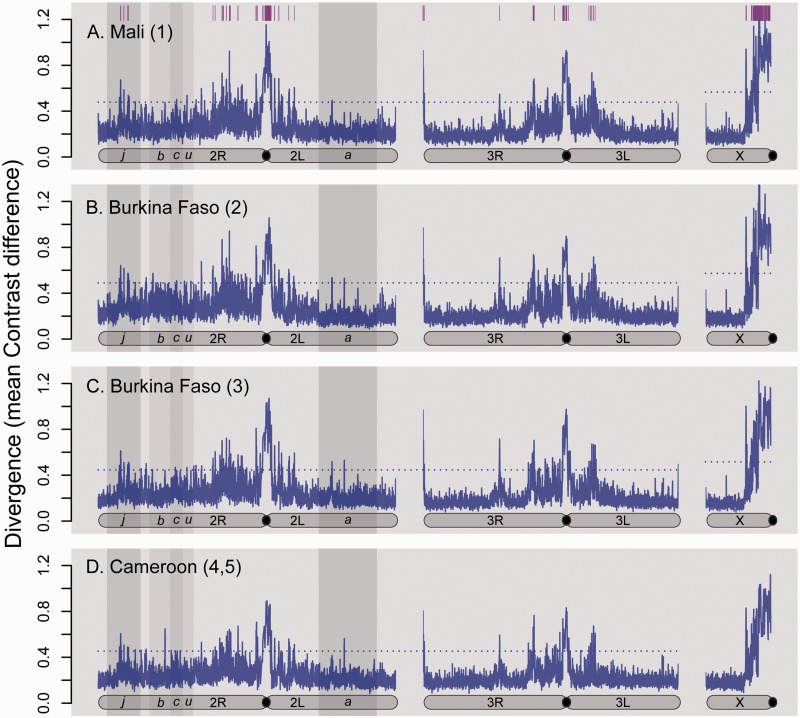
Fig. 2.—
Genome scan of divergence along chromosome arms between sympatric population samples of M and S. Mean difference in Contrast values per nonoverlapping 50-SNP window is plotted: (A) in Mali, for M and S from locality 1; (B, C) in Burkina Faso, for M and S from localities 2 and 3, respectively; and (D) in Cameroon, for M and S from localities 4 and 5. Vertical gray shading behind chromosome 2 marks the regions spanned by polymorphic inversions j, b, c, u, and a. Horizontal dashed line is the threshold for significant divergence. Across the top of panel A, vertical purple lines mark the approximate chromosomal position of 50-SNP windows significantly divergent in all four geographic locations.
Array-Based Genotyping
DNA was extracted from individual female mosquitoes using the DNeasy kit (QIAGEN). The quantity and concentration of DNA in each extraction was determined on a SpectraMax M2 microplate reader (Molecular Devices) using Quant-iT PicoGreen dsDNA reagents (Invitrogen). Separate pools of M and S genomic DNA were made from 20 female mosquitoes of each form for all four of the paired collecting sites. A previous study using the A. gambiae 400K SNP array demonstrated a very high correlation between results from aggregate individual hybridizations and pooled hybridizations of 20 individuals (Pearson’s r2 = 0.96 [Neafsey et al. 2010]). We therefore adopted the pooled assay design for economy. Each of 20 individuals contributed 250 ng of DNA to their respective pool. The pooled mosquito DNA samples were processed following a modified version of the standard human 500K Affymetrix array protocol as previously described in Neafsey et al. (2010). When the DNA quantity from individual mosquitoes was insufficient (locality 2 for both M and S samples; fig. 1), whole-genome amplification was performed before pooling using the REPLI-g Mini kit (QIAGEN). Pooled samples were hybridized to a custom Affymetrix SNP genotyping array using the methods described in Neafsey et al. (2010).
Briefly, the SNP array assays variable SNPs in the A. gambiae genome identified through capillary sequencing of M and S colonies from Mali (Lawniczak et al. 2010). A subset of 400,151 SNPs was chosen for inclusion on the array based primarily on uniformity of SNP coverage across the genome and conformity with design parameters of Affymetrix. (As 80 of the 400,151 SNPs on the array are Y-linked, these were not analyzed in this study given that samples consist of only female mosquitoes; the analysis, therefore, included 400,071 SNPs on the X and autosomes.) SNPs within 15 bp of each other or insertion/deletions were excluded from the array. The median distance between assayed SNPs is ∼300 bp, and 98% of annotated genes contain at least one assayed SNP. A total of 59,878 SNPs occur in coding sequence. It is important to note that because the SNPs on the genotyping array were identified from M and S A. gambiae colonies established from Mali, SNPs may not be geographically representative, and, thus, our data may suffer from a degree of ascertainment bias. As the failure to detect hidden allelic variation is unlikely to result in false signals of genomic divergence, results we report about the shared architecture of M–S genomic divergence across collecting sites should be robust to ascertainment bias. However, one possible manifestation of ascertainment bias would be the underestimation of M–S divergence (and within-taxon divergence) in populations geographically distant from Mali. The extent to which this underestimation may have affected our results is unknown in the absence of resequencing data from the relevant geographic locations.
Raw hybridization data were subject to robust multi-array analysis background correction and quantile normalization using Affymetrix Power Tools. Following Neafsey et al. (2010), we used the Contrast statistic generated by BRLMM-P (Rabbee and Speed 2006) to compare hybridization results between pooled samples. Briefly, the Contrast statistic is a measure of the relative hybridization signal intensity of alternative alleles (Sa and Sb) in a pool, given by the expression (Sa − Sb)/(Sa + Sb) when the correction coefficient (k) = 1. Contrast value data for this study are deposited in VectorBase (www.vectorbase.org, last accessed November 20, 2012).
Outlier Window Analysis
To assess large-scale patterns of genomic differentiation, we calculated the mean difference in Contrast values between pools of M and S mosquitoes at each paired site in nonoverlapping windows of 50 adjacent SNPs. This resulted in a total of 7,998 windows being analyzed across the genome: 4,102 on chromosome 2; 3,066 on chromosome 3; and 830 on the X. The mean physical distance spanned by 50 adjacent SNPs was 28.2 kb (range = 9.1–280 kb, standard deviation = 16.6 kb). Individual windows were tested for significant differences in Contrast values between M and S at each collecting site by nonparametric, Monte Carlo bootstrapping to generate an expected probability distribution to compare to observed values. A total of 10 million iterations were performed to generate the expected distribution by randomly resampling 50 individual SNP assays from across the genome. Separate analyses were conducted for the X chromosome and the autosomes. We used a significance threshold alpha value of 0.05 adjusted for the number of windows examined by Bonferroni correction to identify individual windows that diverged substantially from expectation.
Genetic Distance Trees
We calculated pairwise distances between each population pair as the mean of the absolute value of Contrast value differences at each SNP. On the basis of a matrix of these mean Contrast value differences, we constructed neighbor-joining trees using the APE package in R (Paradis et al. 2004). Bootstrapping (1,000 replicates) was performed on the trees using a custom R script; bootstrap support was summarized using CONSENSE within PHYLIP (Felsenstein 2004). The trees were used to examine two important issues concerning M–S diversification: 1) whether the pattern of genetic divergence for all SNPs resulted in mutually exclusive clustering of M and S, implying the collective evolution of each form across the study area and 2) whether patterns of divergence for a subset of SNPs in low divergence regions provided any evidence for introgression.
To assess the first question of taxonomic clustering of M and S, we constructed genetic distance trees based on all 400,071 SNPs and a subset of SNPs from high divergence regions of the genome. High divergence regions were defined as the 188 significantly diverged 50-SNP windows (9,400 SNPs) common to all four population pairs. (Among this set of SNPs from high divergence regions, only ∼29% were located in centromere-associated “speciation islands” as delimited by White et al. [2010, p. 2284]). We then examined the trees to determine whether M and S populations could be resolved into two distinct genetic clusters across the sampling locations in West and Central Africa. Alternatively, local M and S populations may differ significantly, but all M and S populations may not form mutually exclusive clusters across the study area.
To examine the second question of gene flow, we note that a subset of SNPs has characteristics that make them a priori candidates for introgression, one of which is occurrence in low divergence regions of the genome. Low divergence regions were empirically defined as the 138 50-SNP windows (6,900 SNPs) whose mean Contrast value differences fell in the bottom 15% of the distributions for all four localities. Among these 6,900 SNPs from low divergence regions, the subset that represent synonymous substitutions in coding regions (659 SNPs) are most likely to be neutral and, thus, the best candidates for introgression between M and S. Consequently, genetic distance trees based on these SNP subsets provide a means to test for possible gene flow. The prediction is that if gene flow is ongoing and has occurred at a sufficient level to homogenize allele frequencies between local M and S populations, then genetic distance trees based on these SNP subsets should group M and S populations by geographic proximity and not by form. Synonymous SNPs may still be indirectly affected by physical linkage to other SNPs experiencing form-related selection. However, this concern is mitigated by the short (∼300 bp) median genomic distance between SNPs assayed on the array (Neafsey et al. 2010) and the generally short physical scale (<300 bp) of linkage disequilibrium in A. gambiae (Harris et al. 2010; Neafsey et al. 2010; Weetman et al. 2010).
Results
Pattern of Genome-Wide Divergence
Figure 2 depicts mean Contrast values for nonoverlapping 50 SNP windows plotted along chromosomes for pooled M versus S comparisons at each of the four pairs of collection sites across West and Central Africa. At all four sites, the most pronounced peaks of M–S genomic divergence were associated with the centromere-proximal regions of chromosomes 2, 3, and the X, as previously found. However, each plot also revealed numerous other windows of significant divergence, occurring at irregular intervals along all three A. gambiae chromosomes.
Approximately 300 significantly diverged 50-SNP outlier windows were found for each of the four paired M–S comparisons out of a total of 7,998 windows tested at each geographic location. The majority of these outlier windows were significant across multiple localities (all four sites = 188/427 total outlier windows, 44%; three of four sites = 70/427, 16.4%; and two of four sites = 60/427, 14%) implying a degree of consistency in the geographic pattern of genomic divergence between M and S. This consistency was confirmed by Spearman Rank correlation analyses of M–S Contrast value differences for windows compared across geographic locations. Genome-wide divergence patterns were more similar among locations in West Africa (Spearman’s rs = 0.74–0.77) than for comparisons between West and Central African locations (Spearman’s rs = 0.65–0.69). However, all correlations were highly significant (P < 2.2E-16).
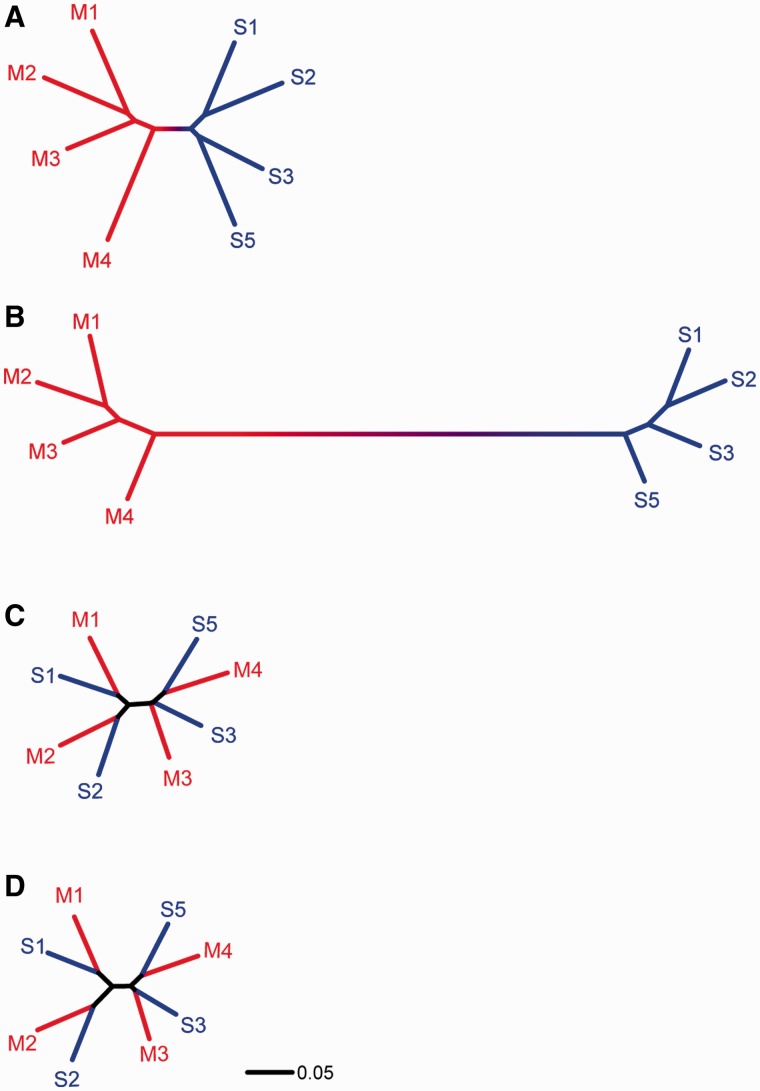
The geographic consistency of genomic differentiation between M and S across locations was further reflected in the Neighbor-joining genetic distance tree. Figure 3A shows the relationships among M and S populations based on mean Contrast value differences across all 400,071 SNPs. Populations of M and S formed two discrete and highly significant 100% bootstrap-supported genetic clusters in the tree. Thus, whole-genome differentiation distinguished M from S across their range of overlap in West and Central Africa, implying that they are evolving collectively across the study area (Rieseberg and Burke 2001). Using only the subset of 9,400 SNPs for tree construction from significantly differentiated genomic regions that were consistently associated with M–S divergence, the difference between M and S was drawn in even starker contrast (fig. 3B).
Fig. 3.—
Genetic distance trees for M and S populations based on alternative genome partitions: (A) all 400,071 SNPs; (B) the 9,400 SNPs from 188 windows consistently associated with significant M–S divergence across the study area; (C) the 6,900 SNPs from 138 windows of consistently low divergence; and (D) the 659 synonymous SNPs from 138 windows of consistently low divergence. M and S populations are represented in red and blue, respectively, and numbered according to the locality in figure 1 where they were collected. All nodes are supported by 100% of 1,000 bootstrap replicates except the node connecting M4 and S5 in panels C and D, which was supported by 94% and 92% of the bootstrap replicates, respectively.
Evidence for Introgression
To address the issue of introgression, we constructed a genetic distance tree based on subsets of SNPs from genomic regions that consistently ranked in the bottom 15% of divergence across all four localities (see Materials and Methods) and compared this to the tree based on all 400,071 assayed SNPs (fig. 3). In contrast to both the whole-genome tree (fig. 3A) and the tree based on SNPs in consistently high divergence genomic regions (fig. 3B), the genetic distance trees based on SNPs from low divergence regions (fig. 3C) and synonymous SNPs from low divergence regions (fig. 3D) grouped local M and S populations together, differentiating populations by geographic proximity and not by form, as predicted if they have recently been subject to introgression. Further, introgression is expected to differentially involve the genetic exchange of neutral SNPs rather than those associated with ecological adaptation and reproductive isolation. A manifestation of this should be decreased genetic distance for neutral SNPs (due primarily to drift) compared with divergently selected SNPs. This prediction is consistent with the substantially shorter branch lengths in figure 3D versus figure 3B, assuming that synonymous SNPs in low divergence regions represent mainly neutral variants and that high divergence regions are likely to contain some divergently selected SNPs.
Discussion
Ecological selection is assumed to be the driver of M–S speciation. In West Africa, the main ecological distinction between the presumed ancestral S form and the derived M form of A. gambiae (Costantini et al. 2009) is the larval habitat; M breeds in irrigated rice fields and thus may have originated in association with rice domestication in Africa. African rice (Oryza glaberrima) was domesticated from a wild ancestor ∼2,000–3,000 years ago in the inland delta floodplains of the upper Niger River in Mali, with subsequent spreading along Sahelian rivers to two secondary domestication centers, one in coastal Gambia, Casamance (Senegal), and Guinea Bissau; the other in the Guinea forest between Sierra Leone and the western Ivory Coast (Porteres 1976; Li et al. 2011). However, aside from speculation about the parallel origin of M with African rice, little is actually known of the timing of M–S divergence, their geographic origins, biogeographic history of contact, or demography (Crawford and Lazzaro 2010). This gap in our knowledge complicates interpretation of the population genomic patterns obtained from genome scans of M and S. One of the most relevant unknowns in the context of speciation genomics concerns whether M and S began diverging in allopatry and subsequently came into secondary contact or whether they have been in contact and exchanging genes since the initiation of ecological divergence. Neither this study nor other studies reporting on admixed individuals (Marsden et al. 2011; Weetman et al. 2012) or introgressed alleles (Djogbenou et al. 2008; Etang et al. 2009; White et al. 2011; Choi and Townson 2012) bear on this question; inference of recent introgressive hybridization between M and S informs only about current gene exchange and not about its history or the geographic context of initial divergence. As such, although the genomic distribution of M–S divergence can be a useful guide both to the location of genes underpinning adaptive divergence between M and S and how divergence is maintained in the face of current gene flow, it does not speak to the question of how initial divergence may have built up in the genome in the presence of gene flow.
When the A. gambiae M and S genomes were sequenced from polymorphic and newly established colonies from Mali, the ensuing population genomic analysis concluded that M–S divergence was widespread beyond isolated “speciation islands” surrounding the centromeres (Lawniczak et al. 2010). Although genetic drift resulting from colonization and laboratory maintenance could have augmented divergence between these colonies relative to natural populations of M and S, the same general pattern of widespread genomic divergence was observed using the 400K SNP genotyping array to map genomic divergence in natural population samples of M and S from the same part of Mali (Neafsey et al. 2010). Two subsequent studies of M and S from other parts of Africa have confirmed some consistent M–S divergence outside of centromeric regions, but their resolution was limited by genotyping at only ∼900 SNPs (Weetman et al. 2010, 2012). Our genome-wide survey of 400,071 SNPs between paired populations of M and S in West and Central Africa confirms the geographic consistency of the more widespread nature of M–S genomic divergence. As noted elsewhere, widespread genomic divergence is not surprising in view of low gene densities in the centromere-proximal regions and the multifarious nature of phenotypic differentiation between M and S (White et al. 2010; Weetman et al. 2012). Our study also confirms that the topology of M–S divergence is not uniform. Many regions of elevated mean divergence can be found interspersed with lower divergence regions. Given the generally low levels of linkage disequilibrium characteristic of the A. gambiae genome outside of inversions and centromeric regions, and the dense spacing of SNPs on the array, this suggests that many loci throughout the genome may be independently subject to divergent selection between M and S. Moreover, it is likely that M–S divergence is even more extensive than we have been able to demonstrate given the relatively course resolution of our conservative, window-based analysis. Some genes contributing to differential adaptation may not be detectable as peaks on such genome scans, particularly if the beneficial mutation(s) arose from standing variation and/or the effect sizes are very small; more modest changes in allele frequency may nonetheless be significant for adaptive events in natural populations (Pritchard and Di Rienzo 2010).
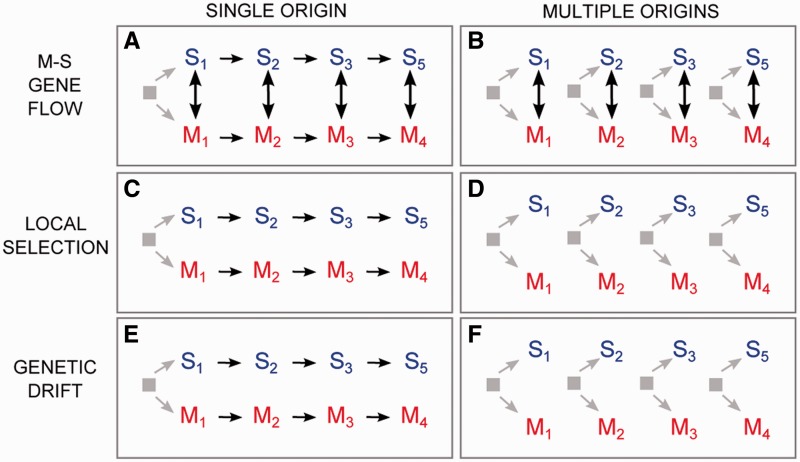
Our analysis of genome regions characterized by consistently low M–S divergence across the study area was aimed to assess whether evolutionarily significant levels of introgression are occurring between M and S, and thus whether these incipient species can withstand low levels of introgression without exhibiting the collapse of differentiation observed in Guinea Bissau (Marsden et al. 2011; Weetman et al. 2012). Specifically, the clustering of geographically proximate M and S populations based on SNPs from low divergence windows (fig. 3C) and the subset of synonymous SNPs from those windows (fig. 3D) are consistent with low levels of recurrent gene exchange homogenizing neutral variation between local populations. Independent support for M–S gene flow previously inferred from limited population and/or genomic sampling includes apparent backcross progeny detected in natural populations (White et al. 2010), allelic introgression (Djogbenou et al. 2008; Etang et al. 2009; White et al. 2011; Choi and Townson 2012), and identification of individual mosquitoes of mixed ancestry (Marsden et al. 2011; Weetman et al. 2012). Taken together, the weight of evidence therefore strongly supports introgression. Alternative scenarios cannot be completely discounted to explain how synonymous SNPs might cluster local M and S populations. These scenarios involve different combinations of: 1) whether selection or genetic drift acting on shared ancestral polymorphism is the primary cause underlying M–S similarity and 2) whether the M form arose once in allopatry from S or multiple times from different local S populations (fig. 4). However, random genetic drift is not expected to cause a repeated pattern of clustering by geographic location. Moreover, both the generally low levels of linkage disequilibrium across much of the A. gambiae genome—which should minimize associations between focal synonymous SNPs and nearby adaptive variants, and the absence of extensive barriers to mosquito migration in this part of Africa—which should erase the genetic signature distinguishing local M populations, render these alternatives unlikely. Indeed, both indirect and direct estimates of gene flow in A. gambiae M and S suggest high rates of intraform migration, shallow population structure, and few restrictions to gene flow across the entire African continent (Taylor et al. 2001; Lehmann et al. 2003; Tripet et al. 2005). Although there are indications of population subdivision within M between West and Central Africa (Slotman et al. 2007; Lee et al. 2009), the potential for rapid exchange of beneficial alleles between these locations has been demonstrated by the spread of insecticide resistance alleles (Djegbe et al. 2011). All indications therefore point to at least some gene flow between M and S sufficient to allow for adaptive gene flow and for neutral alleles to be more similar in frequency between local M and S populations than more geographically distant populations. More definitive evidence awaits a new wave of genome sequencing of individual mosquitoes, made possible by falling costs and increasing throughput of next-generation sequencing technology. Although economical at present, a limitation of the pooled sample approach used in our current SNP genotyping study is an inability to determine multilocus genotypes for individual mosquitoes to conduct a comprehensive test for introgression. Genome sequencing data from individual mosquitoes will enable an estimation of the amount and timing of current and potentially even long-term gene flow, although methods for the latter have proven problematic and need further development (Strasburg and Rieseberg 2011).
Fig. 4.—
Alternative scenarios that could explain the clustering of M and S populations by geographic proximity rather than by form in the genetic distance tree based on synonymous SNPs from low divergence regions of the genome (fig. 3D). Note that the geographic directionality of population splitting and spreading was arbitrarily chosen and is for illustrative purposes only.
Although only S occurs in East Africa, the present-day distributions of M and S are broadly and sometimes even strictly sympatric across much of West and portions of Central Africa (della Torre et al. 2005; Costantini et al. 2009; Simard et al. 2009). The implication is that there has likely been ample opportunity for interform gene flow over a prolonged period of time, whether it has occurred continuously from the onset of ecological divergence or only following secondary contact. Despite compelling evidence for introgressive hybridization between M and S, genetic exchange in the regions sampled for this study has been insufficient to reverse genome-wide divergence, whose architecture is largely consistent across the study area and reflected in M and S populations being distinguishable as separate genetic clusters in a tree based on all 400,071 SNPs. Although differentiation is genome wide, it occurs in a heterogeneous pattern whereby regions that are consistently associated with M–S divergence are interspersed with regions that appear to be introgressing between local M and S populations. Thus, the paradox raised by Hahn et al. (2012) remains: how is it possible to maintain the strong form-specific association between alleles at unlinked markers in the face of recombination and introgressive hybridization between M and S? One possibility is that sporadic local climatic changes can lead to a temporary relaxation of ecologically based postmating barriers, long enough to allow substantial introgression and recombination before environmental conditions return to normal and again impose strong divergent selection, which would purge unsuccessful allelic combinations. In addition, local climatic conditions can affect both the relative frequency of M and S between locales and their level of sympatry, suggesting that the degree of hybridization may fluctuate according to the extent of population contact (Kamdem et al. 2012). These phenomena may explain not only the occasional outbreaks of M–S hybrids where they typically are very rare (Pombi M, Ayala D, Simard F, Besansky N, Costantini C, unpublished data) but also the unusually high rates of M–S hybridization and introgression in westernmost West Africa, if this region is experiencing climate and/or environmental change resulting in the breakdown of reproductive barriers between M and S.
Although our study has provided some insight into the genomic architecture of M–S divergence, two additional formidable challenges remain for understanding ecological adaptation and speciation in A. gambiae. One is the identification of phenotypic traits with differential fitness consequences for M and S in the field. Some progress has been made in this area (Diabate et al. 2005, 2008; Lehmann and Diabate 2008; Gimonneau et al. 2010, 2012b), but much more is needed. The second is the demonstration that particular alleles of candidate genes actually affect the fitness of the phenotypic traits. Both challenges have rarely been met in any system (Barrett and Hoekstra 2011).
In conclusion, the M and S forms of A. gambiae now appear to represent a later stage of speciation than previously appreciated. Much research remains to be done to resolve how M and S evolved to reach this state. However, the foundation exists to build a unified natural history of speciation genomics for A. gambiae in which the ecological basis for reproductive isolation is connected to underlying physiology and genetic causative mechanisms, and the consequences of these associations for the organization and evolution of patterns of genome-wide divergence resolved.
Acknowledgments
The authors thank the entomological teams of MRTC, CNRFP, and OCEAC for fieldwork. They acknowledge members of the Biological Samples Platform and Genetic Analysis Platform of the Broad Institute for sample handling and SNP array data acquisition, and the assistance of E. Lund with sample organization. They are grateful to M. Hahn for critical review and discussion that improved an earlier version of this manuscript. Development of the genome-wide SNP array for A. gambiae was supported by funding from multiple sources including Burroughs Wellcome Fund Request 1008238, the Broad Institute Director's Fund, the Harvard School of Public Health Department of Immunology and Infectious Diseases under direction of D.F. Wirth, Wellcome Trust Programme Grant 077229/Z/05/Z to F.C. Kafatos and G.K. Christophides, and the DeLuca Professorship from Boston College to M.A.T.M. This work was supported by National Institutes of Health grants (R01 AI63508 and R01 AI76584) to N.J.B. and the University of Notre Dame Arthur J. Schmidt Graduate Fellowship to K.R.R.
Literature Cited
- Barrett RD, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Caputo B, et al. Anopheles gambiae complex along the Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar J. 2008;7:182. doi: 10.1186/1475-2875-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B, et al. The “far-west” of Anopheles gambiae molecular forms. PLoS One. 2011;6:e16415. doi: 10.1371/journal.pone.0016415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Townson H. Evidence for X-linked introgression between molecular forms of Anopheles gambiae from Angola. Med Vet Entomol. 2012;26:218–227. doi: 10.1111/j.1365-2915.2011.00977.x. [DOI] [PubMed] [Google Scholar]
- Costantini C, et al. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JE, Lazzaro BP. The demographic histories of the M and S molecular forms of Anopheles gambiae s.s. Mol Biol Evol. 2010;27:1739–1744. doi: 10.1093/molbev/msq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- della Torre A, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10:9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Millogo N, Lehmann T. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae) J Med Entomol. 2007;44:60–64. doi: 10.1603/0022-2585(2007)44[60:eteopi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Diabate A, et al. Larval development of the molecular forms of Anopheles gambiae (Diptera: Culicidae) in different habitats: a transplantation experiment. J Med Entomol. 2005;42:548–553. doi: 10.1093/jmedent/42.4.548. [DOI] [PubMed] [Google Scholar]
- Diabate A, et al. Evidence for divergent selection between the molecular forms of Anopheles gambiae: role of predation. BMC Evol Biol. 2008;8:5. doi: 10.1186/1471-2148-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabate A, et al. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae. Proc Biol Sci. 2009;276:4215–4222. doi: 10.1098/rspb.2009.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djegbe I, et al. Dynamics of insecticide resistance in malaria vectors in Benin: first evidence of the presence of L1014S kdr mutation in Anopheles gambiae from West Africa. Malar J. 2011;10:261. doi: 10.1186/1475-2875-10-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djogbenou L, et al. Evidence of introgression of the ace-1(R) mutation and of the ace-1 duplication in West African Anopheles gambiae s.s. PLoS One. 2008;3:e2172. doi: 10.1371/journal.pone.0002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etang J, et al. Polymorphism of intron-1 in the voltage-gated sodium channel gene of Anopheles gambiae s.s. populations from Cameroon with emphasis on insecticide knockdown resistance mutations. Mol Ecol. 2009;18:3076–3086. doi: 10.1111/j.1365-294X.2009.04256.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6 distributed by the author. Seattle: Department of Genome Sciences, University of Washington. [Google Scholar]
- Gimonneau G, et al. A behavioral mechanism underlying ecological divergence in the malaria mosquito Anopheles gambiae. Behav Ecol. 2010;21:1087–1092. doi: 10.1093/beheco/arq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, et al. Larval habitat segregation between the molecular forms of the mosquito Anopheles gambiae in a rice field area of Burkina Faso, West Africa. Med Vet Entomol. 2012a;26:9–17. doi: 10.1111/j.1365-2915.2011.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, et al. Behavioural responses of Anopheles gambiae sensu stricto M and S molecular form larvae to an aquatic predator in Burkina Faso. Parasit Vectors. 2012b;5:65. doi: 10.1186/1756-3305-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, White BJ, Muir CJ, Besansky NJ. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Philos Trans R Soc Lond B Biol Sci. 2012;367:374–384. doi: 10.1098/rstb.2011.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C, Rousset F, Morlais I, Fontenille D, Cohuet A. Low linkage disequilibrium in wild Anopheles gambiae s.l. populations. BMC Genet. 2010;11:81. doi: 10.1186/1471-2156-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem C, et al. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS One. 2012;7:e39453. doi: 10.1371/journal.pone.0039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, et al. Ecological and genetic relationships of the Forest-M form among chromosomal and molecular forms of the malaria vector Anopheles gambiae sensu stricto. Malar J. 2009;8:75. doi: 10.1186/1475-2875-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8:737–746. doi: 10.1016/j.meegid.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, et al. Microgeographic structure of Anopheles gambiae in western Kenya based on mtDNA and microsatellite loci. Mol Ecol. 1997;6:243–253. doi: 10.1046/j.1365-294x.1997.00177.x. [DOI] [PubMed] [Google Scholar]
- Lehmann T, et al. Population structure of Anopheles gambiae in Africa. J Hered. 2003;94:133–147. doi: 10.1093/jhered/esg024. [DOI] [PubMed] [Google Scholar]
- Li ZM, Zheng XM, Ge S. Genetic diversity and domestication history of African rice (Oryza glaberrima) as inferred from multiple gene sequences. Theor Appl Genet. 2011;123:21–31. doi: 10.1007/s00122-011-1563-2. [DOI] [PubMed] [Google Scholar]
- Marsden C, et al. Asymmetric introgression between the M and S molecular forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridisation. Mol Ecol. 2011;20:4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science. 2010;330:514–517. doi: 10.1126/science.1193036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity. 2009;103:439–444. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Feder JL. Genomic divergence during speciation: causes and consequences. Philos Trans R Soc Lond B Biol Sci. 2012;367:332–342. doi: 10.1098/rstb.2011.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Oliveira E, et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45:1057–1063. doi: 10.1603/0022-2585(2008)45[1057:hlohbm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pennetier C, Warren B, Dabire KR, Russell IJ, Gibson G. “Singing on the wing” as a mechanism for species recognition in the malarial mosquito Anopheles gambiae. Curr Biol. 2010;20:131–136. doi: 10.1016/j.cub.2009.11.040. [DOI] [PubMed] [Google Scholar]
- Porteres R. African cereals: Eleusine, fonio, black fonio, teff, Brachiaria, Paspalum, Pennisetum and African rice. In: Harlan JR, de Wet JMJ, Stemler ABL, editors. Origins of African plant domestication. The Hague (The Netherlands): Mouton Publishers; 1976. pp. 409–452. [Google Scholar]
- Pritchard JK, Di Rienzo A. Adaptation—not by sweeps alone. Nat Rev Genet. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Burke JM. The biological reality of species: gene flow, selection, and collective evolution. Taxon. 2001;50:47–67. [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecol Lett. 2005;8:336–352. [Google Scholar]
- Sanford MR, et al. Morphological differentiation may mediate mate-choice between incipient species of Anopheles gambiae s.s. PLoS One. 2011;6:e27920. doi: 10.1371/journal.pone.0027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, della Torre A, Caccone A. Short report: a new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am J Trop Med Hyg. 2004;70:604–606. [PubMed] [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Simard F, et al. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Slotman MA, et al. Evidence for subdivision within the M molecular form of Anopheles gambiae. Mol Ecol. 2007;16:639–649. doi: 10.1111/j.1365-294X.2006.03172.x. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. Interpreting the estimated timing of migration events between hybridizing species. Mol Ecol. 2011;20:2353–2366. doi: 10.1111/j.1365-294X.2011.05048.x. [DOI] [PubMed] [Google Scholar]
- Taylor C, et al. Gene flow among populations of the malaria vector, Anopheles gambiae, in Mali, West Africa. Genetics. 2001;157:743–750. doi: 10.1093/genetics/157.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet F, Dolo G, Lanzaro GC. Multilevel analyses of genetic differentiation in Anopheles gambiae s.s. reveal patterns of gene flow important for malaria-fighting mosquito projects. Genetics. 2005;169:313–324. doi: 10.1534/genetics.104.026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet F, et al. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol Ecol. 2001;10:1725–1732. doi: 10.1046/j.0962-1083.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW. Genomic islands of speciation or genomic islands and speciation? Mol Ecol. 2010;19:848–850. doi: 10.1111/j.1365-294X.2010.04532.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. Natural selection in action during speciation. Proc Natl Acad Sci U S A. 2009;106:9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, et al. Association mapping of insecticide resistance in wild Anopheles gambiae populations: major variants identified in a low-linkage disequilbrium genome. PLoS One. 2010;5:e13140. doi: 10.1371/journal.pone.0013140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Wilding CS, Steen K, Pinto J, Donnelly MJ. Gene flow-dependent genomic divergence between Anopheles gambiae M and S forms. Mol Biol Evol. 2012;29:279–291. doi: 10.1093/molbev/msr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010;19:925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci U S A. 2011;108:244–249. doi: 10.1073/pnas.1013648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]