Abstract
Micro RNAs (miRs) are small non-coding RNAs aberrantly expressed in human tumors. Here, we aim to identify miRs whose deregulated expression leads to the activation of oncogenic pathways in human gastric cancers (GCs). Thirty nine out of 123 tumoral and matched uninvolved peritumoral gastric specimens from three independent European subsets of patients were analyzed for the expression of 851 human miRs using Agilent Platform. The remaining 84 samples were used to validate miRs differentially expressed between tumoral and matched peritumoral specimens by qPCR. miR-204 falls into a group of eight miRs differentially expressed between tumoral and peritumoral samples. Downregulation of miR-204 has prognostic value and correlates with increased staining of Bcl-2 protein in tumoral specimens. Ectopic expression of miR-204 inhibited colony forming ability, migration and tumor engraftment of GC cells. miR-204 targeted Bcl-2 messenger RNA and increased responsiveness of GC cells to 5-fluorouracil and oxaliplatin treatment. Ectopic expression of Bcl-2 protein counteracted miR-204 pro-apoptotic activity in response to 5-fluorouracil. Altogether, these findings suggest that modulation of aberrant expression of miR-204, which in turn releases oncogenic Bcl-2 protein activity might hold promise for preventive and therapeutic strategies of GC.
Keywords: microRNA expression profile, prognostic value, Bcl-2
Gastric cancer (GC) is the second leading cause of cancer-related death worldwide.1 Improvement in diagnostic techniques and peri-operative management has resulted in good long-term survival for patients with early GC. Surgical resection is still the primary intervention for localized gastric tumors. However, patients with advanced disease frequently develop recurrent disease with nodal and hematogenous metastasis and peritoneal dissemination. These patients exhibit very poor survival rates. Molecular analysis coupled with genome wide approaches have identified various genetic alterations related to gastric tumorigenesis and progression. To date markers for tumorigenesis and progression of GC have not yet been discovered, and specific therapeutic targets have not been identified.2
Micro RNAs (miRs), a class of non-coding small RNAs exert a pivotal role in several biological processes ranging from development and differentiation to apoptosis and proliferation. miRs control gene expression tightly by degrading messenger RNAs (mRNAs) and inhibiting translation.3 Growing evidence show that miRs can act as tumor suppressors or oncogenes. miRs are aberrantly expressed in tumors when compared with matched or unmatched peritumoral samples.4, 5, 6, 7 Altered miRNA expression in tumors frequently correlates with progression and prognosis of human tumors. Antagomirs inhibit in vitro and in vivo pro-tumorigenic activities of oncogenic miRs,8 while reconstitution of tumor suppressor miRs promotes antitumoral activities. This clearly highlights miRs as molecular targets whose modulation might hold therapeutic promise.9
Here, we document by profiling simultaneously the expression of 851 human miRs that gastric tumors exhibit selected differentially expressed miRs when compared with their matched peritumoral samples in an European collection. Among those miRs, we identify miR-204 that is statistically significantly downregulated in GC samples derived from three independent subsets of patients (n=123) compared with their respective matched peritumoral tissues. Notably, ectopic expression of miR-204 impairs the ability of cultured gastric cells to form colonies and to migrate. In vivo engraftment of GTL-16 GC cells is significantly reduced by stable expression of miR-204. Polyclonal mixed populations of GTL-16 cells stably expressing miR-204 are more prone to the killing of 5-fluorouracil and oxaliplatin treatment than control cells. miRs exert their biological effects through the targeting of selected mRNAs. We show that downregulation of miR-204 in GC specimens correlates with increased staining for Bcl-2 protein. miR-204 targets the 3′-untranslated region (UTR) of Bcl-2 and reduces Bcl-2 protein expression. Interestingly, Bcl-2 overexpression counteracted miR-204 apoptotic activity in response to 5-fluorouracil. We also show that miR-204 downregulation is a prognostic factor in GC patients. Collectively, these findings underline tumor suppression activity of miR-204 in GCs whose inactivation release oncogenic Bcl-2 protein activity. This might contribute, at least in part, to the poor response of gastric tumors to conventional anticancer therapy.
Results
Altered miR expression pattern in primary human GCs
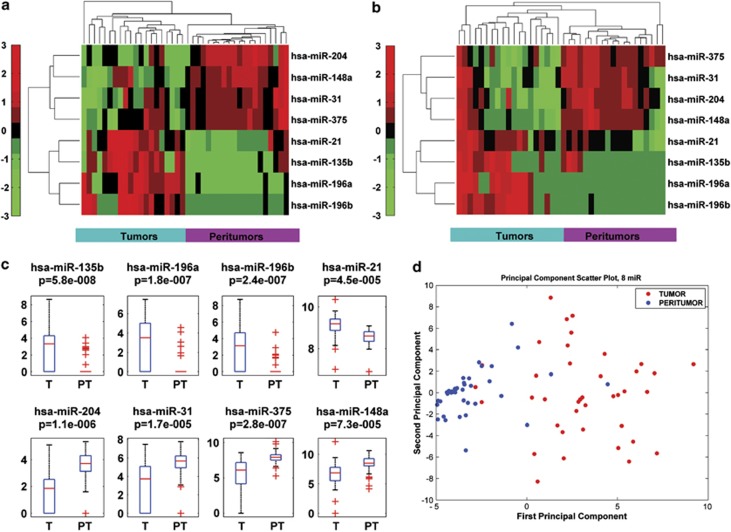
We interrogated simultaneously the expression of 851 human miRs of 39 GC specimens and of their matched peritumoral tissues using the Agilent microarray platform. The first matched samples were obtained from ‘Regina Elena' National Cancer Institute (RENCI) and San Vincenzo Hospital (SVH) subset of 39 patients (see Study Population section in Material and Methods) (Supplementary Tables 1 and 2), and analyzed by unsupervised cluster analysis. This analytical approach allowed us to identify a signature of miRs (eight miRs) that was significantly modulated between tumoral and peritumoral tissues in both subsets of patients (Figures 1a and b). The signature was based on ranking the statistics of different tests (paired and unpaired) with P-value <0.05 and measuring the area under receiver operating characteristic (ROC) curve. Significance of AUC was fixed to 0.7. By applying the same setting parameters to each subset of patients, we found that 62 (RENCI) and 41 (SVH) miRs were modulated, respectively (Supplementary Figures 1a and b and Supplementary Tables 3 and 4). A group of 16 miRs was commonly modulated in both subsets of patients (Supplementary Figures 1 c–e and Supplementary Table 5). Mean area under curve ((AUC)>0.79) between the two subsets was used to identify a signature that included the eight common modulated miRs, which most efficiently characterized the difference between tumoral and peritumoral tissues (miR-204, miR-148a, miR-31, miR-375, miR-21, miR-135b, miR-196a, miR-196b) (Figures 1a and b). The significance level of the difference between signal distributions of the eight selected miRs within the 39 analyzed samples was determined with supervised statistical test (Figure 1c). Unsupervised principal component analysis (PCA) showed that the eight miRs signature discriminate the group of tumor samples from that of matched peritumoral tissues. Peritumoral tissues display a more homogeneous distribution of clustered variables, while as expected, tumor samples exhibit a higher levels of heterogeneity (Figure 1d). In order to validate the eight miR signature, we performed the qPCR analysis on 6 out of the 8 miRs. This analysis confirmed the microarray profiling data (Supplementary Figures 2a–f). Collectively, these findings contribute to identify a signature of eight miRs, which is commonly modulated in gastric tumor samples derived from two independent subsets of patients.
Figure 1.
Signature of the best eight common miRNAs differentiate tumor and peritumor samples in GC. Unsupervised hierarchical clustering on selected miRs signature separately for the two subset of patients. (a and b) Red points represent signals with higher intensity while green points indicate signals with lower intensity level in RENCI cohort of samples (a) and SVH cohort of samples (b). Clustering basing on Euclidean distance is performed for each miRs (rows) and for each samples (columns). (c) Supervised statistical test was used to determinate significance level of the difference between signal distributions of the 39 samples. (d) Unsupervised PCA confirmed the ability of the signature to separate groups of tumoral samples from peritumoral samples. First component represents inter-group variability (axis x) while on the second component is plotted the intra-group variability (axis y)
miR-204 is downregulated in gastric tumor samples
There is growing experimental evidence that downregulation of large sets of miRs has a pivotal role in tumorigenesis independently from the type of the analyzed tumor.10 Indeed, downregulated miRs frequently couple with loss of tumor suppressor activity. In line with this evidence, we found that 4 (miR-204; miR-148a; miR-31; miR-375) out of 8 miRs, which were differentially expressed in our analysis, were selectively downregulated in tumor tissues. Among the four downregulated miRs, we focus on miR-204 because to date, the involvement of its downregulation in gastric tumorigenesis is still the most unknown.11, 12 Thus, we extended a new set of qPCR analysis to those patients (n=84) who were not included in the first microarray analysis (Supplementary Table 6). Our analysis revealed that miR-204 is consistently and statistically (P-value= 5.34*10−7) downregulated in a significant manner in gastric tumor specimens compared with matched peritumoral tissues from all the three analyzed subsets (Figures 2a–c). The significance level of the difference in the downregulation of miR-204 expression obtained by either microarray profiling or qPCR analysis was normalized as folds of modulation between tumor and peritumoral tissues for each of the analyzed subset of patients.
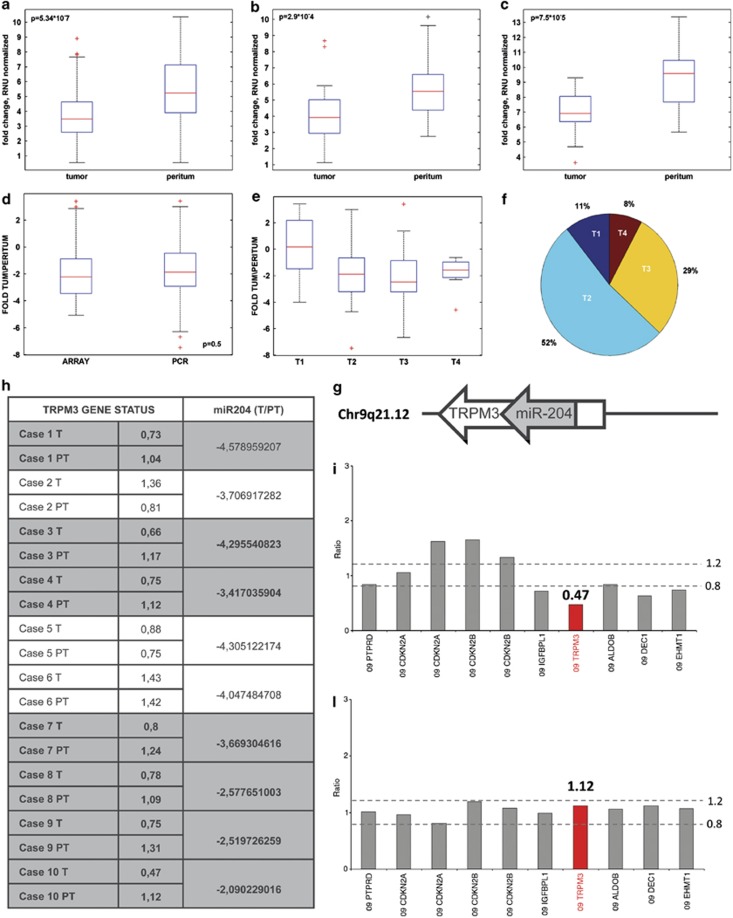
Figure 2.
miR-204 expression correlates with tumor stage (T) data. Expression level of miR-204 in 64 RENCI paired samples, (b) in 16 SVH paired samples and (c) in 12 patients from a third independent cohort of samples SAH (S. Andrea Hospital). (d) Fold change between tumoral and peritumoral tissue on miR-204 were calculated for 111 samples (RENCI and SVH subset of patients). Fold change boxplot distribution of samples processed using different techniques and collected from different institutes indicates a similar distribution of the miR after normalization. (e) Boxplot of the comparison of T1 tumor stage versus T2, T3, T4 stage using fold change level of miR-204 signal. (f) Pie chart of tumor stage distribution of 103 known T-values (RENCI and SVH). (g) Schematic representation of the miR-204 and its host gene TRPM3. (h) Table which compares TRPM3 gene status quantitatively evaluated by multiplex ligation-dependent probe amplification with miR-204 downregulation in 10 primary GC and in the normal-matched peritumoral tissues. (i–l) Multiplex ligation-dependent probe amplification exemplificative histograms showing the heterozygous deletion of TRPM3 gene in a gastric carcinoma as compared with the matched normal peritumoral tissue (TRPM3 gene ratio in the tumor 0.47 versus 1.12 in the normal peritumoral tissue)
First, we did not find statistically significant (P=0.5) difference between microarray profiling and qPCR analysis, thereby providing additional robustness to our results (Figure 2d). Second, when we compared the mean level of miR-204 downregulation by GC stages T1–T4, we found that there was a statistically significant difference between the miR-204 expression levels in advanced gastric tumors (stage T2–T4) versus the observed levels in T1 (Figures 2e–f). No significant association was found with nodal status and histotype (diffuse versus intestinal) of the analyzed GC specimens (Supplementary Figures 2g and h). miR-204 is intragenic miRNA and it is located within the transient receptor potential melastatin-3 (TRPM3) gene, a gene belonging to the family of transient receptor potential (TRP) channels (Figure 2g).13 The latter resides in the long arm of chromosome 9 that is frequently deleted in human cancers.14, 15, 16 This prompted us to investigate whether the deletion of that region caused miR-204 downregulation in gastric tumoral tissues. To this end, we evaluated the presence of TRPM3 gene loss using a PCR-based test termed Multiplex Ligation-dependent Probe Amplification. We found that 7 out of 10 analyzed GCs (70%) exhibiting miR-204 downregulation carried an heterozygous deletion of TRPM3 gene (<0.8) when compared with their matched peritumoral tissues (between 0.8 and 1.2) (Figures 2h–l). Altogether, these findings document that miR-204 downregulation is significantly more pronounced in advanced gastric tumors. This downregulation, at least in part, occurs through the deletion of the chromosomal region containing miR-204 host gene.
miR-204 expression impairs the tumorigenic potential of gastric tumor cells
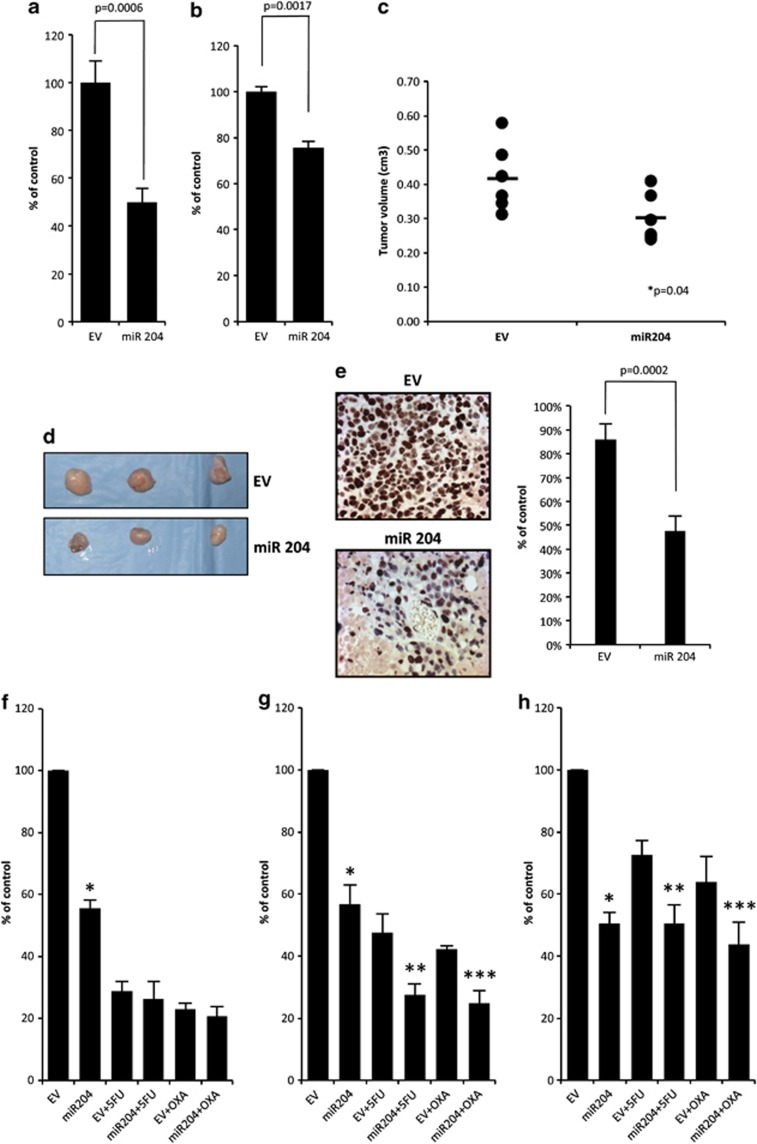
Downregulation of miR expression contributes strongly to tumorigenesis.10, 17 This prompted us to investigate whether reconstitution of miR-204 expression can promote tumor suppressor effects in vitro and in vivo. We found that mixed polyclonal population of GTL-16 or N87 GC cell lines overexpressing miR-204 (GTL-16/miR-204; N87/miR-204) exhibited a statistically significant reduced ability to form colonies when compared with control cells (GTL-16/EV; N87/EV) (Figure 3a and Supplementary Figure 3a). Wound healing assays reveled that GTL-16/miR-204 migrated less efficiently (P=0.0017) than GTL-16/EV counterparts (Figure 3b and Supplementary Figure 3b). Of note, in vivo tumor engraftment of GTL-16/miR-204 cells injected in CD1 mice was significantly (P=0.04) reduced compared with control cells (Figures 3c and d and Supplementary Figure 3c). Furthermore, the analysis of the proliferation index by Ki67 in the excised tumors revealed that there was a statistically meaningful reduction in tumors derived from GTL-16/miR-204 in comparison with GTL-16/EV (Figure 3e).
Figure 3.
miR-204 overexpression affects cell migration, colony forming ability and chemoresistance. (a) Colony assay of GTL-16 cells stably transfected with pCMV-MIR-204 (miR-204) and pCMV-MIR vector (EV). An average of three independent experiments is reported. Error bars represent mean±S.D. (b) Wound healing assay shows that miR-204 inhibits cell migration in GTL-16 cells. Quantity of migrated cells presents an average from three experiments independently. (c) Tumor volumes measured at day 28 after the first injection. The central bold lines denote mean values, the vertical lines±S.E.M. (n=6 mice in each group) and the circles data points. P-values was calculated by 2-samples t-test and indicated in the figure. (d) Images of three representative tumors from mice bearing GTL-16 stable expressing miR-204 or empty vector. (e) Left: positive immunohistochemical staining of paraffin-embedded xenografts indicates cellular proliferation of GTL-16 stable cells. miR-204 overexpressing cells show a reduced proliferation rate compared with EV. Three sections of each of the six tumors per group have been scored. ( × 400, counterstained with hematoxylin). Right: histogram shows the percentage of positive cells from 6 tumors/group. The difference between GTL-16 miR-204 and GTL-16 EV xenograft's proliferation rate is statistically significant (P=0.0002). (f–h) Colony formation assay of GTL-16 stable cells overexpressing miR-204 or the empty vector treated at decreasing concentrations of 5-fluorouracil (5-FU) or oxaliplatin (OXA) drugs. Histogram showing the average colony counts from 5-FU=6 μM, OXA=1 μg/ml (f), 5-FU=3 μM, OXA=500 ng/ml (g), 5-FU=2 mM, OXA=250 ng/ml (h). An average of three independent experiments is reported
Finally, we also found that GTL-16/miR-204 cells were more responsive to IC50 amount of 5-fluorouracil or oxaliplatin treatment than GTL-16/EV cells (Figure 3f). A dilution 1 : 2 and 1 : 4 of IC50 made the contribution of miR-204 to the killing effect of both anticancer drugs significantly (P<0.05) more pronounced (Figures 3g and h).
Altogether, these findings have several implications: (a) miR-204 exerts antitumoral effects in vitro and in vivo; (b) downregulation of miR-204 expression in GC specimens might result in the loss of its tumor suppressor activity; thereby contributing to gastric tumorigenesis; (c) modulation of miR-204 expression might bear therapeutic potential for the treatment of gastric tumors.
Downregulation of miR-204 releases aberrant expression of Bcl-2 protein
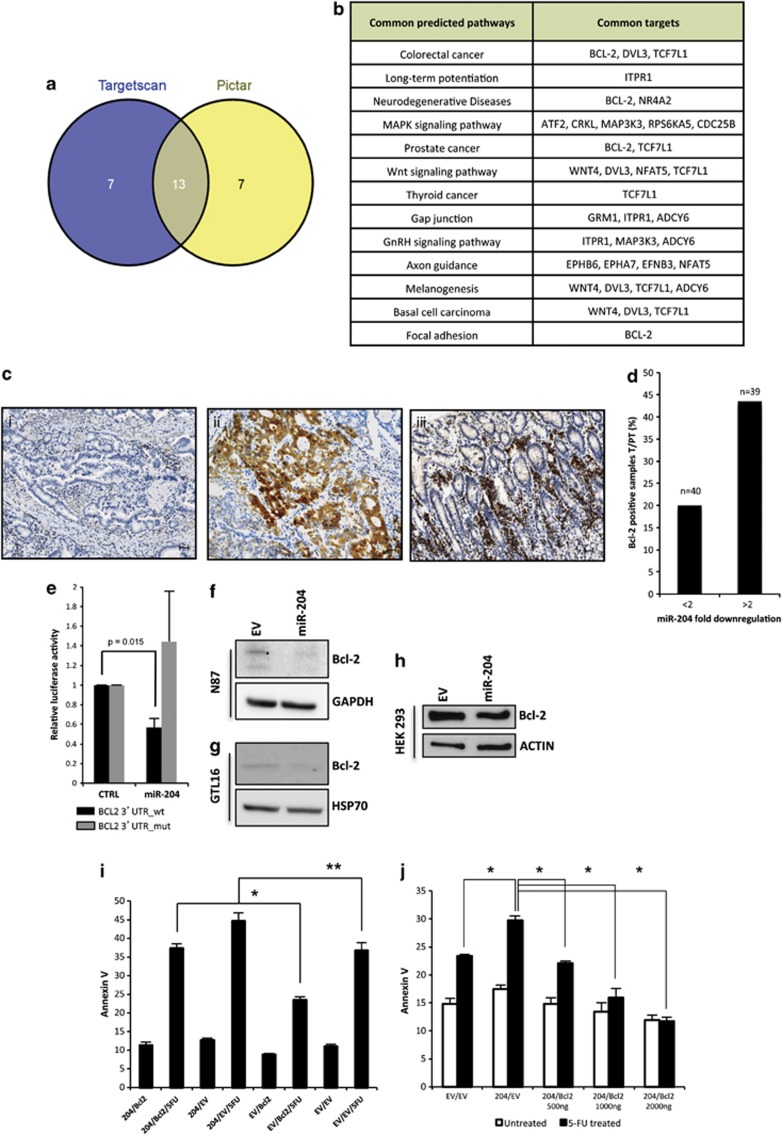
miRs act by degrading mRNAs and inhibiting translation of target genes. Thus, downregulation of a specific miR might result on the aberrant expression of proteins whose mRNAs escape to the tight control of miRs. To search for mRNAs putative targets of miR-204 we performed a computer-assisted analysis with Diana mirPath with the aid of Pictar and Targetscan programs.18 Bcl-2, a very well-known inhibitor of apoptosis, emerges as a putative target mRNA in 4 out of 13 identified pathways (Figures 4a and b and Supplementary Table 7). Furthermore, Bcl-2 as putative target of miR-204 was identified by 7 out of 10 target prediction tools (Supplementary Figure 3d).19 Interestingly none of the other three downregulated miRNAs out of eight miRNAs signature was found to target Bcl-2 (Supplementary Figure 3d). This prompted us to investigate whether downregulation of miR-204 releases aberrant expression of Bcl-2 protein in gastric tumor specimens. Immunohistochemical analysis of Bcl-2 expression in the 79 GC specimens and their matched peritumoral tissues from patients recruited at RENCI were performed by the local Department of Pathology. Immunostaining revealed that Bcl-2: (a) was consistently negative in peritumoral normal gastric mucosa; (b) only 20% of the 40 gastric carcinomas exhibiting a downregulation of miR-204 lower than twofolds, compared with peritumoral tissues, displayed Bcl-2-positive staining; (c) in contrast, 43.6% of the 39 gastric carcinomas exhibiting a downregulation of miR-204 higher than twofolds compared with peritumoral tissues, presented a strong Bcl-2 protein expression (Figures 4c and d). These findings mirror that aberrant expression of Bcl-2 in gastric tumors associates with downregulation of miR-204.
Figure 4.
miR-204 downregulates the expression of Bcl-2 protein. (a) Venn-diagramm displays 13 common pathway shared by TargetScan and Pictar. (b) Table of 13 common pathway and common targets according to TargetScan and Pictar programms. (c) Immunohistochemistry of Bcl-2 protein in representative tissue samples: a representative case of Bcl-2-negative gastric carcinoma (I), a representative case of Bcl-2-positive gastric carcinoma displays a cytoplasmic immunostaining in more than 50% of tumor cells (II), the correspondent autologous morphologically uninvolved peritumoral tissue is consistently bcl-2 negative, the internal control of infiltrating lymphocytes are positive (III). Scale bar 30 μm. (d) Percentage of Bcl-2-positive samples plotted against miR-204 fold downregulation comparing tumoral versus peritumoral samples. In x axis, it is represented the miR-204 fold downregulation, while in y axis the percentage of Bcl-2-positive samples. (e) Expression vectors carrying a luciferase reporter followed by the first 541 bp of 3′-UTR regions of Bcl-2 in their wild-type form (black bars) or mutated in the miR-204 complementary sequence (gray bars), were transfected in H1299 cells in the presence of pCMV-miR-204 or pCMV-EV. Normalized luciferase activity values from three independent experiments in triplicate are shown. (f) Immunoblotting of Bcl-2 protein in N87 cell line stably transfected either with pCMV-MIR vector (EV) or pCMV-miR-204 (miR-204). (g) Immunoblotting of Bcl-2 protein in GTL-16 cell line stably transfected either with pCMV-MIR vector (EV) or pCMV-miR-204 (miR-204). (h) Immunoblotting of Bcl-2 protein in HEK293 cells transfected with either pCMV-miR-204 or pCMV-EV and pCDNA3-Bcl-2 plasmid. (i) GTL-16 gastric cells stably expressing either miR204 or the EV and transiently transfected with Bcl-2 and the EV were stained with Annexin V. Apoptosis was determined using flow cytometry as described in Materials and Methods. The values obtained from Annexin V assays represent the means±S.D. for three independent experiments. *P=0.0008; **P=0.03. (l) 293T cells were cotransfected with miR204 and Bcl-2 and their corresponding EV, and treated with 5-fluorouracil (3 μM). Twenty-four hours after treatment, apoptosis was determined by staining with Annexin V. Histograms report the means±S.D. for three independent experiments. *P<0.005
miR-204-mediated targeting of Bcl-2 expression promotes apoptosis of GC cells
To investigate whether the contribution of miR-204 to the enhanced killing of GTL-16 upon treatment with 5-fluorouracil or oxaliplatin occurs, at least in part, through the targeting of Bcl-2 expression, we inserted a fragment of 541 base pair of the Bcl-2 3′-UTR containing three miR-204 binding sequences downstream of the luciferase coding regions (Supplementary Figure 3e–f). Ectopic expression of miR-204 reduced the relative luciferase activity when compared with cells transduced with control vector (Figure 4e). This effect was not evidenced in the presence of a miR-204 complementary sequence mutated version of Bcl-2 3′-UTR (Figure 4e). Furthermore, overexpression of miR-204 diminished endogenous Bcl-2 protein levels in both N87 and GTL-16 gastric cell lines (Figures 4f and g). Similarly, miR-204 reduced the expression of exogenously expressed Bcl-2 protein in HEK 293T cells (Figure 4h). Mixed populations of GTL-16 gastric cells stably overexpressing miR-204 were more prone to 5-fluorouracil-induced apoptosis than control cells (Figure 4i). Interestingly, Bcl-2 ectopic expression reduced the apoptotic rate of miR-204 overexpressing GTL-16 cells upon treatment with 5-fluorouracil (Figure 4l). The protective effect of Blc-2 protein expression was more evident in the GTL-16 control cells (Figure 4i). In agreement with these findings, increasing amounts of ectopically expressed Bcl-2 protein counteracted the apoptotic effects induced by miR-204 overexpression upon 5-fluorouracil treatment (Figure 4l). Altogether, these findings highlight that (a) Bcl-2 mRNA is target of miR-204; (b) miR-204 pro-apoptotic activity in response to anticancer treatment might occur through the targeting of Bcl-2 expression.
miR-204 is a prognostic factor for GCs
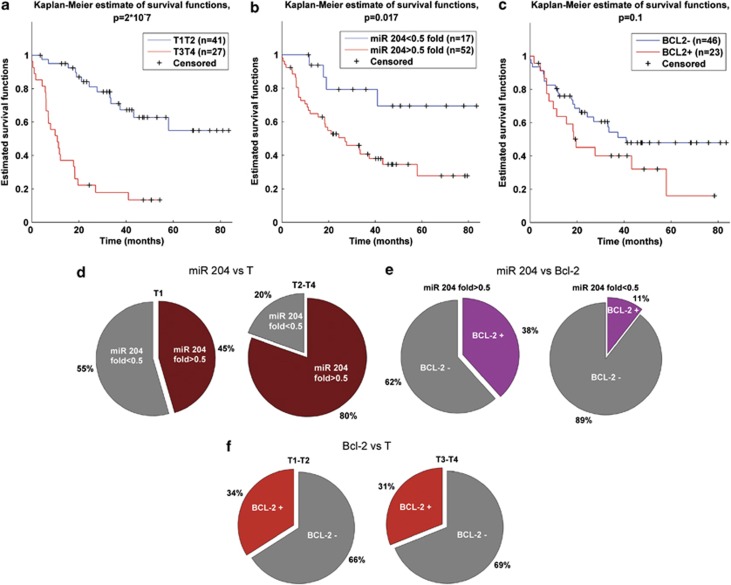
At a median follow-up of 22 months (range 0-83 months), a total of 42 out of 69 GC patients with complete follow-up data were still alive. We first evaluated whether the tumor stage (pT) of the disease, as we could expect, had a prognostic value in our series of GC patients. We conducted this analysis on 69 out of the 79 patients recruited at the RENCI being 10 patients excluded because of missing follow-up data. We observed that the overall survival of patients bearing a pT3–pT4 tumors was statistically significant lower than that of pT1–pT2 tumors (P<0.001 Figure 5a). Furthermore, we analyzed whether the tumor downregulation of miR-204 may have a significant impact on patient survival. We classified our series of GC patients by two levels of miR-204 downregulation: patients with miR-204 downregulation lower than 0.5 fold and patients with miR-204 downregulation equal/more than 0.5. Kaplan–Meier curves showed that the latter category of patients had the worst survival in comparison with patients classified in the first category. The difference between the two set of patients was statistically significant (P=0.017; Figure 5b). Then we analyzed the impact of Bcl-2 protein expression on patient survival. In this case, we did not observe any statistically significant difference between Bcl-2-positive and Bcl-2-negative patients (P=0.1; Figure 5c).
Figure 5.
(a) Kaplan–Meier survival curves of patients at stage T1–T2 versus patients at stage T3–T4. (b) Kaplan–Meier survival curves of subgroups of patients with miR-204-fold higher and lower than 0.5. (c) Kaplan–Meier survival curves of patients with Bcl-2 status positive versus negative. In all analysis the differences between estimated curves are evaluated by log-rank test. (d) Pie charts miR-204 fold distribution of 103 patients with defined T status. The distributions are calculated considering subgroups of patients at stage T1 and stage T2–T3–T4. (e) Pie charts of Bcl-2 distribution of 79 patients with defined Bcl-2 status. The distributions are calculated considering subgroups of patients with miR-204 fold higher and lower 0.5. (f) Pie charts of Bcl-2 distribution of 79 patients with defined Bcl-2 status. The distributions are calculated considering subgroups of patients at stage T1–T2 and stage T3–T4
To complete our observation, we evaluated whether the miR-204 level was related to the GC pT (Figure 5d and Supplementary Table S8a). We showed that in pT1 tumors the different expression level of miR-204 (lower than 0.5, higher than 0.5 fold) was similarly distributed. In contrast, miR-204 downregulation higher than 0.5 was prevalent (P=0.01) in pT2–pT4 tumors (Figure 5d). No significant differences in miR-204 fold expression level distributions were observed for group of patients classified by nodal stadium, tumor histotype, age and sex (Supplementary Figure 4a–d). Figure 5e indicated that the IHC expression of Bcl-2 was lower in the category of patients exhibiting miR-204 downregulation lower than 0.5 fold (P=0.03). There was no difference in Bcl-2 positivity with respect to the pT (P=0.82 Figure 5f).
The impact on survival of each variable was investigated by Cox univariate regression model and log-rank test between curves. Variables with P-values higher than 0.1 were considered significant and used to build multivariate Cox regression model. Despite of correlation between cancer pT and miR-204 downregulation our results suggest the independence of miR-204 deregulation as prognostic factor (Table 1).
Table 1. The impact on survival of different variables of interest were evaluated by log-rank test between survival curves and Cox regression analysis.
| Survival (months) | No. of patients | P-value (log-rank) | HR (CI) univariate | HR (CI) multivariate | |
|---|---|---|---|---|---|
| miR-204 fold<0.5 | NR | 17 | 0.017 | 3.4 (1.2–9.7) | 3.9 (1.3–11.8) |
| miR-204 fold>0.5 | 24.8 | 52 | |||
| Bcl-2 negative | 34.7 | 46 | 0.1 | 1.7 (0.9–3.3) | 1.5 (0.8–3.1) |
| Bcl-2 positive | 16.9 | 23 | |||
| T1–T2 | NR* | 41 | 2 × 10−7 | 6 (3–12.3) | 7.8 (3.7–16.7) |
| T3–T4 | 10.3 | 27 | |||
| N0 | 42 | 30 | 0.2 | 1.6 (0.8–3.2) | |
| N+ | 20.5 | 39 | |||
| Female | NR | 34 | 0.29 | 1.4 (0.7–2.9) | |
| Male | 27 | 35 | |||
| Diffuse | 33.6 | 19 | 0.6 | 0.7 (0.3–1.5) | |
| Intestinale | 51.3 | 46 | |||
| Age<68 years | 37 | 37 | 0.9 | 1 (0.5–1.9) | |
| Age>68 years | 33.2 | 32 |
Abbreviation: NR, not reached Hazard risk of each variable with confidence interval is reported for univariate model and multivariate model was used to investigate confounding components among variables with P-value <0.1
Discussion
In the present study, we performed genome wide analysis to interrogate miR expression profile of gastric tumor specimens compared with their matched peritumoral tissues. Previous work showed that differentially expressed patterns of miRs correlate with specific tumor types and unique miRs associate with progression and prognosis of tumors, including GCs.20 There is growing experimental evidence that downregulation of large sets of miRs has a pivotal role in tumorigenesis independently from the type of the analyzed tumor.10 Indeed, downregulated miRs frequently correspond with loss of tumor suppressor activity. Here, we found that 4 (miR-204; miR-148a; miR-31; miR-375) out of 8 miRs, which were differentially expressed in our analysis, were selectively downregulated in tumor tissues. To date, the involvement of miR-204 downregulation to gastric tumorigenesis is rather unknown.11, 12 Of note, we documented the reconstitution of miR-204 expression impairs in vitro and in vivo the tumor potential of GC cell lines. Ectopic expression of miR-204 inhibited colony forming ability, migration and tumor engraftment of GC cell lines. These findings strongly indicate that downregulation of miR-204 in gastric tumors might coincide with loss of tumor suppressor activity and consequently favor gastric transformation.
miRs exert their biological effects through the selective targeting of mRNAs whose derived proteins are either up- or downregulated. As miR-204 is specifically downregulated in GC specimens the expression of its putative targets might be upregulated. Computer-assisted analysis to search for miR-204 putative mRNA targets that were not shared with miR-148a, miR-31, and miR-375 identified bcl-2 gene (Supplementary Figure 3d), a very well-known inhibitor of apoptosis, which confers increased chemoresistance to many types of cancer cells.21 Here, we provide evidence validating Bcl-2 as mRNA target of miR-204. First, we found that Bcl-2 protein staining is strongly increased in gastric tumors specimens compared with matched peritumoral tissues. Second, we document that miR-204 reduces Bcl-2 protein expression by targeting specific sequences within the 3′-UTR of Bcl-2 transcript. Third, ectopic expression of miR-204 reduces Bcl-2 protein levels in gastric tumor cell lines. Fourth, ectopic expression of Bcl-2 protein counteracted miR-204-mediated apoptosis in response to anticancer treatment. Previous reports have shown that miR-181b, miR-200bc/429 cluster, miR-497, miR-15 and miR-16 target Bcl-2 expression in GC cell lines by modulating multidrug resistance.22, 23, 24, 25 Xenograft tumors derived from GC cell lines overexpressing Bcl-2 protein exhibited enhanced peritoneal dissemination compared with control transfectants.26 Notably, ectopic expression of miR-204 significantly potentiates the killing effects induced by either oxaliplatin or 5-fluorouracil on GC cell lines when compared with control counterparts. Altogether, these findings mirror Bcl-2 protein, whose expression emerges to be tightly controlled by diverse miRs, as an important component of the complex molecular network underlying poor response of gastric tumors to anticancer treatment.
Surgical treatment for GCs is based on locoregional staging. Limited resections are considered the best practice in early cancer. D2 radical gastrectomy is the standard procedure for GCs. Neoadjuvant pre-operative chemotherapy and extended surgery are recommended for far advanced tumors. Of note, we evidenced that downregulation of miR-204 significantly associates with the tumor (T) stage of the analyzed GC samples. GC patients at stage T1 exhibit lower downregulation of mirR-204 than those with more advanced stage. Both T stage and downregulation of miR-204 have prognostic value for the analyzed gastric samples. This emphasizes that (a) miR-204 might be an useful molecular marker for the staging of GCs; (b) the combination of hystopathological (TNM) and molecular (miR-204, Bcl-2, p53 status, ErbB2, c-myc and others) features might strongly contribute to a rather accurate staging of gastric tumors. Preliminary findings suggest that 9q21.12 chromosome portion, where miR-204 is located within intron 6 of human TRPM3, is selectively deleted in tumoral tissue when compared with matched peritumoral samples. A rather attractive challenge would be that of assessing whether a gradient of miR-204 downregulation occurs within T1 gastric patients and is able to predict which of them will advance or not. The impact of Bcl-2 aberrant protein expression on the survival and disease progression of GC patients is rather controversial. Recent evidence shows that Bcl-2 expression of gastric tumor patients analyzed in 501 patients who underwent curative D2 gastrectomy was related to good survival and was an independent prognostic factor for GC patients.27 In agreement with these findings we also document that Bcl-2 expression might be an independent prognostic factor in the European GC patients. Unlike previous evidence we show that GC patients with Bcl-2-positive staining exhibit shorter survival than Bcl-2-negative patients. This apparent discrepancy might contribute to decipher, at the molecular level, the differences existing between Asiatic and European GC patients, namely the incidence of early GC and the striking differences in the outcomes, stages and treatments being equally in the hemispheres. Indeed, miR expression profiling of GC patients from two independent subsets of Japanese patients has identified a signature of miRs including 22 upregulated and 13 downregulated.20 Some of them (miR-148a, miR-181a/b, miR-92, miR-93, miR-375, miR-21, miR-25, miR-135b, miR-425, miR-29c and miR-30a/b/c) are also differentially expressed in our analysis, while downregulation of miR-204, which has prognostic value, appears to be specific of the three independent subsets of European patients in our study. The recruitment of additional subsets of European GC patients might prove further or disprove that the modulation of miR-204 is specific of European GC tumors. There is growing evidence that miRs differentially expressed in tumors can be detected in plasma. Tsujiura et al.28 reported that circulating miRs can be evidenced in the plasma of GC patients. They specifically show that the plasma concentration of miR-17-5p, miR-21, miR-106a, miR-106b was higher in GC patients compared with health volunteers.28 Extensive profiling of circulating miRs as counterparts of those differentially expressed in gastric tumors might turn very useful in staging and monitoring GC patients. miR-204 downregulation bears potential of being analyzed aiming at this purpose.
Notwithstanding the improvements in quality of surgery and multimodality treatments, GC prognosis is poorly developed, as it is still the second leading cause of cancer-related death worldwide. While a significant therapeutic success has been obtained for early GC, very little is available for tackling advanced and disseminated gastric tumors. Our findings contributed to the identification of molecular alterations in GCs whose management might hold therapeutic potential.
Materials and Methods
Study population
Because of the rare condition, in this study we included 123 GC patients subsequently enrolled at three different hospitals in Italy: RENCI in Rome (RENCI) (n=79) (Supplementary Table 1), the SVH in Taormina (n=32) (Supplementary Table 2) and the S. Andrea Hospital (SAH) in Rome (n=12) (Supplementary Table 6). We evaluated 111 GCs from formalin-fixed and paraffin-embedded tissues (FFPE) and 12 GCs, recruited at the SAH, from frozen fresh tissues. Patients recruited in the study between 2005 and 2008 had a median follow-up of 22 months. However, the 12 patients from SAH were recruited in 2011 and did not have significant follow-up to be included in the longitudinal section of the study. From each patient, we collected tissue samples from the tumor lesion and from the uninvolved peritumoral mucosa, which was defined as the portion of tissue located at 2 cm from the resection margin of the tumor lesion.
In order to identify the miRs differentially expressed between the tumoral and the peritumoral areas, we conducted the first step of the microarray analysis on 20 patients from the RENCI and 19 from the SVH for a total of 39 tumors and peritumoral matched samples. Subsequently, we validated the results on the remaining 84 patients (of the initial 123): 59 from RENCI, 13 from SVH and 12 from the SAH. The follow-up of RENCI patients has been carried out through the individual checking of the vital status.
The ethical committee of each institute has approved the study.
Cell cultures and treatments
Human cell lines GTL-16, N87, HEK293 and H1299 were grown in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 ug/ml) at 37 °C in a balanced air humidified incubator with 5% CO2.
For chemoresistance experiments, cells were treated with oxaliplatin (ELOXATIN from Sanofi Aventis, Paris, France) and 5-fluorouracil (Fluorouracil from Teva Italia S.r.l.) at the indicated concentration for 2 weeks.
Plasmid and transfection
The full length 3′-UTR of the human Bcl-2 genes was amplified and inserted downstream of the Renilla luciferase gene into the NotI sites of the dual luciferase reporter plasmid psiCHECK-2 (Promega, Madison, WI, USA). All constructs were verified by sequencing. Bcl-2 mutant was made with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) using the following primers:
Bcl-2 del122-129 Fw (5′-GGCAAAACGTCGAATCAGCTATTTACTGCAATATCATTTATTTTTTACATTATTAAGAA-3′)
Bcl-2 del 122-129 Rv (5′-TTCTTAATAATGTAAAAAATAAATGATATTGCAGTAAATAGCTGATTCGACGTTTTGCC-3′)
The mutant was sequenced to confirm the mutated product. H1299 cells were transfected using Lipofectamine 2000 (Invitrogen) with 25 ng of psiCHECK-2 reporter vector containing the 3′-UTR of putative target gene, together with 375 ng of pCMV-MIR-204 (miR-204) or pCMV-MIR vector (EV), from OriGene Technologies (Rockville, MD, USA) in 24-well plates. Firefly and Renilla luciferase activities were measured 48 h post transfection using the Dual Luciferase Reporter Assay System (Promega) in the GloMax 96 Microplate Luminometer (Promega). Firefly luciferase was used to normalize the Renilla luciferase.
For stable cell lines, GTL-16 and N87 were transfected with the pCMV-MIR-204 (miR-204) and pCMV-MIR vector (EV) from OriGene Technologies using Lipofectamine 2000 (Invitrogen) following the manufacturer's instruction. The day after transfection the standard media was replaced with media containing G418 (Invitrogen) at a concentration of 400 μg/ml. Cells were maintained in selection for 2 weeks.
Wound healing assay
GTL-16 GC cell lines stably transfected with pCMV-MIR-204 (miR-204) and pCMV-MIR vector (EV) were grown to 80% confluence in 6-well tissue culture plates and wounded with a sterile 20 μl pipet tip to remove cells by two perpendicular linear scrapes. The progression of migration was photographed immediately and at 24–48 h after wounding.
Colony formation assay
GTL-16 and N87 cells, stably transfected with pCMV-MIR-204 (miR-204) and pCMV-MIR vector (EV) from OriGene Technologies were seeded at 500 and 1000 cells/well, respectively, into 6-well dishes (CORNING-COSTAR, Tewksbury, MA, USA). Cells were stained with crystal violet and colonies counted after 10 and 14 days later, respectively.
Lysate preparation and immunoblotting analyses
Cells were lysed in buffer with 10 mM Tris-HCl pH.8. 9 M UREA, 0.2% NP-40 (Igepal AC-630). Fresh DTT at final concentration of 200 mM was added prior use. Extracts were centrifuged at 14000 × g r.p.m. for 15 min to remove cell debris. Protein concentrations were determined by colorimetric assay (Bio-Rad, Hercules, CA, USA). Western blotting was performed using the following primary antibodies: mouse monoclonal anti-Actin (Sigma-Aldrich A228, St. Louis, MO, USA), mouse monoclonal anti-Bcl-2 (Santa-Cruz Biotechnologies, Santa Cruz, CA, USA). Secondary antibodies used were goat anti-mouse, conjugated to horseradish peroxidase (Amersham Biosciences, Piscataway, NJ, USA). Immunostained bands were detected by chemiluminescent method (Pierce, Rockford, IL, USA).
cDNA synthesis and RQ–PCR
RQ–PCR quantification of miRNA expression was performed using TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. RTq-PCR quantification of miRNA expression was performed using TaqMan MicroRNA Assays (Applied Biosystems) according to the manufacturer's protocol RNU6B was used as endogenous controls to standardize miRNA expression. All reactions were performed in duplicate.
Immunohistochemistry
Bcl-2 was analyzed by immunohistochemistry using the anti-Bcl-2 MoAb 124 (Dako, Milan, Italy) in a series of 79 gastric carcinoma patients subjected to surgery at the Regina Elena Cancer Institute (Rome, Italy) between 2005 and 2008. Bcl-2 was recorded as positive when tumor cells exhibited a strong homogeneous cytoplasmic immunoreaction in <30% of neoplastic cells. FFPE 5-mm sections of mice xenografts derived from GTL-16 stable cells were stained with anti- Ki-67 antibody (1 : 200, VENTANA, Roche, Indianapolis, IN, USA).
Target prediction
In silico putative target prediction of selected miRs was conducted matching the results of different web server tools. Putative target were individuated by Diana mirPath (http://diana.cslab.ece.ntua.gr/pathways) using Targetscan 5 and Pictar.
RNA extraction, labeling and microarray hybridization
RNA from FFPE samples was extracted using the miRneasy FFPE kit (QIAGEN, Gaithersburg, MD, USA) following the manufacturer's instructions. The concentration and purity of total RNA were assessed using a Nanodrop TM 1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Total RNA (100 ng) was labeled and hybridized to Human miRNA Microarray V2 (Agilent). Scanning and image analysis were performed using the Agilent DNA Microarray Scanner (P/N G2565BA) equipped with extended dynamic range (XDR, Agilent Technologies, Santa Cruz, CA, USA) software according to the Agilent miRNA Microarray System with miRNA Complete Labeling and Hyb Kit Protocol manual. Feature Extraction Software (Version 10.5; Agilent Technologies) was used for data extraction from raw microarray image files using the miRNA_105_Dec08 FE protocol.
GC xenografts
Four-weeks old CD1 nude mice (n=6/each group, Charles River) were subcutaneously injected with GTL-16 GC cell lines stably transfected with pCMV-MIR-204 (miR-204) and pCMV-MIR vector (EV) from OriGene Technologies (2 × 106 cells/mouse) and tumor volume was evaluated twice a week. Animals were killed and xenograft excised after 1 month from the injection. Tumor volume (cm3) was calculated as follows: 0.5 × D12 × D2, where D1 and D2 are the larger and smaller diameters measured by caliper. All tumorigenicity assays were carried out according to the guidelines set by the Internal Ethical Committee.
Apoptosis assay
The extent of apoptosis was evaluated by Annexin V staining. GTL-16 gastric cells stably expressing miR204 were transfected with three different concentrations of the pCDNA3-Bcl-2 vector (500 ng, 1 μg, 2 μg). Forty-eight hours post transfection cells were stained with FITC-conjugated Annexin V antibody (AbCam, Cambridge, UK) and analyzed by flow cytometry. The same setting was used for 293T human embryonic kidney cells cotransfected with pCMV-MIR-204 (miR204) and pCDNA3-Bcl-2 and treated with 5-fluorouracil (4 μM) for 24 h. Counterstaining with propidium iodide (20 μg/ml) (Sigma-Aldrich) was used to check for membrane integrity.
Bioinformatic and statistical analysis
Microarray data analysis:
Preprocessing. The signal of 851 human miRs was processed on Agilent microarray platform. Signals were verified for quality control and extracted by Agilent Feature Extraction 10.7.3.1 software (Agilent Technologies) and processed by MATLAB (The MathWorks Inc., Natick, MA, USA) in house-built routines. All values lower than 3 were considered below detection and thresholded to 1. The arrays were quantile-normalized forcing each slide to assume the same mean distribution and log2-transformed.
Features selection. miRs signature was obtained basing on ranking the statistics of different tests with P-value <0.05, measuring area under ROC curve, setting a significant level for AUC to 0.7. Specifically, non-parametric Wilcoxon test and T-test was used for comparison of subgroups of patients considering both paired and unpaired samples. Same setting parameters were used independently on the two subset of patients, and most deregulated miRs replicated on both set were used as resulting signature. miRs of the signature were ranked basing on the mean AUC of each miR between subset of patients. Unsupervised and supervised classification techniques such as PCA and Hierarchical Clustering were used to distingue different tissues (tumoral and peritumoral) from the same patient basing on a restricted signature of these miRs. Differences in the signal distribution and distance between intensity levels of subgroups of samples were tested overall the 39-matched samples using a restricted signature composed by the first 8 miRs with the best mean AUC ranked from the 16 common features.
Features validation. Real-Time PCR (RT-PCR) was used as alternative technique to validate 6 out of 8 miRs of the signature on a representative group of samples. In particular, we focused our investigation on miR-204, one of the most downregulated feature in both the case studies. We collected 92-matched samples also including samples from a third institute: 64 patients (tumoral and peritumoral paraffin-embedded tissues) from RENCI cohort, 16 patients (tumoral and peritumoral paraffin-embedded tissues) from SVH cohort, and 12 patients with tumoral and peritumoral frozen tissues from SAH in Rome. Eight patients out of the 92 were already studied on Agilent platform. Overall 123 cases, counting tumoral tissue and peritumoral tissues, were analyzed as described by RT-PCR or Agilent microarray platform.
The RT-PCR signals of miR-204 were normalized by RNU6B. In order to investigate correlation between miRNA's signal and clinical patient's outcome, we calculated fold change between tumoral and peritumoral samples in order to merge signals generated from different platform.
Clinical variables investigation. The log-rank test was used to assess differences between subgroups. A multivariate Cox proportional hazard model was also developed using stepwise regression (forward selection) with predictive variables with a P-value <0.1 in the univariate analyses. χ2-test with Yates correction was used to evaluate significance of contingence tables between variables. Significance was assessed at a level of 5%.
Acknowledgments
This work was funded by Scientific direction office ‘Regina Elena' National Cancer Institute and AIRC (Italian Association for Cancer Research): grant numbers 08/30 R/89 and 09/8706.
Glossary
- T
tumor
- PT
peritumor
- GC
gastric cancer
- miR
microRNA
- RENCI
Regina Elena National Cancer Institute
- SVH
San Vincenzo Hospital
- SAH
S. Andrea Hospital
- AUC
area under curve
- PCA
principal component analysis
- MLPA
multiplex ligation-dependent probe amplification
- TRPM3
transient receptor potential melastatin-3
- FFPE
formalin-fixed paraffin-embedded tissue
- EV
empty vector
- 5-FU
5 fluorouracil
- UTR
untranslated region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Candi
Supplementary Material
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim KM, Cheong JH, Noh SH. Current management and future strategies of gastric cancer. Yonsei Med J. 2012;53:248–257. doi: 10.3349/ymj.2012.53.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Donzelli S, Fontemaggi G, Fazi F, Di Agostino S, Padula F, Biagioni F, et al. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19:1038–1048. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Liu Z, Sall A, Yang D. MicroRNA: an emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Lam EK, Wang X, Shin VY, Zhang S, Morrison H, Sun J, et al. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3:209–218. [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberwinkler J. TRPM3, a biophysical enigma. Biochem Soc Trans. 2007;35 (Part 1:89–90. doi: 10.1042/BST0350089. [DOI] [PubMed] [Google Scholar]
- Jiao YF, Sugai T, Habano W, Uesugi N, Takagane A, Nakamura S. Clinicopathological significance of loss of heterozygosity in intestinal- and solid-type gastric carcinomas: a comprehensive study using the crypt isolation technique. Mod Pathol. 2006;19:548–555. doi: 10.1038/modpathol.3800561. [DOI] [PubMed] [Google Scholar]
- Staub E, Grone J, Mennerich D, Ropcke S, Klamann I, Hinzmann B, et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol Cancer. 2006;5:37. doi: 10.1186/1476-4598-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandar V, Lakshman Kumar B, Sasikala K, Manikantan P, Sangeetha R, Mohana Devi S. Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer patients. J Zhejiang Univ Sci B. 2007;8:638–646. doi: 10.1631/jzus.2007.B0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belka C, Budach W. Anti-apoptotic Bcl-2 proteins: structure, function and relevance for radiation biology. Int J Radiat Biol. 2002;78:643–658. doi: 10.1080/09553000210137680. [DOI] [PubMed] [Google Scholar]
- Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J, et al. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 2012;69:723–731. doi: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, et al. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29:384–391. doi: 10.1007/s12032-010-9797-4. [DOI] [PubMed] [Google Scholar]
- Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer. 2010;127:2520–2529. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- Yawata A, Adachi M, Okuda H, Naishiro Y, Takamura T, Hareyama M, et al. Prolonged cell survival enhances peritoneal dissemination of gastric cancer cells. Oncogene. 1998;16:2681–2686. doi: 10.1038/sj.onc.1201792. [DOI] [PubMed] [Google Scholar]
- Liu X, Cai H, Huang H, Long Z, Shi Y, Wang Y. The prognostic significance of apoptosis-related biological markers in Chinese gastric cancer patients. PLoS One. 2011;6:e29670. doi: 10.1371/journal.pone.0029670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.