Abstract
Amino-terminal signal sequences target nascent secretory and membrane proteins to the endoplasmic reticulum for translocation. Subsequent interactions between the signal sequence and components of the translocation machinery at the endoplasmic reticulum are thought to be important for the productive engagement of the translocon by the ribosome-nascent chain complex. However, it is not clear whether all signal sequences carry out these posttargeting steps identically, or if there are differences in the interactions directed by one signal sequence versus another. In this study, we find substantial differences in the ability of signal sequences from different substrates to mediate closure of the ribosome–translocon junction early in translocation. We also show that these differences in some cases necessitate functional coordination between the signal sequence and mature domain for faithful translocation. Accordingly, the translocation of some proteins is sensitive to replacement of their signal sequences. In a particularly dramatic example, the topology of the prion protein was found to depend highly on the choice of signal sequence used to direct its translocation. Taken together, our results reveal an unanticipated degree of substrate-specific functionality encoded in N-terminal signal sequences.
It is commonly believed that the sole function of the N-terminal signal sequence of a nascent secretory or membrane protein is to facilitate its segregation from cytosolic proteins. Although that is still the principal function attributed to the signal sequence, it is becoming clear that its role in protein translocation is more complex (1). Signal sequences are involved in targeting of nascent proteins to their sites of translocation at the endoplasmic reticulum (ER) membrane (2, 3), initiating a stable interaction between the ribosome and translocon (4–6), and providing a ligand for the opening of the translocation channel (7–9). It is thought that these events are all carried out, in succession, via interactions between the signal sequence and proteins in both the cytosol and ER membrane.
Shortly after its synthesis but even before its complete emergence from the ribosome, the signal sequence is bound by the nascent polypeptide-associated complex (NAC). When the signal emerges from the ribosome, NAC appears to be displaced from the nascent chain by the signal recognition particle (SRP) (10). The nascent chain is subsequently transferred, via the SRP receptor at the ER, to the protein translocation channel (11, 12). For the model secretory protein preprolactin (pPrl), all of these events of protein targeting occur by the time ≈35 aa are synthesized beyond the signal sequence (4, 8, 13).
During the synthesis of the next ≈10 aa an interaction between the signal sequence and the Sec61 complex, the primary constituent of the translocation channel (14, 15), is thought to mediate a change in the ribosome–translocon interaction. This change results in the formation of a tight seal between the ribosome and translocon such that the nascent chain becomes shielded from the cytosol (4, 16) and resistant to extraction by high salt (4, 5, 10, 13). Although these events are coincident with a close juxtaposition between the signal sequence and the translocating-chain associated membrane protein (TRAM), the exact role of this protein in translocation remains unclear (15, 17–19). Shortly thereafter (by ≈70 total aa), the translocation channel is opened toward the ER lumen, providing a continuously sealed conduit from the peptidyl transferase center within the ribosome to the luminal aperture of the translocon (8). The growing chain is then vectorially transferred into the ER lumen. The timing of these events has been carefully mapped for pPrl, and in this case, appears to be precisely coordinated such that the mature region of the nascent chain is essentially never exposed to the cytosol.
At present, it is unclear whether the signal sequences of different proteins differ significantly in how they carry out each of these steps. However, variations on the above paradigm seem likely given the enormously diverse set of sequences that serve as signals for targeting and translocation (20), and the complex interactions of these signals with both cytosolic and ER proteins (10, 15, 17, 21–23). In this study, we have focused on the critical, but poorly understood, posttargeting steps of signal sequence function. By comparing the ribosome–translocon junction at this stage in the translocation of multiple substrates, we have discovered significant differences in the posttargeting function of different signal sequences. More remarkable, however, was the finding that for some proteins, altering these signal-mediated posttargeting steps can have significant consequences for their translocation. Thus, these functional differences between signal sequences are not simply random variations reflective of a degenerate sequence motif, but instead may represent physiologically relevant substrate-specific differences that are critical for proper protein biogenesis.
Materials and Methods
Plasmid Constructions.
All constructs are in the pSP64 vector (Promega). Plasmid HG201 encoding preIgG heavy chain (pIgG) (24) was provided by T. Rapoport (Harvard Medical School, Boston). Plasmids encoding pPrl and preβ-lactamase (pβL) have been described (25). To replace the signal sequence of any coding region, a restriction site was introduced by PCR mutagenesis immediately beyond the site of signal cleavage (except for signal-PrP constructs, which used an existing PflM1 site). Subsequently, the sequence between a restriction site preceding the start codon and the introduced restriction site was replaced with foreign sequences generated by either PCR or synthetic oligonucleotides. The pβL signal sequence used in this study contained an Asp at codon 2 rather than Ser. This replacement has no noticeable effect on pβL translocation or βL-PrP topology (data not shown). The constructs in Fig. 5B were generated by insertion of foreign sequences into a PstI site located immediately before the signal cleavage site of IgG-Prl. Inserts encoding residues 16–69 and 16–132 of IgG or 31–147 of pPrl (the stuffer sequence) were generated by PCR. All constructs were verified by automated sequencing.
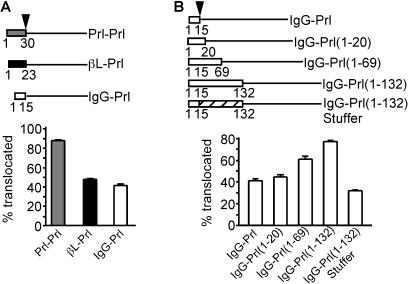
Figure 5.
Role of the ribosome–translocon junction in pPrl translocation. Above each panel is a schematic diagram of each construct. The arrowhead represents the location of signal peptide cleavage. The pPrl mature region is denoted by the solid line, and sequences from pβL and pIgG are shown by filled or open boxes, respectively. The pPrl signal sequence is shown as a gray box. The numbers below each construct represent the amino acids taken from pPrl, pβL, or pIgG. The stippled box for IgG-Prl(1–132)Stuffer represents the stuffer amino acids. The translocation efficiency was determined as the percentage of chains achieving a PK-protected, signal sequence-cleaved fate during translocation into microsomal membranes, ± SEM for three trials.
Cell-Free Translation and Translocation Assays.
In vitro transcription with SP6 RNA polymerase, translation in rabbit reticulocyte lysate, translocation into canine pancreatic rough microsomal membranes (RM), proteolysis with proteinase K (PK), and immunoprecipitations have been described (26). Translation reactions were at 32°C for 20–30 min, and proteolysis reactions were with 0.5 mg/ml PK for 30 min on ice. Where translocation intermediates were analyzed in a protease protection assay, RM were isolated from translation reactions by sedimentation through a 0.5 M sucrose cushion containing 50 mM Hepes (pH 7.4), 5 mM MgCl2, and either 100 mM KOAc (physiologic salt buffer-PSB) or 500 mM KOAc (high-salt buffer), as indicated in the figure legends. The membranes were resuspended in PSB before proteolysis. Analysis of high-salt resistant binding to RM by flotation through a sucrose step gradient (see Fig. 3A) has been described (27). For Fig. 6C, a translation reaction in the absence of RM was synchronized by addition of 75 μM aurintricarboxylic acid (Sigma) at 5 min. Aliquots were removed to ice at staggered intervals, and RM were added to 0.1 eq/ul and returned to 32o for 30 min. Samples were analyzed for prion protein (PrP) topology by proteolysis and immunoprecipitation with the 3F4 anti-PrP antibody (ref. 28; provided by S. Prusiner, University of California, San Francisco) as described (29). SDS/PAGE was on 15% Tris/glycine gels, 15% Tris/tricine gels, or 10% NuPAGE Bis-Tris gels (NOVEX, San Diego). Densitometric quantitation of autoradiographs (digitized by using an Agfa Arcus II flatbed scanner) was performed by using Adobe photoshop software.
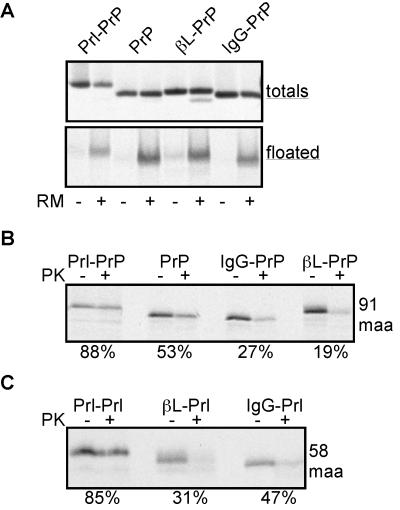
Figure 3.
Analysis of high-salt resistant translocation intermediates. (A) Translocation intermediates of 91 maa of Prl-PrP, PrP, βL-PrP, and IgG-PrP were translated in the presence or absence of microsomes, and an aliquot was set aside (totals) while another aliquot was floated through a sucrose step gradient containing 0.5 M KOAc. The floated material, containing the microsomal membranes, was removed from the top of the gradient and analyzed by autoradiography (floated). (B and C) Translocation intermediates of 91 maa of Prl-PrP, PrP, βL-PrP, and IgG-PrP, or of 58 maa for Prl-Prl, βL-Prl, and IgG-Prl, were translated, the high-salt resistant intermediates were isolated by sedimentation through a sucrose cushion containing 0.5 M KOAc, and the samples were analyzed by proteolysis. The percent of nascent chains protected from protease digestion is indicated below the autoradiograph.
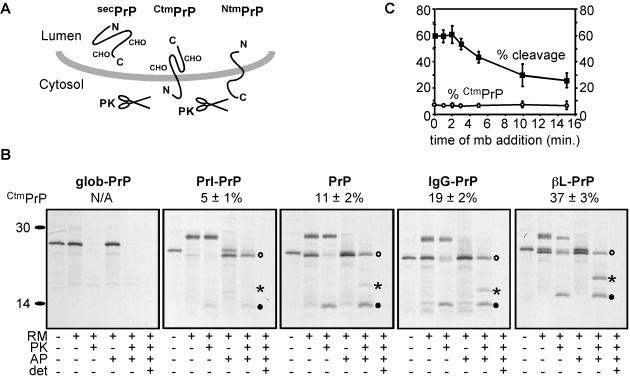
Figure 6.
Effects on topology of replacing the PrP signal sequence. (A) The three topologic forms generated by PrP translocation are shown. Glycosylation of secPrP and CtmPrP is indicated. NtmPrP is unglycosylated because the sites reside in the cytosolically disposed C-terminal domain. (B) Topology of glob-PrP, Prl-PrP, PrP, IgG-PrP, and βL-PrP. Transcripts from each construct were translated in the presence or absence of RM and a peptide inhibitor of glycosylation (AP). After translation, equal aliquots of the translated material were left untreated or treated with PK in the presence or absence of Triton X-100 as in Fig. 1A. After protease digestion, NtmPrP is seen as a nonglycosylated 14-kDa fragment whereas CtmPrP migrates as either a glycosylated 25-kDa band or 19 kDa when glycosylation is inhibited. secPrP is fully protected from protease digestion. The positions of secPrP (○), CtmPrP (*), and NtmPrP (●) are indicated for the nonglycosylated species. The percentage of translocated chains in the CtmPrP topology for each construct from three experiments was quantitated by densitometry of the autoradiographs (mean ± SEM), and is shown above each panel. (C) Microsomal membranes were added at various times from 0 to 15 min after initiation of a synchronized translation reaction of PrP. After the completion of translation, topology was assessed by the protease protection assay (detailed in B) and quantitated to determine the percentage of total translation product that was processed by signal peptidase (■), and of translocated chains synthesized in the CtmPrP topology (○). The mean ± SEM from three experiments is plotted.
Results and Discussion
Closure of the Ribosome–Translocon Junction Is Substrate-Specific.
The detailed characterization of the steps in pPrl translocation has provided a paradigm for understanding the sequential events of targeting, assembly of a functional ribosome-nascent chain-translocon complex, and subsequent translocation. To begin exploring potential variations on the framework defined by pPrl, we analyzed steps in the translocation of two other proteins, pβL and pIgG. Fig. 1A shows that upon translation in rabbit reticulocyte lysate in the presence of equal concentrations of canine pancreatic microsomal membranes, all three of these proteins are efficiently translocated into the lumen. This conclusion is evidenced by signal sequence cleavage (seen with pPrl and pβL), glycosylation (seen with pIgG), and complete protection from protease digestion in the absence, but not presence, of detergent. Thus, each of these proteins contains all of the information necessary for efficient targeting to and subsequent translocation across the ER membrane.
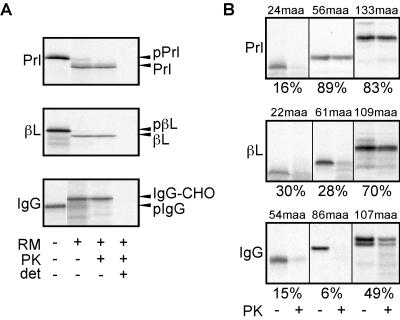
Figure 1.
Analysis of pPrl, pβL, and pIgG translocation. (A) Full-length transcripts encoding native pPrl, pβL, or pIgG were translated in the presence or absence of microsomal membranes (RM), and equal aliquots were set aside or treated with PK in the presence or absence of Triton X-100 (det). The positions of precursor (pPrl, pβL, pIgG), processed (Prl, βL), and glycosylated (IgG-CHO) species are indicated to the right. (B) Translocation intermediates of the indicated lengths (in maa synthesized beyond the signal sequence) were assembled, and the targeted nascent chains were analyzed by proteolysis. The percentage of protease protected chains is shown below each panel.
To assess the state of the nascent chain at different points during translocation, we prepared translocation intermediates of defined lengths for each of these proteins by translation and translocation of truncated messages of different lengths lacking a stop codon. The assembled translocation intermediates then were separated from untargeted nascent chains by sedimentation and were analyzed by a protease protection assay to probe the state of the ribosome–translocon junction (16). A loose or open junction renders the nascent chain susceptible to digestion from cytosolic protease, whereas a closed junction provides effective protection from digestion (4, 16, 26). Consistent with previous studies (4, 10) when only 24 mature amino acids (maa) beyond the signal sequence have been synthesized, pPrl has not yet achieved a tight seal between the ribosome and translocon and is thus accessible to digestion by cytosolically added protease (Fig. 1B). However, at later points in synthesis (by 56 and 133 maa), it is well shielded from protease digestion by the ribosome–translocon junction (Fig. 1B; refs. 4, 16, 26, and 30).
A similar analysis of pβL and pIgG revealed that significantly longer translocation intermediates were accessible to protease than was observed with pPrl. At translocation intermediates of 61 maa of pβL (84 total aa) and 86 maa of pIgG (101 total aa), very few (<30%) of the targeted nascent chains were protected from protease digestion (Fig. 1B). We found instead that pβL and pIgG achieved a state of protease protection, although still not as complete as seen for pPrl, only at lengths more than 50 maa beyond that observed for pPrl. Because the three signal peptides we analyzed differed in length (30, 23, and 15 aa for pPrl, pβL, and pIgG, respectively), it was possible that at early truncation points, the apparent differences we observed in positioning of the nascent chains with respect to the translocon are due solely to differences in overall length. However, the differences in protease accessibility were seen even when pβL and pIgG intermediates that were seven and 15 residues longer (to normalize overall length) were analyzed (data not shown). Thus, small differences in overall length due to the sizes of the signal peptides cannot account for the substantial differences in protease accessibility observed. Similarly, the differences in protease accessibility could not be due to differences in cotranslational glycosylation because at the truncation points analyzed, no potential sites for N-linked glycosylation have been synthesized (the single site in pIgG is at maa 302, whereas pPrl and pβL are not glycoproteins). Taken together, these results suggest that pβL and pIgG differ from pPrl at early phases during their translocation across the ER membrane. Whereas pPrl chains are accessible to the cytosol for a very brief period of chain growth, the pβL and pIgG chains remain accessible for substantially longer during their translocation.
Signal Sequences Encode Differential Closure of the Ribosome–Translocon Junction.
The prolonged exposure of pβL and pIgG chains during their translocation could be explained in one of two ways. The first possibility is that the signal sequences of pβL and pIgG may not facilitate closure of the ribosome–translocon junction in the same manner described for the pPrl signal sequence. Alternatively, it is possible that the signal sequences behave similarly, but regions in the pβL and pIgG mature domains cause a reopening of the ribosome–translocon junction, as has been described in response to the synthesis of either a transmembrane domain (30, 31) or pause transfer sequences (26). To distinguish between these possibilities, we examined translocation intermediates of chimeras containing various combinations of signal sequences and mature domains of these proteins.
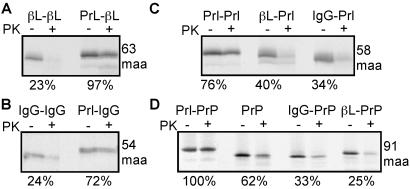
We found that fusion of the pPrl signal sequence to either the pβL or pIgG mature domains (Prl-βL and Prl-IgG, respectively) resulted in the protease protection of early translocation intermediates (63 maa and 54 maa, respectively) of these constructs (Fig. 2 A and B). By contrast, fusion of either the pβL or pIgG signal sequence to the pPrl mature domain (βL-Prl and IgG-Prl, respectively) resulted in protease accessibility of translocation intermediates containing 58 maa (Fig. 2C). These results suggest that the signal sequence, and not the mature domain, is the primary determinant of whether the ribosome–translocon junction is open or closed early in translocation.
Figure 2.
Signal sequences determine early closure of the ribosome–translocon junction. Translocation intermediates of various combinations of signal sequences fused to different mature domains (A, βL; B, IgG; C, Prl; D, PrP) were analyzed by protease protection as in Fig. 1B. The length of the translocation intermediates (in maa) is indicated to the right. The percent of nascent chains protected from protease digestion is indicated below each panel.
To confirm these results, we fused each of these signal sequences to an unrelated protein, PrP. PrP contains a 22-residue N-terminal signal sequence, a potential transmembrane domain from residues 113 to 135, N-linked glycosylation sites at residues 181 and 197, and a C-terminal signal for glycolipid anchor addition (32). Analysis of early translocation intermediates of these chimeric constructs (at 91 maa, before synthesis of the transmembrane domain or glycosylation sites) revealed that, as with the other substrates analyzed, the nascent chain was well protected from PK when the pPrl signal sequence was used but poorly protected when the pβL and pIgG signal sequences were used (Fig. 2D). Thus, different signal sequences vary intrinsically in their respective abilities to mediate the formation of a closed ribosome–translocon junction early in translocation.
In the case of pPrl, formation of a tightly sealed ribosome–translocon junction is accompanied by binding of the nascent chain to the translocon in a salt-resistant manner by the synthesis of ≈40 maa (4, 5, 10, 13). Although this binding state is thought to reflect a productive interaction with the translocon, salt-resistant and protease-resistant binding states can be dissociated (33). To determine whether protease-sensitive chains bearing the pβL and pIgG signal sequences were productively associated with the translocon, we compared the abilities of PrP nascent chains with different signal sequences to be recovered after floatation through a sucrose step gradient containing 0.5 M potassium acetate. We found that all substrates were equally well recovered, suggesting that each signal sequence mediates effective engagement of the translocon (Fig. 3A). When these intermediates were analyzed by proteolysis, we found that IgG-PrP and βL-PrP remained substantially accessible to PK whereas Prl-PrP chains were largely protected (Fig. 3B, compare with Fig. 2D). Similar differences also were observed for salt-resistant translocation intermediates of Prl-Prl, βL-Prl, and IgG-Prl (Fig. 3C, compare with Fig. 2C).
The pPrl, pβL, and pIgG signal sequences differ with respect to timing of their cleavage (see also ref. 19). However, signal cleavage did not correlate temporally with the acquisition of either salt- or protease-resistant binding. Substrates containing each of these signals acquired salt-resistant binding at points before cleavage (e.g., see Fig. 3A, where none of the substrates are signal cleaved). By contrast, acquisition of protease-resistant binding could be observed before, roughly concomitant with, or after signal cleavage. For example, Fig. 1 shows that pPrl acquires protease resistance (at 56 maa) before signal cleavage (at 133 maa) whereas pβL has its signal cleaved by the time it has acquired protease resistance (at 109 maa). For pIgG, ≈30% of nascent chains are signal-cleaved at a point when protease resistant binding is first observed (at 107 maa). In Figs. 2 and 3, none of the translocation intermediates shown have their signal sequences cleaved, yet substantial differences can be seen in their relative levels of protease resistant binding. Thus, it appears that for the substrates analyzed here, cotranslational signal cleavage is not a prerequisite for or an immediate consequence of salt- or protease-resistant interactions between the nascent chain and the translocon.
These results argue that, despite effectively mediating nascent chain targeting to and engagement of the translocon, the signal sequences of pβL and pIgG result in cytosolic disposition of their associated nascent chains. Together, the data lead to the surprising conclusion that the signal sequences of pPrl, PrP, pβL, and pIgG can be functionally discriminated at a posttargeting, posthigh-salt resistant step in translocation that involves formation of the ribosome–translocon junction.
A Role for the Mature Domain in Regulating the Ribosome–Translocon Junction.
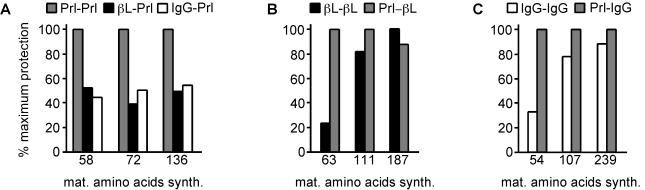
Although translocation of pβL and pIgG begins with an open ribosome–translocon junction, the junction becomes sealed to a greater extent as translocation continues (Fig. 1B). We wondered whether this closure is an inevitable consequence of increased chain synthesis, or a feature specific to the pβL and pIgG mature domains. To address this question, we compared translocation intermediates of each of the signal sequences (from pPrl, pβL and pIgG) fused to the pPrl mature domain at three lengths (58, 72, and 136 maa, Fig. 4A). We found, as noted in Fig. 2C, that there was approximately a 50% reduction in the protection from PK for βL-Prl and IgG-Prl, compared with Prl-Prl, at an early point of 58 maa. However, in contrast to native pβL and pIgG (Fig. 1B), βL-Prl and IgG-Prl failed to achieve enhanced protection from PK after further chain synthesis (Fig. 4A). Thus the pPrl mature domain does not allow for closure of an initially open ribosome–translocon junction. Conversely, when the signal sequences of pβL and pIgG were replaced with the pPrl signal sequence, early translocation intermediates (of 63 and 54 maa, respectively) became well protected (Figs. 2 A and B and 4 B and C). However, at longer chain lengths, these differences largely normalized, with βL-βL and IgG-IgG showing comparable levels of protease protection relative to Prl-βL and Prl-IgG, respectively (Fig. 4 B and C). Taken together, these data suggest that the mature domains of proteins such as pβL and pIgG are able to facilitate closure of an initially open ribosome–translocon junction (Figs. 1B and 4 B and C).
Figure 4.
Effect of the mature domain on closure of the ribosome–translocon junction. Translocation intermediates of various signal sequences fused to pPrl (A), pβL (B), or pIgG (C) were analyzed for protease protection. The length of each translocation intermediate, in maa, is indicated below the graphs. Plotted are the values for protection from protease relative to the maximal level of protection achieved for any sample at that truncation point.
Consequences of Inappropriate Regulation of the Ribosome–Translocon Junction.
The fact that pβL and pIgG have signal sequences that elicit an open ribosome–translocon junction and mature domains which facilitate later closure of the junction suggested that mismatching of signal sequences and mature domains with respect to this characteristic might have adverse consequences for protein translocation. To test this idea, we directly compared the translocation efficiencies of pPrl containing the pPrl, pβL, or pIgG signal sequences. Remarkably, our analysis revealed that βL-Prl and IgG-Prl translocate substantially less efficiently than Prl-Prl (Fig. 5A). Under conditions where pPrl translocation efficiency approached 90%, less than 50% of βL-Prl and IgG-Prl were translocated.
It seemed plausible that the absence of information to close the junction in the pPrl mature domain is the basis for the poor translocation efficiency of βL-Prl and IgG-Prl. To test this hypothesis, we added either the first 5, 54, or 117 aa of the mature region of pIgG to the N terminus of the IgG-Prl construct, immediately after the signal sequence [IgG-Prl(1–20), IgG-Prl(1–69), and IgG-Prl(1–132), respectively] (Fig. 5B). Although the addition of 5 aa had no detectable effect on translocation, inserting either the first 54 or 117 aa of pIgG substantially increased the efficiency of IgG-Prl translocation. The translocation efficiency of the Prl-IgG(1–132) construct approached that of Prl-Prl (compare Fig. 5 B to A). By contrast, insertion of amino acids from pPrl [IgG-Prl(1–132)Stuffer] did not increase translocation efficiency, indicating that the stimulation seen with the pIgG insertions was not due simply to increasing the overall length of the protein. Taken together, these data support the hypothesis that the mature region of pIgG, by facilitating the closure of an initially open ribosome–translocon junction, allows efficient translocation of the protein.
To further explore the hypothesis that altered regulation of the ribosome–translocon junction by signal sequences could have a significant impact on protein translocation, we analyzed the behavior of a more complex protein, PrP. When translocated across the ER, PrP has been shown to adopt three distinct topologic forms (Fig. 6 A and B and refs. 29, 34, and 35). One of these forms, termed secPrP, is fully translocated across the ER membrane. The other two forms of PrP are made as singly spanning membrane proteins in opposite orientations with either the N or C terminus in the ER lumen (termed NtmPrP and CtmPrP, respectively), the latter of which is associated with both spontaneous and transmissible prion disease (29, 36). Ordinarily, PrP is synthesized predominantly in the secPrP and NtmPrP forms with a small, but significant, amount of CtmPrP (≈11%, see Fig. 6B).
To determine whether posttargeting recognition of the PrP signal sequence at the translocon plays a role in topology, we followed the biogenesis of native PrP, Prl-PrP, IgG-PrP, and βL-PrP. We found that PrP topology could indeed be influenced by different signal sequences, with levels of CtmPrP synthesis corresponding to the relative effect of each signal sequence on the ribosome–translocon junction (Fig. 6B). The pPrl signal sequence, which closes the ribosome–translocon junction (Fig. 3B), resulted in significantly less CtmPrP compared with native PrP, with an increased amount of secPrP (Fig. 6B). By contrast, both the pIgG and pβL signal sequences markedly increased CtmPrP synthesis (Fig. 6B) while also leading to an open ribosome–translocon junction (Fig. 3B). As expected, lack of a functional signal sequence (glob-PrP, in which the PrP signal sequence is replaced by 22 aa of the cytosolic protein globin) resulted in a lack of translocation.
To rule out the possibility that the effects of different signal sequences on PrP topology were attributable to differences in their targeting properties rather than their impact on the ribosome–translocon junction, we manipulated the kinetics of PrP targeting and monitored the resultant topology. Synchronized translation reactions lacking microsomal membranes were provided microsomes at successively later times. Because the signal recognition particle does not effectively arrest translation in reticulocyte lysate (37), progressively longer chain lengths will have been synthesized before being given the opportunity to interact with the membranes. As expected, the efficiency of overall translocation (as assessed by the efficiency of signal sequence cleavage) decreased at later time points due to an increasing percentage of chains that had presumably completed translation before productive targeting. Yet regardless of the duration for which translation was allowed to occur before the addition of microsomes, the ratio of topologic forms remained constant (the percentage of translocated chains in the CtmPrP topology is plotted in Fig. 6C). Similar experiments revealed that PrP topology also was not substantially affected by varying the concentration of microsomal membranes (and hence functional translocons) in the translation reactions (data not shown).
Taken together these results argue that different signal sequences impact PrP topology not via differences in targeting, which all of these signal sequences appear to carry out effectively for both their own (Fig. 1A) and a heterologous (Fig. 3A) substrate, but by differential regulation of the ribosome–translocon junction (Fig. 2D).
Conclusions and Perspective.
In this study, we have compared the early stages in the translocation of multiple substrates to determine the extent to which these stages may differ from one protein to another. We have found significant substrate-specific variation at a posttargeting step, after the formation of salt-resistant contacts, at a stage when the tight seal between the ribosome and translocon is formed. Based on the action of their signal sequences, some proteins, such as pPrl, establish a sealed ribosome–translocon junction very rapidly after targeting, and remain shielded from the cytosol as translation continues. Conversely, the signal sequences of pβL and pIgG cause these proteins to remain exposed to the cytosol for significantly longer, and so the mature domains of these proteins contain information that acts to eventually close the junction. Alternatively, the mature region may instead contain a membrane-spanning domain that converts chains with an open junction into transmembrane chains of the CtmPrP type.
The role for signal sequences described here provides another example of sequence-specific alteration of nascent chain environment, an emerging theme in the study of translocational regulation. In addition to the roles played by signal sequences shown here and elsewhere (23), regulation of the ribosome–translocon junction has been observed during and after the synthesis of transmembrane domains (30, 31) and transiently for large domains of secretory proteins (26). However, it is not yet clear which ER factors participate in this process or how the nascent chain elicits changes in the architecture and function of the translocon. Although the translocating-chain-associated membrane protein (TRAM) has been implicated in certain aspects of signal sequence recognition (19) and regulation of the ribosome–translocon junction (27), no precise role for TRAM has yet emerged. Biochemical depletion and reconstitution of individual translocon components will be needed to identify the factors responsible for differential recognition of signal sequences and regulation of the ribosome–translocon junction.
The consequences of regulation of the ribosome–translocon junction by signal sequences are particularly noteworthy in the case of PrP, wherein the effect of altering the state of the junction leads to changes in final topology. Previous studies have shown that alteration of PrP topogenesis can have dramatic effects on the development of neurodegenerative disease in transgenic mice (29, 36). In particular, increases in CtmPrP can result in development of neurodegeneration, whereas decreases in CtmPrP may result in decreased susceptibility to transmissible prion disease (36). Thus, at least in this case regulation of the ribosome–translocon junction may prove to be of tremendous pathophysiological importance.
Although the consequences of altered recognition of signal sequences at the ER are apparent for pPrl and PrP, the consequences for other substrates may be more difficult to discern and may go beyond merely changes in the association between the ribosome and translocon. Signal sequences might influence ER-associated degradation, transport kinetics, protein–protein interactions, or even final protein conformation. In any case, while many signal sequences may suffice to simply transport a protein across the ER membrane, only a subset of these may faithfully initiate the folding and maturation events needed for optimal function.
Acknowledgments
We thank A. Calayag, M. Zimmerman, J. Kuszinski, and M. Corpuz for technical assistance, C. M. Ott and M. R. St. Jean for critical comments, and T. A. Rapoport for pSP HG201. This work was supported by grants from the National Institutes of Health, the Sandler Foundation, and the American Heart Association to V.R.L., the National Cancer Institute to R.S.H., and a Howard Hughes Predoctoral Fellowship in Biological Sciences to D.T.R.
Abbreviations
- ER
endoplasmic reticulum
- pPrl
preprolactin
- pβL
preβ-lactamase
- pIgG
preIgG heavy chain
- PrP
prion protein
- maa
mature amino acids
- RM
canine pancreatic rough microsomes
- PK
proteinase K
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Martoglio B, Dobberstein B. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 2.Blobel G, Dobberstein B. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 4.Jungnickel B, Rapoport T A. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 5.Gilmore R, Blobel G. Cell. 1985;42:497–505. doi: 10.1016/0092-8674(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 6.Crowley K S, Reinhart G D, Johnson A E. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- 7.Simon S M, Blobel G. Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- 8.Crowley K S, Liao S, Worrell V E, Reinhart G D, Johnson A E. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 9.Hanein D, Matlack K E, Jungnickel B, Plath K, Kalies K U, Miller K R, Rapoport T A, Akey C W. Cell. 1996;4:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 10.Wiedmann B, Sakai H, Davis T A, Wiedmann M. Nature (London) 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 11.Gilmore R, Blobel G. Cell. 1983;35:677–685. doi: 10.1016/0092-8674(83)90100-9. [DOI] [PubMed] [Google Scholar]
- 12.Connolly T, Gilmore R. Cell. 1989;57:599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- 13.Wolin S L, Walter P. J Cell Biol. 1993;121:1211–1219. doi: 10.1083/jcb.121.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görlich D, Rapoport T A. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 15.Mothes W, Prehn S, Rapoport T A. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connolly T, Collins P, Gilmore R. J Cell Biol. 1989;108:299–307. doi: 10.1083/jcb.108.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Görlich D, Hartmann E, Prehn S, Rapoport T A. Nature (London) 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 18.High S, Martoglio B, Gorlich D, Andersen S S, Ashford A J, Giner A, Hartmann E, Prehn S, Rapoport T A, Dobberstein B, et al. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- 19.Voigt S, Jungnickel B, Hartmann E, Rapoport T A. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Heijne G. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 21.Widemann M, Kurzchalia T V, Hartmann E, Rapoport T A. Nature (London) 1987;328:830–833. doi: 10.1038/328830a0. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann E, Görlich D, Kostka S, Otto A, Kraft R, Knespel S, Bórger E, Rapoport T A, Prehn S. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- 23.Zheng T, Nicchitta C V. J Biol Chem. 1999;274:36623–36630. doi: 10.1074/jbc.274.51.36623. [DOI] [PubMed] [Google Scholar]
- 24.Darsley M J, Rees A R. EMBO J. 1985;4:393–398. doi: 10.1002/j.1460-2075.1985.tb03641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon K, Perara E, Lingappa V R. J Biol Chem. 1987;104:1165–1172. doi: 10.1083/jcb.104.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde R S, Lingappa V R. Cell. 1996;85:217–228. doi: 10.1016/s0092-8674(00)81098-3. [DOI] [PubMed] [Google Scholar]
- 27.Hegde R S, Voigt S, Rapoport T A, Lingappa V R. Cell. 1998;92:621–631. doi: 10.1016/s0092-8674(00)81130-7. [DOI] [PubMed] [Google Scholar]
- 28.Scott M R, Kohler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegde R S, Mastrianni J A, Scott M R, DeFea K A, Tremblay P, Torchia M, DeArmond S J, Prusiner S B, Lingappa V R. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 30.Mothes W, Heinrich S U, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport T A. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 31.Liao S, Lin J, Do H, Johnson A E. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 32.Hay B, Barry R A, Lieberburg I, Prusiner S B, Lingappa V R. Mol Cell Biol. 1987;7:914–920. doi: 10.1128/mcb.7.2.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicchitta C V, Zheng T. J Cell Biol. 1997;139:1697–1708. doi: 10.1083/jcb.139.7.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart R S, Harris D A. J Biol Chem. 2001;276:2212–2220. doi: 10.1074/jbc.M006763200. [DOI] [PubMed] [Google Scholar]
- 35.Holscher C, Bach U C, Dobberstein B. J Biol Chem. 2001;276:13388–13394. doi: 10.1074/jbc.M007331200. [DOI] [PubMed] [Google Scholar]
- 36.Hegde R S, Tremblay P, Groth D, DeArmond S J, Prusiner S B, Lingappa V R. Nature (London) 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- 37.Wolin S L, Walter P. J Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]