Abstract
Type 1 diabetes (T1D) is an autoimmune disease targeting pancreatic beta cells. Genome-wide association studies and gene expression analysis identified interferon (IFN)-driven gene networks as crucial pathways in the pathogenesis of T1D. IFNs are linked to the response to viral infections and might contribute to the initiation of the autoimmune process in T1D. We presently analyzed the role of ubiquitin-specific peptidase 18 (USP18), an interferon-stimulated gene 15-specific protease, on IFN-induced pancreatic beta cell inflammation and apoptosis. Our findings indicate that USP18 inhibition induces inflammation by increasing the STAT signaling and exacerbates IFN-induced beta cell apoptosis by the mitochondrial pathway of cell death. USP18 regulates activation of three BH3-only proteins, namely, DP5, Bim and PUMA in pancreatic beta cells, suggesting a direct link between regulators of the type I IFN signaling pathway and members of the BCL-2 family. USP18 depletion increases the expression of the T1D candidate gene MDA5, leading to an upregulation of double-stranded RNA-induced chemokine production. These data suggest a cross talk between the type I IFN signaling pathway and a candidate gene for T1D to increase pro-inflammatory responses in beta cells. The present study shows that USP18 is a key regulator of IFN signaling in beta cells and underlines the importance of this pathway in beta cell inflammation and death.
Keywords: USP18, type 1 diabetes, beta cells, type I IFNs, inflammation, apoptosis
Type 1 diabetes (T1D) is a chronic autoimmune disease in which pancreatic beta cells are destroyed by the immune system. The familial clustering of T1D is explained by multiple common gene variants, each providing a modest risk but potentially associated to specific molecular mechanisms.1 Integration of genome-wide association studies (GWAS) with global gene expression analysis have identified interferon (IFN)-driven gene networks, potentially linked to the response to viral infections, as crucial regulating pathways in the pathogenesis of T1D.2 Several of the genes present in these networks are expressed in pancreatic beta cells,3 reinforcing the concept of a ‘dialog' between these cells and the immune system in the early stages of T1D.4
Type I IFNs are important mediators of antiviral responses and their implication in the pathogenesis of T1D is supported by human data.5 IFNs might contribute to the initiation and acceleration of the autoimmune process in T1D by inducing activation of dendritic cells, proliferation of T cells and direct beta cell apoptosis.4, 6 There is a positive correlation between elevated levels of IFNα in blood and T1D associated with enterovirus infections,7 and histological studies indicate that pancreatic beta cells express IFNα in almost all cases of new onset T1D.8 Moreover, observations in diabetes-prone nonobese diabetic mice indicate that neutralizing the type I IFN receptor (IFNAR) or IFNα decreases the prevalence of diabetes.9, 10 The loci that regulate IFN signaling pathways are important for beta cell dysfunction and death, contributing to disease pathogenesis. Thus, we have recently shown that PTPN2 (tyrosine-protein phosphatase non-receptor type 2), a T1D candidate gene, is implicated in IFN-induced beta cell death via regulation of the mitochondrial pathway of cell death.11 Other key mediators of IFN signaling such as the transcription factor STAT1 (signal transducer and activator of transcription factor 1)12 also have an important role in the modulation of beta cell inflammation and death. Thus, knockout of STAT1 gene suppresses development of diabetes in the nonobese diabetic mice13 and protects against streptozotocin-induced diabetes.14 These data suggest that regulators of type I IFN signaling pathways may be potential therapeutic targets for the prevention of T1D.
Against this background, it is important to understand: (1) how is IFN signaling regulated at the pancreatic beta cell level; (2) how can this regulation modulate beta cell death and the generation of signals that contribute to trigger and amplify the local inflammation (insulitis); and (3) how can this regulation cross talk with relevant candidate genes for T1D.
Ubiquitin-specific peptidase 18 (USP18; UBP43) is a major IFN-stimulated gene 15 (ISG15)-specific protease and its activity is crucial for proper regulation of ISG15-conjugated proteins.15 ISG15 is an IFN-stimulated ubiquitin-like protein (Ubl) that conjugates to a number of cellular substrates via an ubiquitination-like process named ‘ISGylation'.16 Both USP18 expression and conjugation of ISG15 are strongly induced by viral infections and type I IFNs,17, 18 suggesting that protein modifications by USP18-regulated ISG15 have a role in responses to viruses and type I IFN signaling.16
We presently analyzed the role of USP18 in type I IFN-induced beta cell inflammation and apoptosis. Our findings indicate that USP18 inhibition increases IFN-induced beta cell apoptosis via the mitochondrial pathway of cell death and increases inflammation by exacerbating the STAT signaling pathway and the expression of the T1D candidate gene MDA5 (melanoma differentiation-associated protein 5). These data clarify the role of USP18 as a master regulator of type I IFN signaling in beta cells and underlines the importance of this pathway in beta cell inflammation and death in T1D.
Results
Cytokines upregulate USP18 expression in INS-1E cells and primary rat beta cells
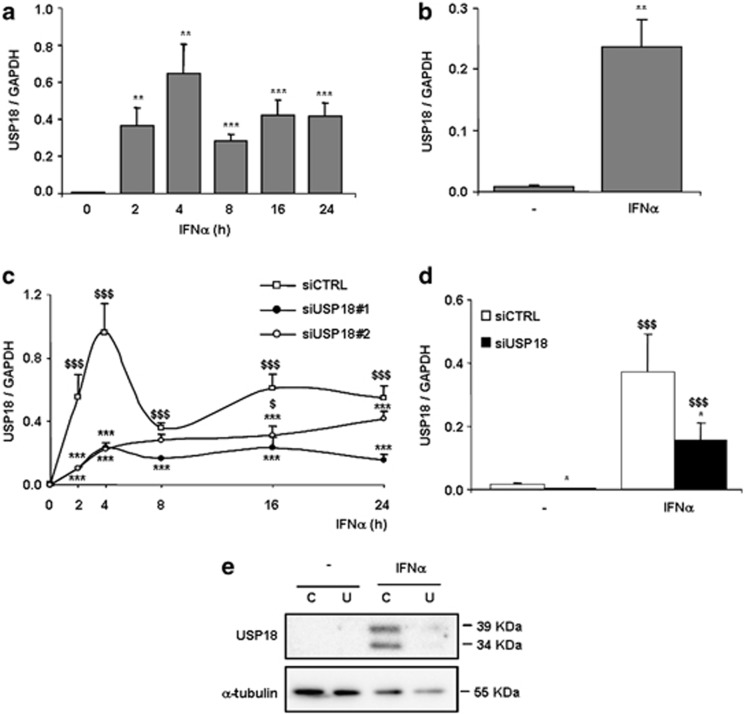
Untreated INS-1E cells showed low USP18 mRNA expression, but IFNα upregulated USP18 expression from 2 to 24 h (Figure 1a). The basal expression of USP18 in primary beta cells was higher than in INS-1E cells, and IFNα treatment increased its expression by nearly 30-fold after 48 h (Figure 1b). We also observed an upregulation in USP18 expression by IFNγ alone or in combination with interleukin (IL)-1β or tumor necrosis factor-α in INS-1E cells (Supplementary Figures S1A–C). During viral infections, beta cells release type I IFNs that act via IFNAR.11 We have previously shown that polyinosinic:polycytidylic acid (PIC; a synthetic viral double-stranded RNA (dsRNA) analog)-induced beta cell death is partially mediated by type I IFNs released by the beta cells themselves.11 Interestingly, a time course of PIC transfection in INS-1E cells indicated that USP18 mRNA expression was already induced after 2 h, increasing progressively up to 24 h (Supplementary Figure S1D).
Figure 1.
IFNα upregulates USP18 mRNA expression in INS-1E cells, primary rat beta cells and dispersed human islet cells, and specific silencing by siRNA inhibits basal and IFNα-induced USP18 expression. (a, b) INS-1E cells (a) or primary rat beta cells (b) were left untreated or treated with IFNα (1000 U/ml) for 2, 4, 8, 16 or 24 h (a) or for 48 h (b). USP18 mRNA expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. Results are means±S.E.M. of 4–7 independent experiments; **P< 0.01 and ***P< 0.001 versus time 0 h (i.e., not treated with cytokines); Student's t test. (c, d) INS-1E cells (c) or primary rat beta cells (d) were transfected with siCTRL or with siRNAs targeting USP18 (USP18#1 and #2). After 48 h of recovery, cells were left untreated or treated with IFNα for 2, 4, 8,16 or 24 h (c) or for 48 h (d). USP18 mRNA expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. Results are means±SEM of 3–6 independent experiments; $P< 0.05 and $$$P< 0.001 versus time 0 h (i.e., not treated with cytokines) transfected with the same siRNA; *P<0.05 and ***P< 0.001 versus siCTRL treated with IFNα at the same time point; ANOVA followed by Student's t test or ratio t test with Bonferroni correction. (e) Human-dispersed islets were tranfected with siCTRL (C) or siUSP18 (U) and treated with IFNα (2000 U/ml) for 48 h. USP18 and α-tubulin (as loading control) were evaluated by western blot. The results are representative of three independent experiments
Two different small interfering RNAs (siRNAs) targeting USP18 (siUSP18#1 and siUSP18#2) efficiently decreased USP18 mRNA expression in untreated condition and after treatment with IFNα at different time points (Figure 1c). In primary rat beta cells, siUSP18#1 inhibited USP18 mRNA expression by 80% and 60%, respectively, in the untreated condition and after 48 h of IFNα treatment (Figure 1d).
In human-dispersed islets, USP18 protein expression was undetectable by Western blot under basal condition (e.g., not treated with IFNα). A 48 h treatment with IFNα, however, upregulated its expression and this was efficiently inhibited (>70%) by a specific siRNA against human USP18 (Figure 1e).
USP18 inhibition activates IFNα-induced STAT signaling pathway and increases pro-inflammatory chemokine production in pancreatic beta cells
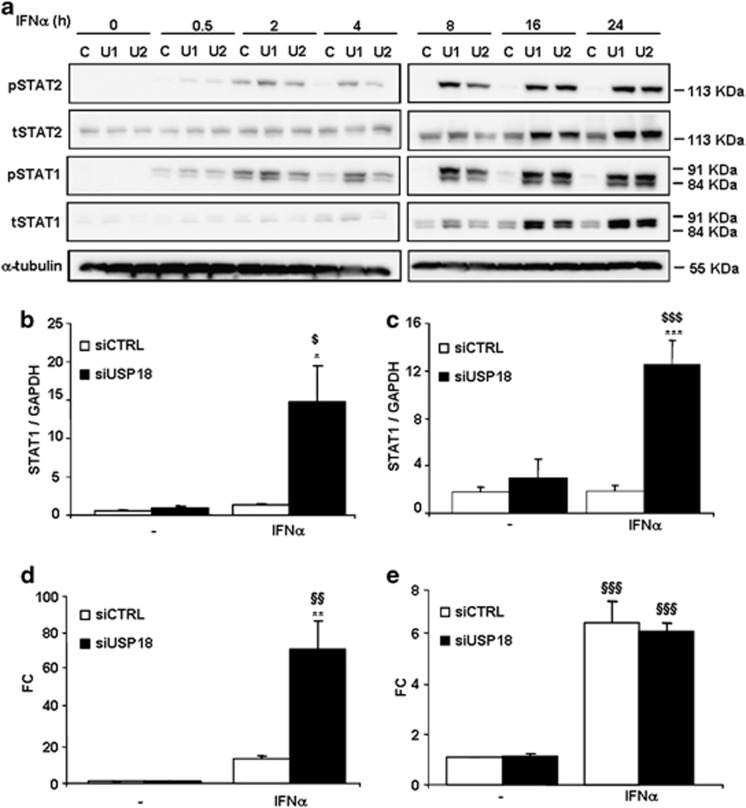
We next examined the effect of USP18 inhibition on the kinetics of IFNα-induced STAT signaling activation. IFNα-induced STAT1 and STAT2 phosphorylation was markedly enhanced in INS-1E cells exposed to siUSP18 after 2–4 h of IFNα treatment, reaching a peak after 8 h that was maintained until 24 h (Figure 2a). USP18 inhibition increased IFNα-induced total STAT1 and STAT2 protein expression after 8 h, and this effect was prolonged up to 24 h after IFNα treatment (Figure 2a). STAT1 mRNA expression was also upregulated after treatment with IFNα in USP18-inhibited INS1-E cells (Figure 2b) and primary rat beta cells (Figure 2c).
Figure 2.
USP18 inhibition increases IFNα-induced STAT signaling pathway activation and ISRE reporter activity. (a) INS-1E cells were transfected with siCTRL (C) or two different siRNAs against USP18 (U1 and U2). After 2 days of recovery, they were left untreated or treated with IFNα (1000 U/ml) for 0.5, 2, 4, 8, 16 or 24 h. Phospho-STAT1, total STAT1, phospho-STAT2, total STAT2 and α-tubulin (used as loading control) were evaluated by western blot. The results are representative of three independent experiments. (b,c) INS-1E cells (b) or primary rat beta cells (c) were transfected with siCTRL or siUSP18 and, after 48 h of recovery, they were left untreated or treated with IFNα for 24 h (b) or 48 h (c). STAT1 expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. The results are means±S.E.M. of 3–4 independent experiments; $P<0.05 and $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; *P<0.05 and ***P<0.001 versus siCTRL treated with IFNα; ANOVA. (d,e) INS-1E cells were transfected with siCTRL or siUSP18 and co-transfected with an ISRE (d) or GAS reporter construct (e) plus a pRL-CMV plasmid (used as internal control). After 12 h of recovery, they were left untreated or treated with IFNα for 16 h and luciferase activities were measured. Results are means±S.E.M. of three independent experiments; $$P<0.01 and $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; **P<0.01 versus siCTRL treated with IFNα; ANOVA
Exposure to IFNα increased IFN-stimulated response element (ISRE) reporter activity by 12-fold in INS-1E cells, and inhibition of USP18 further enhanced this effect by 65-fold (Figure 2d). However, IFNα-induced GAS (IFNγ-activated site) reporter activity was not affected by USP18 knockdown (Figure 2e).
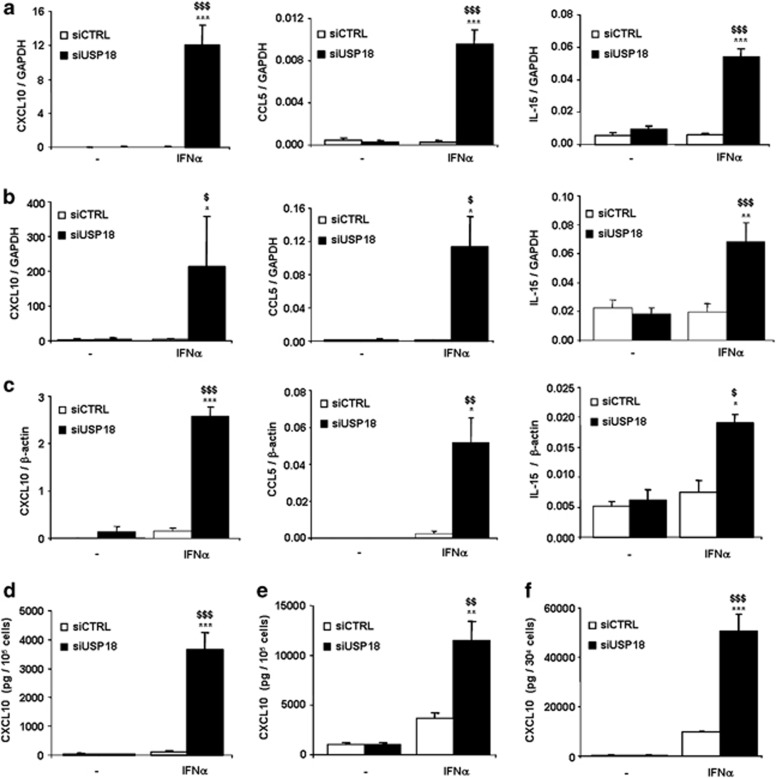
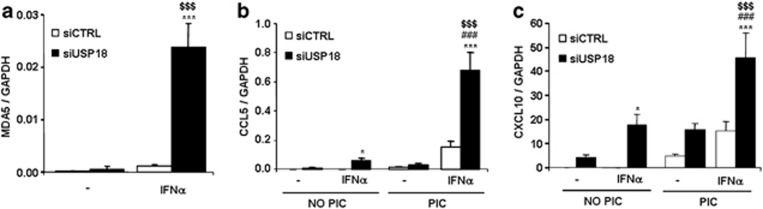
To evaluate the role of USP18 in IFNα-induced chemokine production, we analyzed the mRNA expression of CXCL10 (C-X-C motif chemokine 10), CCL5 (chemokine (C-C motif) ligand 5) and IL-15 in USP18-silenced INS-1E cells (Figure 3a), primary rat beta cells (Figure 3b) and human-dispersed islet cells (Figure 3c). Treatment with IFNα for 24 h or 48 h did not affect chemokine mRNA expression in siCTRL-transfected INS-1E cells, but inhibition of USP18 exacerbated IFNα-induced expression of CXCL10, CCL5 and IL-15 by 280-, 35- and 9-fold, respectively (Figure 3a). These results were confirmed in primary rat beta cells (Figure 3b) in which USP18 inhibition upregulated CXCL10, CCL5 and IL-15 mRNA expression. IFNα-induced CXCL10, CCL5 and IL-15 expression was also upregulated by 18-, 21- and 2.5-fold, respectively, in USP18-silenced dispersed human pancreatic islets (Figure 3c).
Figure 3.
Inhibition of USP18 exacerbates IFNα-induced chemokine expression in INS-1E cells, primary rat beta cells and human-dispersed islet cells. (a–c) INS-1E cells (a), primary rat beta cells (b) and dispersed human islet cells (c) were transfected with siCTRL or siUSP18 and, after 48 h of recovery, they were left untreated or treated with IFNα for 24 h (a) or 48 h (b, c). CXCL10, CCL5 and IL-15 were assayed by RT-PCR and normalized for the housekeeping genes GAPDH (a, b) or β-actin (c). The results are means±S.E.M. of 3–7 independent experiments; $P<0.05, $$P<0.01 and $$$P<0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; *P<0.05, **P<0.01 and ***P<0.001 versus siCTRL treated with IFNα; ANOVA. (d–f) Cells were transfected with siCTRL or siUSP18. After 48 h of recovery, they were treated with IFNα and CXCL10 secretion was evaluated in INS-1E cells (n=4; d), primary rat beta cells (n=3; e) and human-dispersed islet cells (n=4; f) using an ELISA kit; $$P<0.01 and $$$P<0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; **P<0.01 and ***P<0.001 versus siCTRL treated with IFNα; ANOVA
The upregulation of CXCL10 mRNA expression in USP18-inhibited INS-1E, primary rat beta and human-dispersed islet cells was confirmed at the protein level, as USP18-silenced cells secreted higher amounts of CXCL10 compared with control cells after IFNα treatment (Figures 3d–f). These data indicate an inhibitory role of USP18 in the expression of IFNα-induced pro-inflammatory chemokines in pancreatic beta cells.
The increase in chemokine expression in USP18-silenced cells is mediated by the transcription factors STAT1 and STAT2
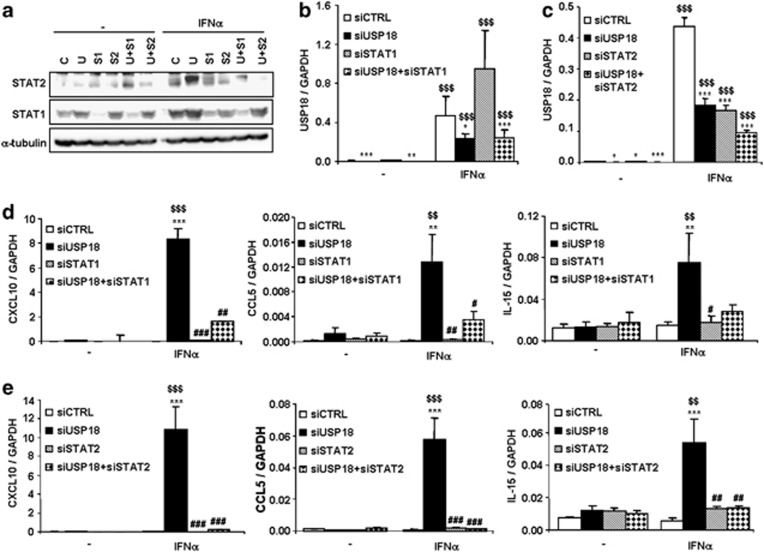
To evaluate the implication of STAT1 and STAT2 in IFNα-induced chemokine expression in USP18-inhibited cells, we performed a double-knockdown approach to silence USP18 in combination with STAT1 or STAT2. As shown in Figure 4a, the individual siRNAs targeting STAT1 or STAT2 efficiently inhibited their target protein alone or in combination with siUSP18. The siRNA targeting USP18 inhibited its mRNA expression by 50–78%, alone or when combined with STAT1 (Figure 4b) or STAT2 (Figure 4c). As shown above (Figure 3), inhibition of USP18 exacerbated IFNα-induced chemokine expression but double knockdown of USP18 and STAT1 (Figure 4d) or STAT2 (Figure 4e) reverted this effect for all the chemokines tested.
Figure 4.
The transcription factors STAT1 and STAT2 mediate IFNα-induced chemokine production in USP18-silenced INS-1E cells. (a–c) INS-1E cells were transfected with siCTRL (C), siUSP18 (U), siSTAT1 (S1) or siSTAT2 (S2) or double transfected with siUSP18+siSTAT1 (U+S1) or siUSP18+siSTAT2 (U+S2) and, after 48 h of recovery, they were left untreated or treated with IFNα for 24 h. (a) Protein expression of STAT1, STAT2 and α-tubulin (as loading control) was determined by western blot analysis to confirm the inhibition efficiency of siRNAs targeting STAT1 and STAT2, alone or in combination with siUSP18. The results are representative of 3 independent experiments. (b, c) Inhibition efficiency of siUSP18 alone or in combination with siSTAT1 (b) or siSTAT2 (c) was determined by USP18 mRNA expression analysis by RT-PCR. Expression results were normalized for the housekeeping gene GAPDH. The results are means±S.E.M. of three independent experiments; $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; *P<0.05, **P<0.01 and ***P<0.001 versus siCTRL; ANOVA. (d,e) INS-1E cells were transfected with siCTRL, siUSP18, siSTAT1 or siSTAT2 or double transfected with siUSP18+siSTAT1 (d) or siUSP18+siSTAT2 (e). After 48 h of recovery, they were left untreated or treated with IFNα for 24 h and CXCL10, CCL5 and IL-15 expression assayed by RT-PCR and normalized for the housekeeping gene GAPDH. The results are means±S.E.M. of three independent experiments; $$P<0.01 and $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; **P<0.01 and ***P<0.001 versus siCTRL treated with IFNα; #P<0.05 ##P<0.01 and ###P<0.001 versus siUSP18 treated with IFNα; ANOVA
Knockdown of USP18 also exacerbated mRNA expression of the transcription factor IRF7 (interferon regulatory factor 7) in USP18-inhibited INS-1E cells (Supplementary Figure S2A). The increase in IRF7 expression in USP18-silenced cells was reverted after double inhibition of USP18 and STAT1 (Supplementary Figure S2B) or STAT2 (Supplementary Figure S2C), suggesting that IRF7 is downstream of the STAT signaling pathway. IRF7 is a major regulator of chemokine production in beta cells.19 To assess whether IRF7 mediates chemokine production in USP18-silenced cells, we silenced USP18 and IRF7 (using a previously validated siRNA against IRF719). Knockdown of USP18 and IRF7 (between 30 and 61%) was confirmed by RT-PCR (Supplementary Figures S2D and E). USP18 inhibition increased IFNα-induced mRNA expression of CXCL10, CCL5 and IL-15; however, double knockdown of USP18 and IRF7 failed to revert this exacerbation in chemokine expression (Supplementary Figure S2F). These results suggest that IRF7 is not a key mediator in IFNα-induced chemokine upregulation in USP18-inhibited cells.
Inhibition of USP18 in INS-1E cells exacerbates IFNα-induced mRNA expression of the T1D candidate gene MDA5
MDA5 (IFIH1) is a T1D candidate gene, and we have previously shown that it has a crucial role in dsRNA-induced chemokine production in pancreatic beta cells.20 IFNα did not induce MDA5 expression in siCTRL-tranfected cells, but MDA5 mRNA was upregulated by 24-fold in USP18-inhibited INS-1E cells after IFNα treatment (Figure 5a). To test the functional significance of MDA5 mRNA upregulation in USP18-inhibited cells, we analyzed CXCL10 and CCL5 mRNA expression under IFNα and PIC (synthetic dsRNA) treatment. Inhibition of USP18 increased IFNα-induced CCL5 and CXCL10 expression by 120- and 280-fold, respectively, and this effect was further exacerbated (by 12- and 2.6-fold) after 10 h of PIC transfection (Figures 5b and c).
Figure 5.
USP18 inhibition upregulates IFNα-induced expression of MDA5 and amplifies PIC-induced chemokine expression. (a) INS-1E cells were transfected with siCTRL or siUSP18 and, after 48 h of recovery, they were left untreated or treated with IFNα for 24 h. MDA5 expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. The results are means±S.E.M. of four independent experiments; $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; ***P<0.001 versus siCTRL treated with IFNα; ANOVA. (b and c) INS-1E cells were transfected as in (a) and after 48 h of recovery they were left untreated or treated with IFNα for 24 h. They were then left untransfected or transfected with PIC for 10 h. CCL5 and CXCL10 were analyzed by RT-PCR and normalized for GAPDH. The results are means±S.E.M. of six independent experiments; $$$P< 0.001 versus siUSP18 transfected with PIC and non-treated with IFNα; *P<0.05 and ***P<0.001 versus siCTRL under the same treatment; ###P<0.001 versus siUSP18 non-transfected with PIC and treated with IFNα; ANOVA
Inhibition of USP18 by specific siRNAs exacerbates IFN- and PIC-induced beta cell apoptosis
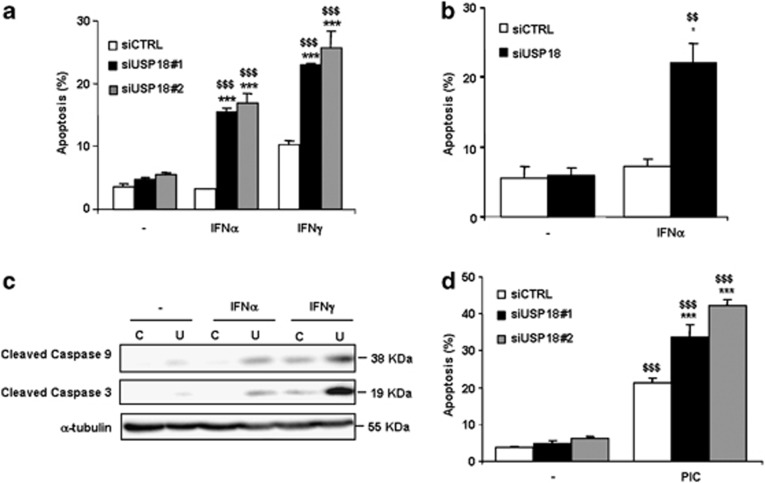
USP18 inhibition by two independent siRNAs increased beta cell apoptosis following exposure to IFNα or IFNγ (Figure 6a), whereas siCTRL has no such effect. USP18 inhibition also increased apoptotic cell death by nearly 4-fold in primary rat beta cells after 48 h of treatment with IFNα (Figure 6b). Inhibition of USP18 in INS-1E cells also increased cleaved caspases-9 and -3 after 24 h of IFNα treatment (Figure 6c), suggesting apoptosis via the mitochondria-driven intrinsic pathway of cell death.
Figure 6.
Inhibition of USP18 exarcebates IFNα-, IFNγ- and PIC-induced cell apoptosis in INS-1E cells and primary rat beta cells. (a, b) INS-1E cells (a) or primary rat beta cells (b) were transfected with siCTRL or siUSP18. After 48 h of recovery, INS-1E cells were left untreated or treated with IFNα (1000 U/ml) or IFNγ (100 U/ml) for 24 h (a) and primary rat beta cells with IFNα for 48 h (b). Apoptosis was measured using HO/PI staining. Results are means±S.E.M. of 3–5 independent experiments; $$P<0.01 and $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; ***P<0.001 versus siCTRL treated with the same cytokine; ANOVA. (c) INS-1E cells were transfected with siCTRL (C) or siUSP18 (U) and exposed to IFNα or IFNγ for 24 h. Expression of cleaved caspases-9 and -3 and α-tubulin (used as loading control) were measured by western blot. Results are representative of three independent experiments. (d) INS-1E cells were transfected as in (a). After 48 h of recovery, they were left untransfected or transfected with PIC for 24 h. Apoptosis was measured using HO/PI staining. Results are means±S.E.M. of three independent experiments; $$$P< 0.001 versus untreated (i.e., not transfected with PIC); ***P<0.001 versus siCTRL transfected with PIC; ANOVA
Transfection with PIC increased beta cell death, and this effect was also exacerbated after inhibition of USP18 by two different siRNAs (Figure 6d).
IFNα-induced beta cell apoptosis in USP18-silenced cells is mediated via three members of the BH3-only protein family
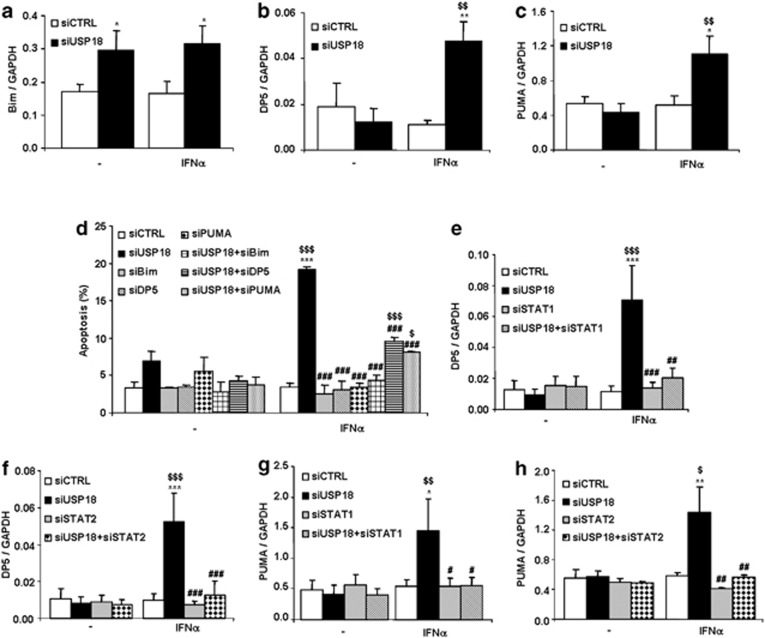
Death protein 5 (DP5), p53 upregulated modulator of apoptosis (PUMA) and BCL2-like protein 11 (Bim) are members of the BCL-2 (B-cell lymphoma 2) family and have been shown to contribute to cytokine-induced beta cell death.11, 21, 22 USP18 inhibition in INS-1E cells induced Bim expression in untreated and IFNα-treated conditions (Figure 7a), whereas DP5 (Figure 7b) and PUMA (Figure 7c) mRNA expression was significantly upregulated when USP18-silenced cells were exposed to IFNα. Post-transcriptional phosphorylation of Bim at a serine residue increases its pro-apoptotic activity in beta cells,11 but USP18 inhibition in INS-1E cells did not increase Bim phosphorylation at serine 65 (data not shown).
Figure 7.
Double knockdown of USP18 and Bim, DP5 or PUMA protects INS-1E cells against IFNα-induced apoptosis. (a–c) INS-1E cells were transfected with siCTRL or siUSP18 and, after 48 h of recovery, they were left untreated or treated for 24 h with IFNα. Expression of Bim (a), DP5 (b) and PUMA (c) were assayed by RT-PCR and normalized for the housekeeping gene GAPDH. Results are means±S.E.M. of 11 independent experiments; $$P<0.01 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; *P<0.05 and **P<0.01 versus siCTRL treated with IFNα; ANOVA. (d) Cells were transfected with siCTRL, siUSP18, siBim, siDP5, siPUMA or combinations of siUSP18 and siBim, siDP5 or siPUMA. After 48 h of recovery, they were left untreated or treated with IFNα for 24 h. Apoptosis was measured using HO/PI staining. Results are means±S.E.M. of three independent experiments; $P<0.05 and $$$P< 0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; ***P<0.001 versus siCTRL treated with IFNα; ###P<0.001 versus siUSP18 treated with IFNα; ANOVA. (e–h) Cells were transfected with siCTRL, siUSP18, siSTAT1 (e and g) or siSTAT2 (f and h) or double transfected with siUSP18+siSTAT1 (e and g) or siUSP18+siSTAT2 (f and h). After 48 h of recovery, they were left untreated or treated with IFNα for 24 h and DP5 or PUMA mRNA expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. The results are means±S.E.M. of 3–5 independent experiments; $P<0.05, $$P<0.01 and $$$P<0.001 versus untreated (i.e., not treated with cytokines) transfected with the same siRNA; *P<0.05, **P<0.01 and ***P<0.001and versus siCTRL treated with IFNα, #P<0.05, ##P<0.01 and ###P<0.001 versus siUSP18 treated with IFNα; ANOVA
To clarify whether Bim, DP5 and PUMA contribute to IFNα-induced beta cell death in USP18-silenced cells, we silenced USP18 and each of the three BH3-only proteins in a double-knockdown approach. All siRNAs used have been previously validated,21, 22, 23 and they presently inhibited all target mRNAs as expected (Supplementary Figure S3). Inhibition of USP18 significantly exacerbated beta cell apoptosis after treatment with IFNα (Figure 7d), confirming the findings from Figure 6a. This effect was counteracted by DP5, PUMA and Bim knockdown, which protected INS-1E cells against IFNα-induced apoptosis in USP18-silenced cells by 47%, 58% and 78%, respectively (Figure 7d).
Upregulation of Bim and DP5 mRNA expression by pro-inflammatory cytokines is partially mediated by STAT1 in beta cells.12, 23 To asses whether STAT1 or STAT2, which are both upregulated by USP18 inhibition (Figure 2), are implicated in the upregulation of Bim, DP5 and PUMA mRNA expression in USP18-silenced cells, we inhibited in parallel USP18 and STAT1 or STAT2 in INS-1E cells by use of specific siRNAs. USP18 depletion significantly upregulated DP5 and PUMA expression after IFNα treatment, and this effect was totally reversed by double knockdown of USP18 and STAT1 (Figures 7e and g) or STAT2 (Figures 7f and h), suggesting that IFNα-induced DP5 and PUMA upregulation in USP18-inhibited cells is controlled by STAT1 and STAT2. Inhibition of STAT1 and STAT2 also induced a decrease in Bim expression following USP18 silencing and IFNα treatment, but this effect was less marked (Supplementary Figure S4).
We further analyzed the implication of STAT1 and STAT2 in IFNα-induced cell death by a double-knockdown approach to inhibit USP18 and STAT1 or STAT2. As shown in Supplementary Figure S5, siSTAT1 (Supplementary Figures S5A and B) and siSTAT2 (Supplementary Figures S5A and B) reverted the potentiating effects of USP18 knockdown in IFNα-induced cell death both in INS-1E (Supplementary Figures S5A and B) and primary rat beta cells (Supplementary Figures S5C and D). These results suggest that STAT signaling pathway has a crucial upstream role in IFNα-induced apoptosis following USP18 silencing.
Discussion
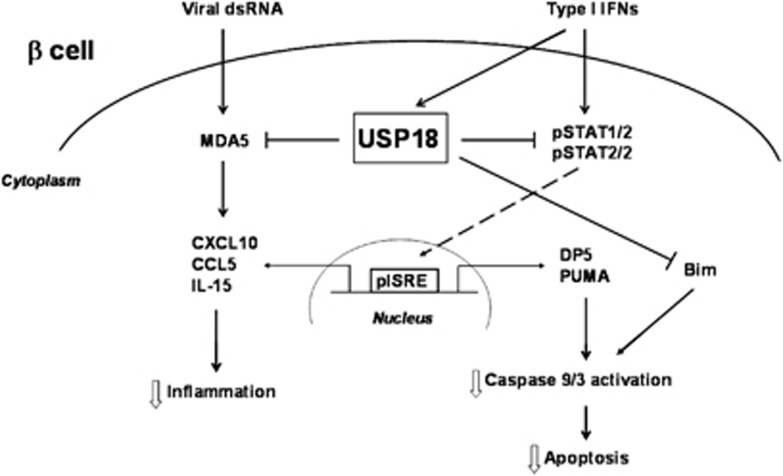
The current study shows that USP18 is a key regulator of type I IFN signaling pathway in pancreatic beta cells and its inhibition contributes to IFN-induced beta cell apoptosis and inflammation (Figure 8). Thus, we show that inhibition of USP18 exacerbates pro-inflammatory chemokine production via induction of the STAT signaling pathway and increases IFN-induced beta cell apoptosis by activation of the mitochondrial-driven pathway of cell death. Moreover, we demonstrate a direct cross talk between a candidate gene for T1D (MDA5) and the USP18-regulated type I IFN signaling pathway that amplifies the viral dsRNA-induced chemokine production.
Figure 8.
A model for the central role of USP18 in the regulation of type I IFN signaling pathways in pancreatic beta cells. Type I IFNs induce USP18 expression leading to a negative feedback on the STAT signaling pathway. This negative feedback decreases IFNα-induced chemokine production and prevents excessive inflammatory response in pancreatic beta cells. USP18 also inhibits STAT-regulated expression of DP5 and PUMA, and, by an unknown mechanism, expression of Bim. Inhibition of DP5, PUMA and Bim decreases caspases 9/3 activation, reducing IFNα-induced beta cell apoptosis. Interestingly, USP18 inhibits the expression of the T1D candidate gene MDA5, potentially decreasing the proinflammatory response of beta cells to a viral infection
USP18 is a 43-kDa protein highly upregulated by type I IFNs, viral infections and bacterial lipopolysaccharide.24, 25 We presently show that IFNα and dsRNA induce USP18 expression in human islets, primary rat beta cells and INS-1E cells, supporting its role in IFN-related signaling pathways and antiviral responses in pancreatic beta cells.
Low and constitutive expression of type I IFNs have been detected in many cell types,26 and their pleiotropic activities are highly regulated in time and space by several mechanisms that coexist to attenuate IFN-initiated STAT signaling.27 Upon binding of type I IFNs to their cell surface receptor (IFNAR), the tyrosine kinases Tyk2 and JAK1 are autophosphorylated and activated, leading to phosphorylation of the transcription factors STAT1 and STAT2.28 In USP18-silenced beta cells (present data), activation of IFNα-induced STAT signaling pathway is exacerbated and leads to an increase in ISRE reporter activity. These results are in line with previous studies in USP18 knockout mice in which IFNα induced long-lasting activation of JAK-STAT signaling29 and increased IFN-inducible gene expression.30
Increased STAT signaling pathway in USP18-inhibited beta cells leads to an upregulation of several pro-inflammatory chemokines, including CCL5, CXCL10 and IL-15. This is in line with previous findings that bone marrow-derived macrophages of USP18 knockout mice have increased chemokine and chemokine receptor expression after exposure to IFNβ.30 Of note, CXCL10 is the most expressed chemokine in USP18-silenced rodent and human beta cells (present data). CXCL10 is a key chemokine expressed in vivo in islets T1D patients,31 and human insulitis is characterized by expression of CXCL10 in endocrine cells, while the infiltrating lymphocytes express the corresponding chemokine receptor CXCR3.32
Chemokine expression in USP18-inhibited cells is regulated by STAT1 and STAT2 (present data). STAT1 has an important role in cytokine-induced inflammation in beta cells by regulating the expression of several pro-inflammatory chemokines, such as CXCL10, CXCL9, CXCL1 and CCL20.12 STAT1 was originally described as a transcription factor essential for type I IFN signaling in other cell types, but recent findings in STAT1 knockout mice indicate the presence of STAT2-mediated antiviral mechanisms.33 We presently observed that STAT2 has a key role in the regulation of chemokine expression in USP18-silenced beta cells, and its inhibition prevents USP18-induced chemokine expression. Unphosphorylated STAT1 and STAT2 are also upregulated by USP18 knockdown, suggesting their possible implication in IFNα-induced chemokine expression. Unphosphorylated STATs may prolong the expression of several IFN-induced genes,34 but whether unphosphorylated STATs have a role in increased chemokine expression in USP18-inhibited pancreatic beta cells needs further investigation.
In recent years, >50 candidate genes have been identified for human T1D;35 many of them are related to type I IFN signaling and antiviral responses.2 The expression of the T1D candidate gene MDA5 in USP18-silenced cells is highly increased after IFNα treatment (present data) and this upregulation amplifies dsRNA-induced chemokine production, suggesting a cross talk between the type I IFN signaling pathway and a candidate gene for T1D that increase pro-inflammatory responses in beta cells. Thus, combination of a viral infection with upregulation of the type I IFN signaling pathway might lead to an excessive immune response against beta cells in genetically susceptible individuals (e.g., individuals with risk alleles in MDA5), increasing the risk of autoimmunity and T1D.
Exacerbation of STAT signaling pathway and chemokine production in USP18-silenced beta cells is accompanied by exacerbation of cell death. In fact, USP18 knockdown contributes to IFN- and PIC-induced caspases-9 and -3 activation and beta cell apoptosis. Activation of caspase-9 suggests triggering of the mitochondrial pathway of cell death, characterized by the lost of mitochondrial membrane integrity, mitochondrial cytochrome c release to the cytosol and activation of caspases-9 and -3.36 Mitochondrial integrity is controlled by interactions between pro- and antiapoptotic members of the Bcl-2 protein family and several of these proteins are crucial regulators of beta cell apoptosis induced by pro-inflammatory cytokines or lipotoxicity.37 In USP18-silenced cells, mRNA expression of the BH3-only proteins DP5 and PUMA is highly enhanced after IFNα treatment, whereas Bim expression is upregulated in USP18-depleted cells in both untreated and IFNα-treated cells. It is noteworthy that upregulation of Bim mRNA expression secondary to USP18 inhibition is not sufficient to trigger beta cell apoptosis in the absence of IFNα. This suggests that increased basal expression of Bim by USP18 knockdown together with signals provided by the other BH3-only proteins, for example, PUMA and DP5, are required to overcome anti-apoptotic signals provided by BCL-2, MCL-1 and BCL-XL and thus trigger beta cell death in USP18-silenced cells. In line with this hypothesis, double knockdown of USP18 and DP5, PUMA or Bim protects against USP18 knockdown-induced beta cell death. This suggests the implication of the intrinsic mitochondrial pathway of cell death in this process and emphasizes the need for multiple pro-apoptotic signals to initiate beta cell apoptosis. DP5 and PUMA expression is regulated by the transcription factors STAT1 and STAT2 in USP18-silenced cells. In fact, double inhibition of USP18 and STAT1 or STAT2 reverted the supportive effect of USP18 knockdown in IFNα-induced beta cell apoptosis. Both DP5 and PUMA have been previously shown to be implicated in cytokine- and ER stress-induced beta cell death,21, 22 and their transcriptional activation has been shown to be regulated by NF-κB22 and STAT1,12 respectively. This is the first study, however, reporting the implication of both STAT1 and STAT2 in IFNα-induced DP5 and PUMA transcriptional regulation.
The current study shows that USP18 has a crucial role in type I IFN signaling in pancreatic beta cells via regulation of the STAT signaling pathway (Figure 8). Under physiological conditions, USP18 inhibits STAT signaling, decreasing IFNα-induced chemokine production and activation of several members of the BH3-only protein family. There is also a cross talk between a candidate gene for T1D (MDA5) and the USP18-regulated type I IFN signaling pathway that amplifies the viral dsRNA-induced chemokine production. These data suggest that a perturbation of the type I IFN signaling in beta cells may induce an excessive inflammatory process and increased activation of proapoptotic signaling pathways, resulting in increased beta cell inflammation and apoptosis following, for instance, a local viral infection. These results further support the implication of type I IFN-driven pathways and antiviral responses in the pathogenesis of T1D and point out the regulators of these pathways as potential targets for pharmacological interventions in T1D.
Materials and Methods
Culture of FACS-purified rat beta cells, human islets and INS-1E cells
Male Wistar rats (Charles River Laboratories, Brussels, Belgium) were housed and used according to the Belgian Regulations for Animal Care and with permission from the local Ethical Committee. Rat islets were isolated by collagenase digestion and handpicked under a stereomicroscope. For beta cell isolation, islets were dispersed and beta cells were purified by FACS (FACSAria, BD Bioscience, San Jose, CA, USA) as described.38 The beta cell preparations used in this study contained 90±3% beta cells (n=6). Purified beta cells were cultured for 48 h in Ham's F-10 medium containing 10 mM glucose, 2 mM glutaMAX, 50uM isobutyl-L-methylxanthine, 5% fetal bovine serum, 0.5% charcoal-absorbed bovine serum albumin (bovine serum albumin fraction V, Boehringer, IN, USA), 50 U/ml penicillin and 50 μg/ml streptomycin. For cytokine treatment, cells were cultured in the same medium but without serum. For siRNA and PIC transfection, bovine serum albumin-free and antibiotic-free medium was used.
Human islets were isolated from four non-diabetic organ donors (age: 61±4 years; body mass index: 28±3 kg/m2) in Pisa, Italy, after approval of the local Ethics Committee. Islets were isolated by enzymatic digestion and density–density gradient purification.39 After overnight recovery in Ham's F-10 containing 6.1 mM glucose, 10% fetal bovine serum, 2 mM GlutaMAX, 50 μM 3-isobutyl-1-methylxanthine, 1% bovine serum albumin, 50 U/ml penicillin and 50 μg/ml streptomycin, islets were dispersed for treatment and viability assays as previously described.40 The percentage of beta cells, examined in the four dispersed islet preparations by staining with anti-insulin antibody (1:1000, Sigma, Bornem, Belgium) and donkey anti-mouse IgG rhodamine (1:200, Lucron Bioproducts, De Pinte, Belgium), was 48±7%. In all experiments, islets from each individual donor were counted as n=1. The results shown are thus mean±SEM of 3–4 independent donors.
The insulin-producing INS-1E cell line (a kind gift from Dr. C Wollheim, Center Medical Universitaire, Geneva, Switzerland) was cultured in RPMI 1640 GlutaMAX-I, 5% fetal bovine serum, 10 mM HEPES, 1 mM Na-pyruvate and 50 μM 2-mercaptoethanol.
RNA interference
The siRNAs used in the present study are listed in Supplementary Table 1. The optimal concentrations (30 nM), and conditions for siRNA transfection was established based on previous dose–response studies.41 Cells were transfected using Lipofectamine RNAiMAX lipid reagent (Invitrogen, Paisley, UK) as described.42 After transfection, cells were cultured for a 48 h recovery period and subsequently exposed to cytokines (see below).
Cytokine exposure and dsRNA transfection
The following cytokine concentrations were used based on dose–response experiments previously performed by our group (11, 38; unpublished data): recombinant rat IFNα (specific activity: 1 × 108 U/mg, PBL Biomedical Laboratories, Piscataway, NJ, USA) at 1000 U/ml, recombinant human IFNα at 2000 U/ml (specific activity: 1.8 × 108 U/mg, PeproTech, Rocky Hill, USA), recombinant rat IFNβ at 1000 U/ml (specific activity: 8 × 107 U/mg, PBL Biomedical Laboratories), recombinant rat IFNγ at 100 U/ml (specific activity: 2 × 107 U/mg, R&D Systems, Abingdon, UK), recombinant human IL-1β (specific activity 1.8 × 107 U/mg; a gift from C.W. Reinolds, National Cancer Institute, Bethesda, MD) at 10 U/ml and recombinant murine tumor necrosis factor-α (specific activity: 2 × 108 U/mg; Innogenetics, Gent, Belgium) at 1000 U/ml.
The synthetic dsRNA analog PIC (Invivogen, San Diego, CA, USA) was used at the final concentration of 1 μg/ml. PIC transfection was performed in the same conditions as described for siRNA but using 0.15 μl of Lipofectamine 2000.43
Assessment of cell viability
The percentage of living, apoptotic and necrotic cells was determined after 15 min incubation with the DNA-binding dyes propidium iodide (PI, 5 μg/ml, Sigma, Bornem, Belgium) and Hoechst 33342 (HO, 5 μg/ml, Sigma) as described.21, 42 Apoptosis was confirmed by western blot analysis of cleaved caspases 9 and 3 (see below).
mRNA extraction and RT-PCR
mRNA extraction and reverse transcription were performed as previously described38 and the real-time PCR amplification was done using SYBR Green and compared with a standard curve.38 Expression values were corrected for the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin; GAPDH expression in rat beta cells and β-actin in human samples are not affected by cytokine treatment.40 The primers used for mRNA expression analysis are listed in Supplementary Table 2.
Evaluation of chemokine accumulation in the medium by ELISA
INS-1E, rat primary beta and human-dispersed islet cells were transfected with siCTRL or siUSP18 and subsequently left untreated or treated with IFNα as described above. Supernatants from INS-1E and primary rat beta cells were collected after 24 h or 48 h of treatment for determination of CXCL10 using the commercially available ELISA kit for rat CXCL10 (Abnova, Taipei City, Taiwan). Supernatants from human-dispersed islet cells were collected after 48 h of treatment with IFNα and determination of CXCL10 was performed using an ELISA kit for Human CXCL10 (R&D Systems, Abingdon, UK).
Western blot analysis
Cells were washed with cold PBS and lysed in Laemmli buffer. Immunoblot analysis was performed with the antibodies listed in Supplementary Table 3 and immunoreactive bands were revealed using the SuperSignal West Femto chemiluminescent substrate (Thermo Scientific, Chicago, IL, USA), detected using a Bio-Rad Molecular Imager ChemiBoc XRS+ with ImageLab software (Bio-Rad, Nazareth Eke, Belgium).
Luciferase reporter assays
INS-1E cells (105) were plated in 24-well plates and transfected with siRNAs as described above. After 12 h of recovery, cells were co-transfected with pRL-CMV encoding Renilla luciferase (Promega, Madison, WI, USA) and either a firefly luciferase promoter-reporter construct containing three GAS consensus sequences (GAS reporter—LR0075, Panomics, Fremont, CA, USA) or four ISRE consensus sequences (ISRE reporter—LR0040). After 24 h of recovery and 16 h of treatment with IFNα, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) and corrected for the luciferase activity of the internal control plasmid, pRL-CMV.44
Statistical analysis
Data are presented as mean±SEM. Comparisons were performed by two-tailed paired Student's t-test, ratio t-test or ANOVA followed by Student's t-test with Bonferroni correction as indicated. A P-value <0.05 was considered statistically significant.
Acknowledgments
This work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS) Belgium, the Communauté Française de Belgique—Actions de Recherche Concertées (ARC) and the European Union (projects Naimit and BetaBat, in the Framework Program 7 of the European Community). IS is the recipient of a postdoctoral fellowship of the Department of Education of the Basque Country. FM was the recipient of a Post-Doctoral Fellowship from FNRS, Belgium. CB is supported by AIRC (IG-10437). We thank Dr. M.L. Colli and Dr. T.C. Nogueira for providing samples and for helpful discussions and Dr. F. Vigneron for help in viability assays. We thank A. Musuaya, S. Mertens and M. Pangerl from the Laboratory of Experimental Medicine, ULB, for excellent technical support.
Glossary
- BCL-2
B-cell lymphoma 2
- Bim
BCL2-like protein 11
- CCL5
chemokine (C-C motif) ligand 5
- CXCL10
C-X-C motif chemokine 10
- DP5
death protein 5
- dsRNA
double-stranded RNA
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GAS
IFNγ-activated site
- GWAS
genome-wide association study
- IFN
Interferon
- IFNAR
type I IFN receptor
- IL
interleukin
- IRF7
interferon regulatory factor 7
- ISG15
interferon-stimulated gene 15
- ISRE
IFN-stimulated response element
- MDA5
melanoma differentiation-associated protein 5
- PIC
polyinosinic:polycytidylic acid
- PUMA
p53 up-regulated modulator of apoptosis
- siRNA
small interfering RNA
- STAT
signal transducer and activator of transcription
- T1D
type 1 diabetes
- USP18
ubiquitin-specific peptidase 18
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5:e1000540. doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik DcL, SammethM, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- Dotta F, Censini S, van Halteren AG, Marselli L, Masini M, Dionisi S, et al. Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci USA. 2007;104:5115–5120. doi: 10.1073/pnas.0700442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Chehadeh W, Weill J, Vantyghem MC, Alm G, Lefebvre J, Wattre P, et al. Increased level of interferon-alpha in blood of patients with insulin-dependent diabetes mellitus: relationship with coxsackievirus B infection. J Infect Dis. 2000;181:1929–1939. doi: 10.1086/315516. [DOI] [PubMed] [Google Scholar]
- Huang X, Yuang J, Goddard A, Foulis A, James RF, Lernmark A, et al. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44:658–664. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- Stewart TA. Neutralizing interferon alpha as a therapeutic approach to autoimmune diseases. Cytokine Growth Factor Rev. 2003;14:139–154. doi: 10.1016/s1359-6101(02)00088-6. [DOI] [PubMed] [Google Scholar]
- Li Q, Xu B, Michie SA, Rubins KH, Schreriber RD, McDevitt HO. Interferon-alpha initiates type 1 diabetes in nonobese diabetic mice. Proc Natl Acad Sci USA. 2008;105:12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin I, Moore F, Colli ML, Gurzov EN, Marselli L, Marchetti P, et al. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic beta-cell apoptosis via regulation of the BH3-only protein Bim. Diabetes. 2011;60:3279–3288. doi: 10.2337/db11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore F, Naamane N, Colli ML, Bouckenooghe T, Ortis F, Gurzov EN, et al. STAT1 is a master regulator of pancreatic beta-cell apoptosis and islet inflammation. J Biol Chem. 2011;286:929–941. doi: 10.1074/jbc.M110.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim HS, Chung KW, Oh SH, Yun JW, Im SH, et al. Essential role for signal transducer and activator of transcription-1 in pancreatic beta-cell death and autoimmune type 1 diabetes of nonobese diabetic mice. Diabetes. 2007;56:2561–2568. doi: 10.2337/db06-1372. [DOI] [PubMed] [Google Scholar]
- Callewaert HI, Gysemans CA, Ladriere L, D'Hertog W, Hagenbrock J, Overbergh L, et al. Deletion of STAT-1 pancreatic islets protects against streptozotocin-induced diabetes and early graft failure but not against late rejection. Diabetes. 2007;56:2169–2173. doi: 10.2337/db07-0052. [DOI] [PubMed] [Google Scholar]
- Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- Ritchie KJ, Zhang DE. ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol. 2004;15:237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf IK, Houmani JL. Sample JT Epstein-Barr virus independent dysregulation of UBP43 expression alters interferon-stimulated gene expression in Burkitt lymphoma. PLoS One. 2009;4:e6023. doi: 10.1371/journal.pone.0006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortis F, Naamane N, Flamez D, Ladriere L, Moore F, Cunha DA, et al. Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes. 2010;59:358–374. doi: 10.2337/db09-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colli ML, Moore F, Gurzov EN, Ortis F, MDA5 EizirikDL. and PTPN2, two candidate genes for type 1 diabetes, modify pancreatic beta-cell responses to the viral by-product double-stranded RNA. Hum Mol Genet. 2010;19:135–146. doi: 10.1093/hmg/ddp474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurzov EN, Ortis F, Cunha DA, Gosset G, Li M, Cardozo AK, et al. Signaling by IL-1beta+IFN-gamma and ER stress converge on DP5/Hrk activation: a novel mechanism for pancreatic beta-cell apoptosis. Cell Death Differ. 2009;16:1539–1550. doi: 10.1038/cdd.2009.99. [DOI] [PubMed] [Google Scholar]
- Gurzov EN, Germano CM, Cunha DA, Ortis F, Vanderwinden JM, Marchetti P, et al. p53 up-regulated modulator of apoptosis (PUMA) activation contributes to pancreatic beta-cell apoptosis induced by proinflammatory cytokines and endoplasmic reticulum stress. J Biol Chem. 2010;285:19910–19920. doi: 10.1074/jbc.M110.122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthson J, Germano CM, Moore F, Maida A, Drucker DJ, Marchetti P, et al. Cytokines tumor necrosis factor-alpha and interferon-gamma induce pancreatic beta-cell apoptosis through STAT1-mediated Bim protein activation. J Biol Chem. 2011;286:39632–39643. doi: 10.1074/jbc.M111.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010;70:655–665. doi: 10.1158/0008-5472.CAN-09-1942. [DOI] [PubMed] [Google Scholar]
- Pierangeli A, Degener AM, Ferreri ML, Riva E, Rizzo B, Turriziani O, et al. Interferon-induced gene expression in cervical mucosa during human papillomavirus infection. Int J Immunopathol Pharmacol. 2011;24:217–223. doi: 10.1177/039463201102400126. [DOI] [PubMed] [Google Scholar]
- Yan M, Luo JK, Ritchie KJ, Sakai I, Takeuchi K, Ren R, et al. Ubp43 regulates BCR-ABL leukemogenesis via the type 1 interferon receptor signaling. Blood. 2007;110:305–312. doi: 10.1182/blood-2006-07-033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Uze G, Pellegrini S. Negative regulation of type I interferon signaling: facts and mechanisms. Cell Mol Biol (Noisy-le-grand) 2006;52:77–87. [PubMed] [Google Scholar]
- Wesoly J, Szweykowska-Kulinska Z, Bluyssen HASTAT. activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim Pol. 2007;54:27–38. [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Wang X, Yan M, Duong FH, Poli V, Hilton DJ, et al. Alpha-Interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol. 2009;29:4841–4851. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Kim JH, Handidu A, Li X, Kim KI, Yan M, et al. Microarray analysis reveals that Type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem Biophys Res Commun. 2007;356:193–199. doi: 10.1016/j.bbrc.2007.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roep BO, Kleijwegt FS, van Halteren AG, Bonato V, Boggi U, Vendrame F, et al. Islet inflammation and CXCL10 in recent-onset type 1 diabetes. Clin Exp Immunol. 2010;159:338–343. doi: 10.1111/j.1365-2249.2009.04087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nishida Y, Aida K, Maruyama T, Shimada A, Suzuki M, et al. Enterovirus infection, CXC chemokine ligand 10 (CXCL10), and CXCR3 circuit: a mechanism of accelerated beta-cell failure in fulminant type 1 diabetes. Diabetes. 2009;58:2285–2291. doi: 10.2337/db09-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci USA. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- Gurzov EN, Eizirik DL. Bcl-2 proteins in diabetes: mitochondrial pathways of beta-cell death and dysfunction. Trends Cell Biol. 2011;21:424–431. doi: 10.1016/j.tcb.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Rasschaert J, Ladriere L, Urbain M, Dogusan Z, Katabua B, Sato S, et al. Toll-like receptor 3 and STAT-1 contribute to double-stranded RNA+ interferon-gamma-induced apoptosis in primary pancreatic beta-cells. J Biol Chem. 2005;280:33984–33991. doi: 10.1074/jbc.M502213200. [DOI] [PubMed] [Google Scholar]
- Lupi R, Dotta F, Marselli L, Del Guerra S, Masini M, Santangelo C, et al. Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that beta-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes. 2002;51:1437–1442. doi: 10.2337/diabetes.51.5.1437. [DOI] [PubMed] [Google Scholar]
- Moore F, Colli ML, Cnop M, Esteve MI, Cardozo AK, Cunha DA, et al. PTPN2, a candidate gene for type 1 diabetes, modulates interferon-gamma-induced pancreatic beta-cell apoptosis. Diabetes. 2009;58:1283–1291. doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore F, Cunha DA, Mulder H, Eizirik DL. Use of RNA interference to investigate cytokine signal transduction in pancreatic beta cells. Methods Mol Biol. 2012;820:179–194. doi: 10.1007/978-1-61779-439-1_11. [DOI] [PubMed] [Google Scholar]
- Moore F, Santin I, Nogueira TC, Gurzov EN, Marselli L, Marchetti P, et al. The transcription factor C/EBP delta has anti-apoptotic and anti-inflammatory roles in pancreatic beta cells. PLoS One. 2012;7:e31062. doi: 10.1371/journal.pone.0031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colli ML, Nogueira TC, Allagnat F, Cunha DA, Gurzov EN, Cardozo AK, et al. Exposure to the viral by-product dsRNA or Coxsackievirus B5 triggers pancreatic beta cell apoptosis via a Bim/ Mcl-1 imbalance. PLoS Pathog. 2011;7:e1002267. doi: 10.1371/journal.ppat.1002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortis F, Pirot P, Naamane N, Kreins AY, Rasschaert J, Moore F, et al. Induction of nuclear factor-kappaB and its downstream genes by TNF-alpha and IL-1beta has a pro-apoptotic role in pancreatic beta cells. Diabetologia. 2008;51:1213–1225. doi: 10.1007/s00125-008-0999-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.